Published online Jun 7, 2020. doi: 10.3748/wjg.v26.i21.2781
Peer-review started: December 29, 2019
First decision: January 19, 2020
Revised: March 30, 2020
Accepted: May 23, 2020
Article in press: May 23, 2020
Published online: June 7, 2020
Processing time: 159 Days and 18.5 Hours
Hepatitis D virus (HDV) is a global health threat with more than 15 million humans affected. Current treatment options are largely unsatisfactory leaving chronically infected humans at high risk to develop liver cirrhosis and hepatocellular carcinoma. HDV is the only human satellite virus known. It encodes only two proteins, and requires Hepatitis B virus (HBV) envelope protein expression for productive virion release and spread of the infection. How HDV could evolve and why HBV was selected as a helper virus remains unknown. Since the discovery of Na+-taurocholate co-transporting polypeptide as the essential uptake receptor for HBV and HDV, we are beginning to understand the interactions of HDV and the immune system. While HBV is mostly regarded a stealth virus, that escapes innate immune recognition, HBV-HDV coinfection is characterized by a strong innate immune response. Cytoplasmic RNA sensor melanoma differentiation antigen 5 has been reported to recognize HDV RNA replication and activate innate immunity. Innate immunity, however, seems not to impair HDV replication while it inhibits HBV. In this review, we describe what is known up-to-date about the interplay between HBV as a helper and HDV’s immune evasion strategy and identify where additional research is required.
Core tip: Hepatitis D virus (HDV) is the only known human satellite virus requiring hepatitis B virus (HBV) coinfection for productive viral release. However, it was recently shown that HDV can be disseminated by viruses other than HBV in experimental setups, so it remains unexplained why HDV chose HBV as a helper virus. As HDV might possibly profit from HBV mediated immunosuppression, we first focus on recent findings on HDV recognition by the innate immune system. Later on, we summarize partially controversial data on immunomodulatory mechanisms of both, HBV and HDV.
- Citation: Jung S, Altstetter SM, Protzer U. Innate immune recognition and modulation in hepatitis D virus infection. World J Gastroenterol 2020; 26(21): 2781-2791
- URL: https://www.wjgnet.com/1007-9327/full/v26/i21/2781.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i21.2781
First identified in 1977 by Rizzetto et al[1], Hepatitis D virus (HDV) represents a unique pathogen that defines it’s stand-alone genus Deltavirus. Eight genotypes of HDV varying in their RNA-genome sequences have been described. As a satellite RNA virus, HDV does not encode its own envelope proteins for packaging of its ribonucleoprotein (RNP) and therefore depends on the envelop glycoproteins of the hepatitis B virus (HBV) for virion assembly, envelopment and transmission. HDV has a broad cell and host tropism and, theoretically, several virus genera can provide help by enveloping the HDV RNP[3]. Clinically, however, HDV infection has so far only been described as coinfection with HBV or as a superinfection of chronic HBV carriers. Both, co- and superinfection may lead to HDV persistence and inflammatory liver disease, called hepatitis D. Currently, World Health Organization estimates that 15-20 million people are infected with HDV worldwide, while others predict up to 70 million carriers.
The pathogenesis of hepatitis D has been recently summarized by Koh et al[6]. Coinfection with HBV and HDV tends to result in both, acute hepatitis B and D at the same time, leading to most severe disease. In a superinfection scenario, HDV profits from pre-existing hepatitis B surface antigen (HBsAg) expression for progeny virus production but at the same time decreases HBV replication rates[7,8]. Chronic hepatitis D is the viral hepatitis form that is most likely to lead to liver cirrhosis and it is associated with a significant risk of hepatocellular carcinoma development and high mortality rates[6]. The reasons for more severe disease progression in HBV-HDV infection compared to HBV monoinfection have not been ultimately resolved. Chimpanzee studies indicate that liver damage by HBV is immune mediated whereas in HDV infection it is mainly cytopathic[9,10]. Direct cytopathic effects and induction of liver fibrosis by HDV antigen (HDAg) were also indicated in in vitro studies[11-14].
Current treatment options rely on interferon alpha and are largely unsatisfactory leaving chronically infected at high risk to develop liver cirrhosis and hepatocellular carcinoma. New treatment options include interferon lambda, a farnesyl transferase inhibitor (Lonafarnib), the entry inhibitor peptide Mycludex B (Bulevirtide), nucleic acid polymers (e.g. REP 2139-Ca) that are applied alone or in combination with interferon and show promising results in phase II clinical trials[6].
The viral genome of HDV is a single-stranded, circular, negative sense RNA with a length of approximately 1680 nucleotides. Due to broad base pairing within the RNA molecule, the genome appears as a double stranded, rod-like structure resembling a plant viroid. During HDV replication, exclusively taking place in the nucleus, three distinct RNAs, which includes the genome, the positive-stranded antigenome, and viral mRNA, are generated by host RNA polymerases. RNA-Pol I drives the transcription of genome to antigenome in the nucleolus, whereas RNA-Pol II is responsible for genome replication using the antigenome as template on the one hand and for transcription of mRNA in the nucleoplasm on the other hand. Genome replication functions via a double rolling-circle mechanism. RNA oligomers of genomic and antigenomic orientation are generated, followed by self-cleavage into monomers through genome and antigenome intrinsic ribozyme activity, respectively.
Although there are several open reading frames within the HDV genome, only a single one is actively transcribed leading to the expression of two isoforms of HDAg. The small HDAg (S-HDAg) is composed of 195 amino acids, and the large HDAg (L-HDAg) is comprised of 214 amino acids. Initially, only S-HDAg is expressed because a termination codon prevents protein translation of L-HDAg. In order to produce the large isoform, the stop-codon (UAG) within the antigenome is mutated into a tryptophan codon (UGG) by the cellular enzyme Adenosine Deaminase Acting on RNA (ADAR1). ADAR1 is an “RNA editor” induced by interferon. Transcription of this modified genome into mRNA extends the open reading frame until the next stop codon is reached, resulting in the translation of L-HDAg harbouring an additional 19 C-terminal amino acids[23,24].
For both isoforms, post-translational modifications play an important role. For S-HDAg it has been shown that phosphorylation of a serine residue enables interactions with the cellular RNA-Pol II, which is essential for HDV replication. Due to its C-terminal elongation, L-HDAg incorporates a nuclear export signal and a prenylation site, which allows farnesylation. The farnesylated form of L-HDAg inhibits HDV replication by masking a conformational epitope present in S-HDAg that is essential for trans-activating HDV RNA replication. The farnesylated L-HDAg is also crucial for virion assembly through its promotion of the interaction of the viral genome with a tryptophan-rich domain in the cytosolic loop of HBsAg. Common arginine rich motifs within S- and L-HDAg allow their mutual binding to RNA, leading to the formation of the so-called RNP complex, which consists of HDV genomic RNA and both HDAg isoforms. The RNP is subsequently exported into the cytoplasm, where virion assembly takes place. Export is likely mediated by nuclear export factor 1 and the cellular RNA export factor REF/Aly. During these different steps of viral replication, HDV induces a pronounced cytokine response and activates a broad range of host defence mechanisms[31-33]. This review focuses on the mode of HDV-detection by cellular pattern recognition receptors (PRRs) and selective modulatory properties of the HDV antigens. Additionally, immune-evasive and immuno-suppressive strategies of HDV, and its coexisting host virus HBV, are discussed.
The immune system of vertebrates acts as protective mechanism against damage on cellular and organism level, and is subdivided into two branches, the innate and the adaptive immune systems[34]. Innate immunity, as the evolutionary older system, is the frontline of host defence which upon infection with a pathogen initiates and fine-tunes pathogen-specific adaptive immunity. For this purpose, innate immunity possesses the capacity to distinguish between self and non-self, as well as different classes of pathogens by recognizing certain structural patterns. This function is enabled by the expression of PRRs that detect distinct pathogen associated molecular patterns, also referred to as “PAMPs”, such as unusually structured or located nucleic acids or characteristic bacterial proteins which are not found in a given cellular compartment under physiological conditions. In the case of viral infections, innate immune sensing mostly depends on characteristic modifications of viral genomes or genome-replication intermediates and mRNA as well as special RNA structures which are normally absent in eukaryotic cells.
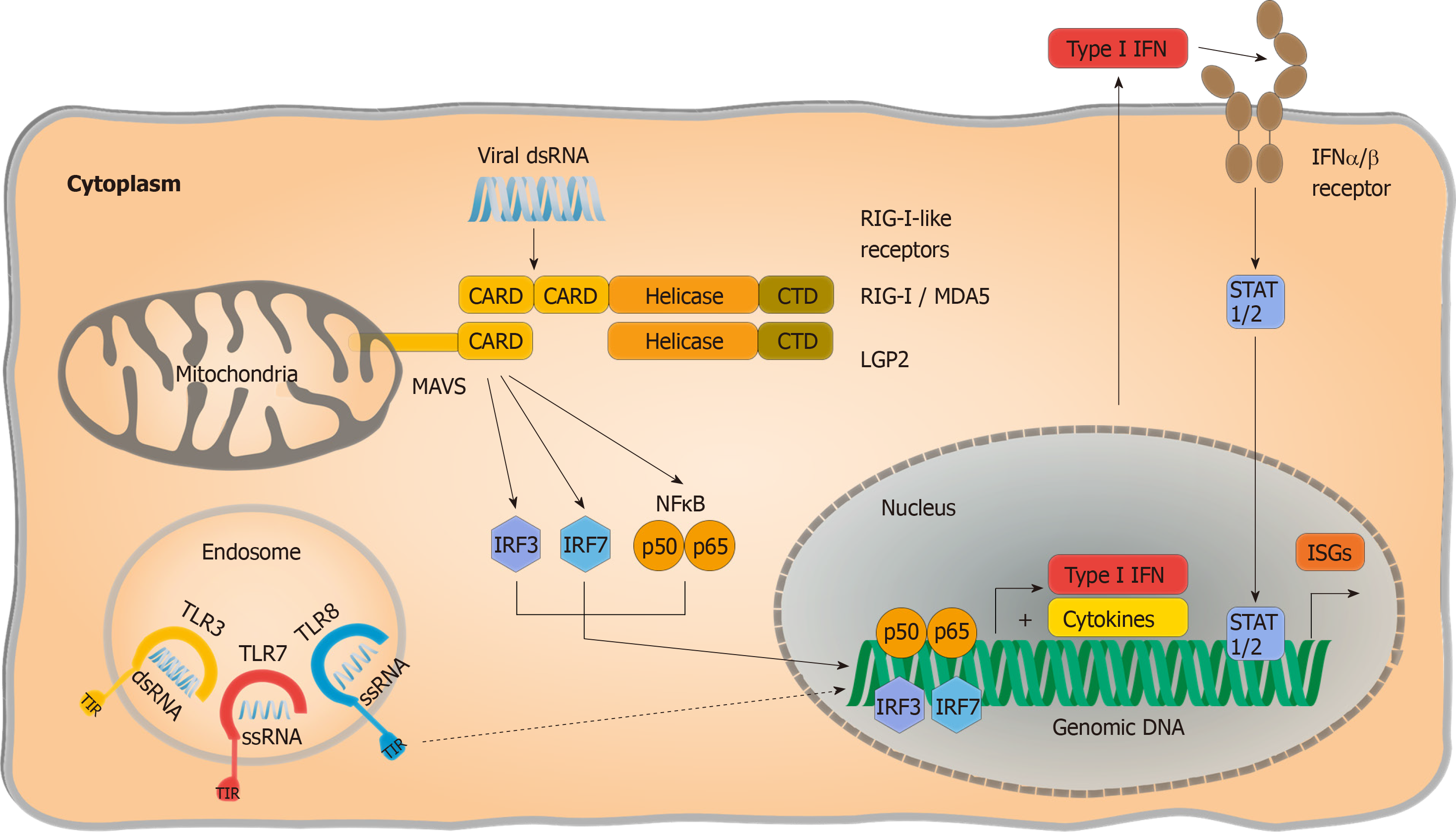
Extensive studies have narrowed viral RNA detection down to two families of PRRs: Endosomal Toll like receptors (TLRs) and cytosolic RIG I like receptors (RLRs) (Figure 1). The latter consists of two activating PRRs, retinoic acid inducible gene 1 (RIG I) and melanoma differentiation associated gene 5 (MDA5), as well as a third signalling-incompetent accessory molecule termed laboratory of genetics and physiology 2. Double-stranded RNA regions are both required for RIG I and MDA5 activation, although MDA5 was reported to bind longer double-stranded RNA whereas RIG I activation is mostly thought to be triggered by shorter double-stranded RNA or hairpin structures with a 5’ phosphorylation. Interaction of RLRs with their specific RNA patterns results in intramolecular conformational changes, exposing their “Caspase activation and recruitment domain” site for interaction with the mitochondrial antiviral signalling (MAVS) protein[37]. Subsequently, MAVS functions as a scaffold and initiates two divergent immune signalling pathways: (1) Proinflammatory cytokine release is provoked in a nuclear factor “kappa-light-chain-enhancer” of activated B-cells (NF-κB) dependent manner; and (2) Phosphorylation and nuclear translocation of “Signal transducer and activator of transcription” (STAT 1/2) induces production of interferon (IFN). IFN signalling activates the expression of interferon-stimulated gene (ISGs) by modulating cellular homeostasis in both autocrine and paracrine manners, resulting in an antiviral state that protects both infected and noninfected cells and suppresses viral replication and progeny virus production.
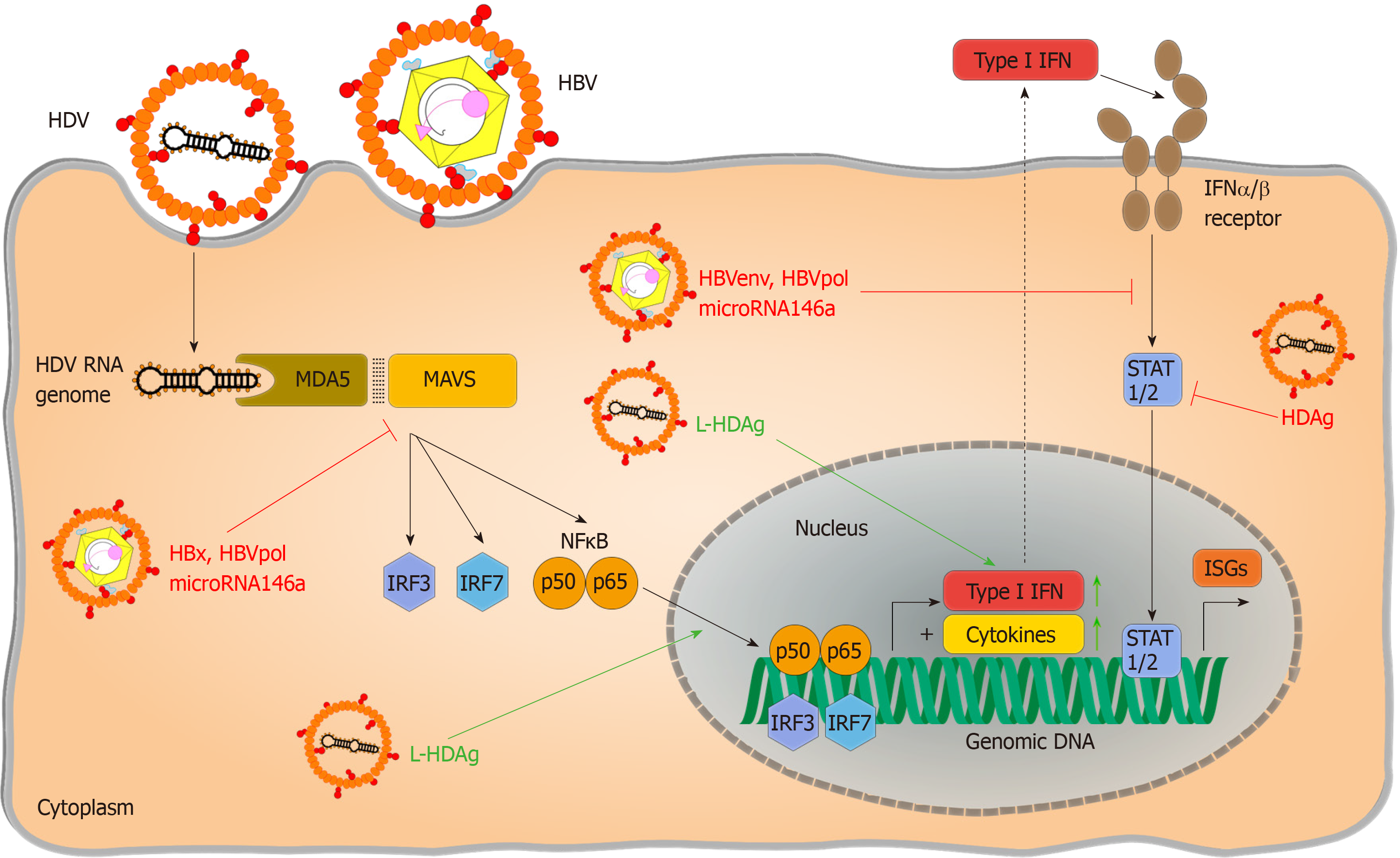
Partial dependence on RLR signalling has been reported by Suárez-Amarán et al[38] in immune pattern recognition of HDV. The authors used adenovirus-associated virus (AAV) to deliver HBV and HDV genomes (AAV-HBV and AAV-HDV) into murine liver cells to circumvent species-specific limitations of viral entry. Both wildtype (wt) and MAVS-knockout (MAVS-ko) mice showed HDV gene expression and replication, but the innate immune response to HDV infection was diminished in MAVS-ko cells. HDV-induced immune activation resulting in type I and type III IFN production was later found to be dependent on MDA5 in both primary human hepatocytes and hepatoma-cell lines[32]. While pattern recognition of HDV RNA is considered the primary source of immune activation, direct induction of IFN-signalling by L-HDAg has also been reported[39]. Nevertheless, pattern recognition of HDV infection has not been conclusively resolved. As residual IFN-responses are still detectable in the absence of RLR-signalling[38], the impact of synergic immune activating pathways require further investigation. Additionally, the nature of HDV-specific molecular patterns that activate PRRs and changes in HBV-induced cellular immunoregulatory pathways are still poorly characterized. Considering that HDV only occurs as a satellite virus and chose HBV as a helper under natural conditions although theoretically a broad variety of viruses could provide their envelops[3], the detailed characterization of pattern recognition should regard potential confounding effects of HBV coinfection.
The impact of HBV infection on innate immunity has been subject to numerous studies and discussions. Numerous studies proved that HBV is sensitive to interferons and other antiviral cytokines in vivo in the liver[40,41], in primary hepatocytes or in HepaRG cells that have maintained their sensitivity to innate immune stimulation[42,43]. Cytokines can block HBV replication at transcriptional and posttranscriptional steps (Summarized in: Xia et al 2017) and affect cccDNA stability by inducing the enzymes that edit and subsequently digest it[45,46]. Up to date, the discussion is ongoing whether HBV can actively interfere with or suppress innate immunity, and thus support HDV persistence.
HBV is primarily regarded a stealth virus, neither activating nor inhibiting an innate immune response during virus replication[47-49]. Macrophages may, however, recognize virus particles early during infection[50,51]. This may be responsible for suppression of HBV replication shortly after infection and allow to prevent early activation of adaptive immunity. Several HBV proteins have been reported to have distinct features resulting in active interference with immune recognition or immune suppression. A number of these studies were done in settings where HBV proteins were overexpressed resulting in controversial discussions about the physiological relevance of the results[42,52-55]. An inhibition of interferon responses by HBV, however, has also been described in mice with humanized livers[56]. An inhibition of interferon responses by HBV, however, has also been described in mice with humanized livers. One would expect these livers to be close to the human physiological situation although HBV replication levels may be higher due to the lack of adaptive immunity and a cross-talk between human hepatocytes and murine non-hepatocytes.
A recent publication showed that HDV can be efficiently disseminated by helper viruses other than HBV from different genera, including flavivirus, vesicular stomatitis virus and the hepatitis C virus in vitro and in mice[3]. HDV particles packaged within a vesicular stomatitis virus envelope were able to overcome liver specificity conferred by the HBV envelope proteins and efficiently infect human embryonic kidney cell derived 293 cells[3]. In the context of tissue-specific pattern recognition, liver tropism may only confer a minor advantage for HDV since dsRNA-detecting TLR3, RIG I and MDA5 functions have been verified despite low protein expression levels in vitro[42,57,58] and in vivo[59].
From an evolutionary standpoint, one would argue that coinfection with HBV must be favourable for HDV, leading to the question of what benefit this confers. One possible explanation could be that HBV does indeed prevent or block innate immunity and that HDV profits from this. Regarding HDV recognition by MDA5, downregulation of MAVS-induced signalling by HBV-encoded X-protein has been proposed[60-64]. Interference has also be reported by the HBV polymerase[65] or by HBV induced microRNA146a[66] – all of which could support the survival of HDV infected cells. Proving this, however, requires additional studies using infection models because there are potential confounding effects from overexpression of HBV proteins in these experiments. The impact of HBV infection on downstream immune pathways also remains controversial. Direct blocking of interferon-signalling by HBV polymerase[52,54,67], HBV envelope protein[68], X-protein[69,70] or microRNA 146a[71] has been reported, which could also benefit HDV infection. These effects may well be subtle as HDV envelopment requires a certain level of interferon activity to allow induction of expression of the interferon-stimulated ADAR1 that is essential for L-HDAg expression. Regardless of the exact mode of HBV-induced immuno-suppression, dependency of HDV on help to survive innate immunity seems likely, given that it only encodes for a single protein.
In addition to exploiting immunosuppressive and immune-evasive mechanisms, HDV possesses some resistance to interferon-mediated antiviral effects. In contrast to HBV, HDV induces interferon signalling in both cell lines and mice without viral replication being affected[31,32,38]. The most prominent interferon-induced protein in HDV infection is ADAR1, which both inhibits viral replication and promotes RNA packaging. ADAR1 exists in two isoforms, constitutively expressed short ADAR1p110 present in the nucleus and IFN-inducible long ADAR1p150 which is both present in the cytoplasm and nucleus. In untreated Huh7-cells, non-inducible ADAR1p110 is mainly responsible for L-HDAg expression, whereas ADAR1p150 can enhance HDV-RNA editing rates up to one-third upon IFN-treatment[24,74,75]. Though ADAR1p150 editing was hypothesized to be partially responsible for the antiviral effects of interferon-α therapy, this effect seems to be limited due to additional regulatory mechanisms.
HDV production appears to remain unaffected or even promoted during proinflammatory cytokine responses. Various groups have reported L-HDAg enhanced NF-κB translocation to the nucleus and the upregulation of proinflammatory genes in response to transfection of HDV-encoding plasmid[76-78]. This might be necessary for viral assembly as L-HDAg translocation from nucleus to cytoplasm was reported to be induced by NF-κB activation. However, all these experiments were performed as transient HDAg overexpressions, which could also induce unfolded protein response in the endoplasmatic reticulum, leading to NF-κB activation[80,81]. These circumstances were caused by unavailability of HDV-susceptible cell lines until the identification of Na+-taurocholate co-transporting polypeptide (NTCP) as an essential factor for HBV/HDV infection[82]. Newly developed infection systems utilizing AAV-HDV, as well as NTCP-expressing cell lines and mouse models, should be used to strengthen previously published results on antiviral activity of cytokines against HDV.
Despite the very limited coding capacity of its genome, HDV has evolved mechanisms to escape immunity (Figure 2). First of all, the HDV genome avoids direct contact with cytoplasmic or endosomal PRRs by replicating in the nucleus, taking advantage of cellular compartmentalization. Furthermore, it forms a circular RNA genome without “open” 5’ or 3’ ends to prevent PRR binding, as circular RNA has been reported not to activate RIG I[83], and an RNP complex reducing the binding of PRRs to virus-characteristic structures. Transfection of HDV-cDNA and HDV-encoding plasmid pSVL(D3) in Huh7 cells reduced STAT-signalling and expression of ISGs in response to IFN-α treatment and inhibited phosphorylation and nuclear translocation of STAT-proteins. This direct inhibition of interferon signalling was hypothesized to account for poor responsiveness to IFN treatment in infected patients. However, these results have not been reproduced in HDV-infection so far and, contradictory to the complete blocking of IFN signalling observed in this study, HDV triggers immune activation via MDA-5[32]. Whether HDV immune recognition by alternate PRR plays a role and which HDV-RNA structures trigger HDV-immune recognition still needs to be identified. It also remains ambiguous if HDV initiates IFN production in infected patients, since as to the authors’ knowledge no data exist on this so far. HDV also offers little for adaptive immunity to attack since there is only two proteins expressed, and S-HDAg and L-HDAg largely overlap in their protein sequence. Thus, few HDV-derived peptides can be presented on infected cells and recognized by T cells. In a systematic screen to define CD8 epitopes, the overall number of epitopes identified was very low compared to other hepatotropic viruses[85]. When sequences of HDV RNA and HLA class I alleles that present epitope peptides to CD8+ T cells in patients with persistent HDV infection were analyzed, HDV variants were identified that can escape T cell-mediated immunity[85,86]. As an RNA virus, HDV genomes are mutated during virus replication allowing immune escape variants to emerge. Hereby, HDV escape from the immune response was associated with uncommon HLA class I alleles, indicating that HDV has evolved, at the population level, to evade recognition by common HLA class I alleles[86,87]. T cell exhaustion doesn’t seem to be a major reason for failure to clear HDV. Activated HDV-specific CD8+ T cells target conserved epitopes and seem to contribute to disease progression. Even memory-like HDV-specific CD8+ T cells remain functional but are unable to clear HDV because of the presence of escape variants[86,87]. Thus, HDV mainly escapes adaptive immunity because there are so few epitopes that may be presented by human HLA haplotype repertoire and recognized by T cells.
As the only known satellite virus known in humans, HDV has chosen HBV as a helper virus although HDV per se is promiscuous. HDV seems to profit from the co-existence of HBV. Whether the advantage conferred is the strict liver tropism of HBV where pattern recognition is tuned down due to the constant exposure to bacterial components, or whether active HBV-induced immunosuppression contributes remains open. HDV shows some capabilities to escape immune responses and also a certain degree of resistance to interferon activity. Detailed studies on the mode of HDV-induced immune regulation and immune activation could contribute valuable key information to target this virus and develop new therapies against this fatal disease.
The authors thank Britta Möhl-Meinke, Anna Kosinska and Aaron Lucko for critically reading the manuscript. Studies in UP’s laboratory are supported by the German Research Foundation via TRR179, the European Union within the Horizon 2020 program via VIROFIGHT and TherVacB consortia, the Helmholtz -Alberta Initiative on Infection Research (HAI-IDR) and the German Center for Infection Research. Jung S received a Postdoc Fellowship of the Helmholtz Association.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: American Society for Microbiology, No. 1894152; and the European Association for the Study of the Liver, No. 1603.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Germany
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A, A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Farshadpour F, Ivanov A, Maggi F, McQuillan GM, Manesis EK S-Editor: Wang JL L-Editor: A E-Editor: Qi LL
| 1. | Rizzetto M, Canese MG, Aricò S, Crivelli O, Trepo C, Bonino F, Verme G. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977;18:997-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 642] [Article Influence: 13.4] [Reference Citation Analysis (1)] |
| 2. | Magnius L, Taylor J, Mason WS, Sureau C, Dény P, Norder H, Ictv Report Consortium. ICTV Virus Taxonomy Profile: Deltavirus. J Gen Virol. 2018;99:1565-1566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 3. | Perez-Vargas J, Amirache F, Boson B, Mialon C, Freitas N, Sureau C, Fusil F, Cosset FL. Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat Commun. 2019;10:2098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (1)] |
| 4. | World Health Organization. Hepatitis D, 2019. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-d. |
| 5. | Mentha N, Clément S, Negro F, Alfaiate D. A review on hepatitis D: From virology to new therapies. J Adv Res. 2019;17:3-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 6. | Koh C, Heller T, Glenn JS. Pathogenesis of and New Therapies for Hepatitis D. Gastroenterology. 2019;156:461-476.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 7. | Liu L. Fields Virology, 6th Edition. Clin Infect Dis. 2014;59:613-613. [RCA] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 8. | Lütgehetmann M, Mancke LV, Volz T, Helbig M, Allweiss L, Bornscheuer T, Pollok JM, Lohse AW, Petersen J, Urban S, Dandri M. Humanized chimeric uPA mouse model for the study of hepatitis B and D virus interactions and preclinical drug evaluation. Hepatology. 2012;55:685-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 183] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 9. | Gilman C, Heller T, Koh C. Chronic hepatitis delta: A state-of-the-art review and new therapies. World J Gastroenterol. 2019;25:4580-4597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (6)] |
| 10. | Smedile A, Farci P, Verme G, Caredda F, Cargnel A, Caporaso N, Dentico P, Trepo C, Opolon P, Gimson A, Vergani D, Williams R, Rizzetto M. Influence of delta infection on severity of hepatitis B. Lancet. 1982;2:945-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 277] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Cole SM, Gowans EJ, Macnaughton TB, Hall PD, Burrell CJ. Direct evidence for cytotoxicity associated with expression of hepatitis delta virus antigen. Hepatology. 1991;13:845-851. [PubMed] |
| 12. | Wang D, Pearlberg J, Liu YT, Ganem D. Deleterious effects of hepatitis delta virus replication on host cell proliferation. J Virol. 2001;75:3600-3604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Chang J, Gudima SO, Tarn C, Nie X, Taylor JM. Development of a novel system to study hepatitis delta virus genome replication. J Virol. 2005;79:8182-8188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Choi SH, Jeong SH, Hwang SB. Large hepatitis delta antigen modulates transforming growth factor-beta signaling cascades: implication of hepatitis delta virus-induced liver fibrosis. Gastroenterology. 2007;132:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 15. | Radjef N, Gordien E, Ivaniushina V, Gault E, Anaïs P, Drugan T, Trinchet JC, Roulot D, Tamby M, Milinkovitch MC, Dény P. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades. J Virol. 2004;78:2537-2544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 175] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 16. | Wang KS, Choo QL, Weiner AJ, Ou JH, Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL, Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986;323:508-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 537] [Cited by in RCA: 531] [Article Influence: 13.6] [Reference Citation Analysis (1)] |
| 17. | Chen PJ, Kalpana G, Goldberg J, Mason W, Werner B, Gerin J, Taylor J. Structure and replication of the genome of the hepatitis delta virus. Proc Natl Acad Sci USA. 1986;83:8774-8778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 252] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 18. | Huang WH, Chen YS, Chen PJ. Nucleolar targeting of hepatitis delta antigen abolishes its ability to initiate viral antigenomic RNA replication. J Virol. 2008;82:692-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Modahl LE, Lai MM. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J Virol. 1998;72:5449-5456. [PubMed] |
| 20. | Macnaughton TB, Wang YJ, Lai MM. Replication of hepatitis delta virus RNA: effect of mutations of the autocatalytic cleavage sites. J Virol. 1993;67:2228-2234. [PubMed] |
| 21. | Gudima S, Chang J, Moraleda G, Azvolinsky A, Taylor J. Parameters of human hepatitis delta virus genome replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J Virol. 2002;76:3709-3719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 22. | Lamers MM, van den Hoogen BG, Haagmans BL. ADAR1: "Editor-in-Chief" of Cytoplasmic Innate Immunity. Front Immunol. 2019;10:1763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 23. | Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 252] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 24. | Wong SK, Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci USA. 2002;99:15118-15123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 150] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 25. | Hong SY, Chen PJ. Phosphorylation of serine 177 of the small hepatitis delta antigen regulates viral antigenomic RNA replication by interacting with the processive RNA polymerase II. J Virol. 2010;84:1430-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Lee CH, Chang SC, Wu CH, Chang MF. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J Biol Chem. 2001;276:8142-8148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Hwang SB, Lai MM. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J Virol. 1994;68:2958-2964. [PubMed] |
| 28. | Sureau C. The role of the HBV envelope proteins in the HDV replication cycle. Curr Top Microbiol Immunol. 2006;307:113-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Rizzetto M. Hepatitis D: thirty years after. J Hepatol. 2009;50:1043-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 30. | Huang HC, Lee CP, Liu HK, Chang MF, Lai YH, Lee YC, Huang C. Cellular Nuclear Export Factors TAP and Aly Are Required for HDAg-L-mediated Assembly of Hepatitis Delta Virus. J Biol Chem. 2016;291:26226-26238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 31. | Alfaiate D, Lucifora J, Abeywickrama-Samarakoon N, Michelet M, Testoni B, Cortay JC, Sureau C, Zoulim F, Dény P, Durantel D. HDV RNA replication is associated with HBV repression and interferon-stimulated genes induction in super-infected hepatocytes. Antiviral Res. 2016;136:19-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 32. | Zhang Z, Filzmayer C, Ni Y, Sültmann H, Mutz P, Hiet MS, Vondran FWR, Bartenschlager R, Urban S. Hepatitis D virus replication is sensed by MDA5 and induces IFN-β/λ responses in hepatocytes. J Hepatol. 2018;69:25-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 33. | Giersch K, Allweiss L, Volz T, Helbig M, Bierwolf J, Lohse AW, Pollok JM, Petersen J, Dandri M, Lütgehetmann M. Hepatitis Delta co-infection in humanized mice leads to pronounced induction of innate immune responses in comparison to HBV mono-infection. J Hepatol. 2015;63:346-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 107] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 34. | Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1476] [Cited by in RCA: 1389] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 35. | Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol. 2016;14:360-373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 338] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 36. | Reikine S, Nguyen JB, Modis Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front Immunol. 2014;5:342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 303] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 37. | Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 343] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 38. | Suárez-Amarán L, Usai C, Di Scala M, Godoy C, Ni Y, Hommel M, Palomo L, Segura V, Olagüe C, Vales A, Ruiz-Ripa A, Buti M, Salido E, Prieto J, Urban S, Rodríguez-Frias F, Aldabe R, González-Aseguinolaza G. A new HDV mouse model identifies mitochondrial antiviral signaling protein (MAVS) as a key player in IFN-β induction. J Hepatol. 2017;67:669-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 39. | Williams V, Brichler S, Radjef N, Lebon P, Goffard A, Hober D, Fagard R, Kremsdorf D, Dény P, Gordien E. Hepatitis delta virus proteins repress hepatitis B virus enhancers and activate the alpha/beta interferon-inducible MxA gene. J Gen Virol. 2009;90:2759-2767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 40. | Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 840] [Cited by in RCA: 840] [Article Influence: 29.0] [Reference Citation Analysis (1)] |
| 41. | Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 949] [Cited by in RCA: 914] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 42. | Luangsay S, Gruffaz M, Isorce N, Testoni B, Michelet M, Faure-Dupuy S, Maadadi S, Ait-Goughoulte M, Parent R, Rivoire M, Javanbakht H, Lucifora J, Durantel D, Zoulim F. Early inhibition of hepatocyte innate responses by hepatitis B virus. J Hepatol. 2015;63:1314-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 43. | Xia Y, Cheng X, Blossey CK, Wisskirchen K, Esser K, Protzer U. Secreted Interferon-Inducible Factors Restrict Hepatitis B and C Virus Entry In Vitro. J Immunol Res. 2017;2017:4828936. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Xia Y, Protzer U. Control of Hepatitis B Virus by Cytokines. Viruses. 2017;9:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 45. | Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 741] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 46. | Xia Y, Stadler D, Lucifora J, Reisinger F, Webb D, Hösel M, Michler T, Wisskirchen K, Cheng X, Zhang K, Chou WM, Wettengel JM, Malo A, Bohne F, Hoffmann D, Eyer F, Thimme R, Falk CS, Thasler WE, Heikenwalder M, Protzer U. Interferon-γ and Tumor Necrosis Factor-α Produced by T Cells Reduce the HBV Persistence Form, cccDNA, Without Cytolysis. Gastroenterology. 2016;150:194-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 266] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 47. | Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369-9380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 348] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 48. | Suslov A, Boldanova T, Wang X, Wieland S, Heim MH. Hepatitis B Virus Does Not Interfere With Innate Immune Responses in the Human Liver. Gastroenterology. 2018;154:1778-1790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 138] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 49. | Mutz P, Metz P, Lempp FA, Bender S, Qu B, Schöneweis K, Seitz S, Tu T, Restuccia A, Frankish J, Dächert C, Schusser B, Koschny R, Polychronidis G, Schemmer P, Hoffmann K, Baumert TF, Binder M, Urban S, Bartenschlager R. HBV Bypasses the Innate Immune Response and Does Not Protect HCV From Antiviral Activity of Interferon. Gastroenterology. 2018;154:1791-1804.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 50. | Hösel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ, Langenkamp A, Falk C, Büning H, Rose-John S, Protzer U. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50:1773-1782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 289] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 51. | Cheng X, Xia Y, Serti E, Block PD, Chung M, Chayama K, Rehermann B, Liang TJ. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology. 2017;66:1779-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 52. | Foster GR, Ackrill AM, Goldin RD, Kerr IM, Thomas HC, Stark GR. Expression of the terminal protein region of hepatitis B virus inhibits cellular responses to interferons alpha and gamma and double-stranded RNA. Proc Natl Acad Sci USA. 1991;88:2888-2892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 108] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 53. | Christen V, Duong F, Bernsmeier C, Sun D, Nassal M, Heim MH. Inhibition of alpha interferon signaling by hepatitis B virus. J Virol. 2007;81:159-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (2)] |
| 54. | Wu M, Xu Y, Lin S, Zhang X, Xiang L, Yuan Z. Hepatitis B virus polymerase inhibits the interferon-inducible MyD88 promoter by blocking nuclear translocation of Stat1. J Gen Virol. 2007;88:3260-3269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (1)] |
| 55. | Lucifora J, Durantel D, Testoni B, Hantz O, Levrero M, Zoulim F. Control of hepatitis B virus replication by innate response of HepaRG cells. Hepatology. 2010;51:63-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 56. | Lütgehetmann M, Bornscheuer T, Volz T, Allweiss L, Bockmann JH, Pollok JM, Lohse AW, Petersen J, Dandri M. Hepatitis B virus limits response of human hepatocytes to interferon-α in chimeric mice. Gastroenterology. 2011;140:2074-2083, 2083.e1-2083.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 57. | Preiss S, Thompson A, Chen X, Rodgers S, Markovska V, Desmond P, Visvanathan K, Li K, Locarnini S, Revill P. Characterization of the innate immune signalling pathways in hepatocyte cell lines. J Viral Hepat. 2008;15:888-900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (1)] |
| 58. | Faure-Dupuy S, Vegna S, Aillot L, Dimier L, Esser K, Broxtermann M, Bonnin M, Bendriss-Vermare N, Rivoire M, Passot G, Lesurtel M, Mabrut JY, Ducerf C, Salvetti A, Protzer U, Zoulim F, Durantel D, Lucifora J. Characterization of Pattern Recognition Receptor Expression and Functionality in Liver Primary Cells and Derived Cell Lines. J Innate Immun. 2018;10:339-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 59. | Ebert G, Poeck H, Lucifora J, Baschuk N, Esser K, Esposito I, Hartmann G, Protzer U. 5' Triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology. 2011;141:696-706, 706.e1-706.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 60. | Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, Jia Y, Yuan Y, Guan K, Xu Y, Cheng X, Zhang Y, Yang X, Wang Y, Wen C, Wu Q, Shi W, Zhong H. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol. 2010;185:1158-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 188] [Article Influence: 12.5] [Reference Citation Analysis (1)] |
| 61. | Jiang J, Tang H. Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein. Protein Cell. 2010;1:1106-1117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 86] [Article Influence: 5.7] [Reference Citation Analysis (4)] |
| 62. | Kumar M, Jung SY, Hodgson AJ, Madden CR, Qin J, Slagle BL. Hepatitis B virus regulatory HBx protein binds to adaptor protein IPS-1 and inhibits the activation of beta interferon. J Virol. 2011;85:987-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 110] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 63. | Wang X, Li Y, Mao A, Li C, Li Y, Tien P. Hepatitis B virus X protein suppresses virus-triggered IRF3 activation and IFN-beta induction by disrupting the VISA-associated complex. Cell Mol Immunol. 2010;7:341-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (2)] |
| 64. | Khan M, Syed GH, Kim SJ, Siddiqui A. Hepatitis B Virus-Induced Parkin-Dependent Recruitment of Linear Ubiquitin Assembly Complex (LUBAC) to Mitochondria and Attenuation of Innate Immunity. PLoS Pathog. 2016;12:e1005693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 65. | Yu S, Chen J, Wu M, Chen H, Kato N, Yuan Z. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3. J Gen Virol. 2010;91:2080-2090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (3)] |
| 66. | Hou Z, Zhang J, Han Q, Su C, Qu J, Xu D, Zhang C, Tian Z. Hepatitis B virus inhibits intrinsic RIG-I and RIG-G immune signaling via inducing miR146a. Sci Rep. 2016;6:26150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 67. | Chen J, Wu M, Zhang X, Zhang W, Zhang Z, Chen L, He J, Zheng Y, Chen C, Wang F, Hu Y, Zhou X, Wang C, Xu Y, Lu M, Yuan Z. Hepatitis B virus polymerase impairs interferon-α-induced STA T activation through inhibition of importin-α5 and protein kinase C-δ. Hepatology. 2013;57:470-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 104] [Article Influence: 8.7] [Reference Citation Analysis (2)] |
| 68. | Xu F, Song H, Li N, Tan G. HBsAg blocks TYPE I IFN induced up-regulation of A3G through inhibition of STAT3. Biochem Biophys Res Commun. 2016;473:219-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 69. | Cho IR, Oh M, Koh SS, Malilas W, Srisuttee R, Jhun BH, Pellegrini S, Fuchs SY, Chung YH. Hepatitis B virus X protein inhibits extracellular IFN-α-mediated signal transduction by downregulation of type I IFN receptor. Int J Mol Med. 2012;29:581-586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 70. | Tsunematsu S, Suda G, Yamasaki K, Kimura M, Izumi T, Umemura M, Ito J, Sato F, Nakai M, Sho T, Morikawa K, Ogawa K, Tanaka Y, Watashi K, Wakita T, Sakamoto N. Hepatitis B virus X protein impairs α-interferon signaling via up-regulation of suppressor of cytokine signaling 3 and protein phosphatase 2A. J Med Virol. 2017;89:267-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 71. | Hou ZH, Han QJ, Zhang C, Tian ZG, Zhang J. miR146a impairs the IFN-induced anti-HBV immune response by downregulating STAT1 in hepatocytes. Liver Int. 2014;34:58-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 72. | Samuel CE. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411:180-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 249] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 73. | Casey JL. Control of ADAR1 editing of hepatitis delta virus RNAs. Curr Top Microbiol Immunol. 2012;353:123-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 74. | Hartwig D, Schoeneich L, Greeve J, Schütte C, Dorn I, Kirchner H, Hennig H. Interferon-alpha stimulation of liver cells enhances hepatitis delta virus RNA editing in early infection. J Hepatol. 2004;41:667-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 75. | Hartwig D, Schütte C, Warnecke J, Dorn I, Hennig H, Kirchner H, Schlenke P. The large form of ADAR 1 is responsible for enhanced hepatitis delta virus RNA editing in interferon-alpha-stimulated host cells. J Viral Hepat. 2006;13:150-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 76. | Park CY, Oh SH, Kang SM, Lim YS, Hwang SB. Hepatitis delta virus large antigen sensitizes to TNF-alpha-induced NF-kappaB signaling. Mol Cells. 2009;28:49-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 77. | Williams V, Brichler S, Khan E, Chami M, Dény P, Kremsdorf D, Gordien E. Large hepatitis delta antigen activates STAT-3 and NF-κB via oxidative stress. J Viral Hepat. 2012;19:744-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 78. | Goto T, Kato N, Ono-Nita SK, Yoshida H, Otsuka M, Shiratori Y, Omata M. Large isoform of hepatitis delta antigen activates serum response factor-associated transcription. J Biol Chem. 2000;275:37311-37316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 79. | Huang IC, Chien CY, Huang CR, Lo SJ. Induction of hepatitis D virus large antigen translocation to the cytoplasm by hepatitis B virus surface antigens correlates with endoplasmic reticulum stress and NF-kappaB activation. J Gen Virol. 2006;87:1715-1723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 80. | Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2260] [Cited by in RCA: 2366] [Article Influence: 118.3] [Reference Citation Analysis (0)] |
| 81. | Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455-462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1641] [Cited by in RCA: 1587] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 82. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1602] [Article Influence: 123.2] [Reference Citation Analysis (1)] |
| 83. | Wesselhoeft RA, Kowalski PS, Parker-Hale FC, Huang Y, Bisaria N, Anderson DG. RNA Circularization Diminishes Immunogenicity and Can Extend Translation Duration In Vivo. Mol Cell. 2019;74:508-520.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 290] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 84. | Pugnale P, Pazienza V, Guilloux K, Negro F. Hepatitis delta virus inhibits alpha interferon signaling. Hepatology. 2009;49:398-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 85. | Karimzadeh H, Kiraithe MM, Kosinska AD, Glaser M, Fiedler M, Oberhardt V, Salimi Alizei E, Hofmann M, Mok JY, Nguyen M, van Esch WJE, Budeus B, Grabowski J, Homs M, Olivero A, Keyvani H, Rodríguez-Frías F, Tabernero D, Buti M, Heinold A, Alavian SM, Bauer T, Schulze Zur Wiesch J, Raziorrouh B, Hoffmann D, Smedile A, Rizzetto M, Wedemeyer H, Timm J, Antes I, Neumann-Haefelin C, Protzer U, Roggendorf M. Amino Acid Substitutions within HLA-B*27-Restricted T Cell Epitopes Prevent Recognition by Hepatitis Delta Virus-Specific CD8+ T Cells. J Virol. 2018;92:e01891-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 86. | Karimzadeh H, Kiraithe MM, Oberhardt V, Salimi Alizei E, Bockmann J, Schulze Zur Wiesch J, Budeus B, Hoffmann D, Wedemeyer H, Cornberg M, Krawczyk A, Rashidi-Alavijeh J, Rodríguez-Frías F, Casillas R, Buti M, Smedile A, Alavian SM, Heinold A, Emmerich F, Panning M, Gostick E, Price DA, Timm J, Hofmann M, Raziorrouh B, Thimme R, Protzer U, Roggendorf M, Neumann-Haefelin C. Mutations in Hepatitis D Virus Allow It to Escape Detection by CD8+ T Cells and Evolve at the Population Level. Gastroenterology. 2019;156:1820-1833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 87. | Kefalakes H, Koh C, Sidney J, Amanakis G, Sette A, Heller T, Rehermann B. Hepatitis D Virus-Specific CD8+ T Cells Have a Memory-Like Phenotype Associated With Viral Immune Escape in Patients With Chronic Hepatitis D Virus Infection. Gastroenterology. 2019;156:1805-1819.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |