Published online May 28, 2020. doi: 10.3748/wjg.v26.i20.2584
Peer-review started: February 28, 2020
First decision: April 9, 2020
Revised: May 8, 2020
Accepted: May 19, 2020
Article in press: May 19, 2020
Published online: May 28, 2020
Processing time: 90 Days and 1.9 Hours
Since it is currently not possible to eradicate hepatitis B virus (HBV) infection with existing treatments, research continues to uncover new therapeutic strategies. HBV core protein, encoded by the HBV core gene (HBC), intervenes in both structural and functional processes, and is a key protein in the HBV life cycle. For this reason, both the protein and the gene could be valuable targets for new therapeutic and diagnostic strategies. Moreover, alterations in the protein sequence could serve as potential markers of disease progression.
To detect, by next-generation sequencing, HBC hyper-conserved regions that could potentially be prognostic factors and targets for new therapies.
Thirty-eight of 45 patients with chronic HBV initially selected were included and grouped according to liver disease stage [chronic hepatitis B infection without liver damage (CHB, n = 16), liver cirrhosis (LC, n = 5), and hepatocellular carcinoma (HCC, n = 17)]. HBV DNA was extracted from patients’ plasma. A region between nucleotide (nt) 1863 and 2483, which includes HBC, was amplified and analyzed by next-generation sequencing (Illumina MiSeq platform). Sequences were genotyped by distance-based discriminant analysis. General and intergroup nt and amino acid (aa) conservation was determined by sliding window analysis. The presence of nt insertion and deletions and/or aa substitutions in the different groups was determined by aligning the sequences with genotype-specific consensus sequences.
Three nt (nt 1900-1929, 2249-2284, 2364-2398) and 2 aa (aa 117-120, 159-167) hyper-conserved regions were shared by all the clinical groups. All groups showed a similar pattern of conservation, except for five nt regions (nt 1946-1992, 2060-2095, 2145-2175, 2230-2250, 2270-2293) and one aa region (aa 140-160), where CHB and LC, respectively, were less conserved (P < 0.05). Some group-specific conserved regions were also observed at both nt (2306-2334 in CHB and 1935-1976 and 2402-2435 in LC) and aa (between aa 98-103 in CHB and 28-30 and 51-54 in LC) levels. No differences in insertion and deletions frequencies were observed. An aa substitution (P79Q) was observed in the HCC group with a median (interquartile range) frequency of 15.82 (0-78.88) vs 0 (0-0) in the other groups (P < 0.05 vs CHB group).
The differentially conserved HBC and HBV core protein regions and the P79Q substitution could be involved in disease progression. The hyper-conserved regions detected could be targets for future therapeutic and diagnostic strategies.
Core tip: New tools for hepatitis B virus infection treatment and follow-up are needed. Hepatitis B virus core protein has a key role in viral replication and persistence. Analysis of viral quasispecies by next-generation sequencing can identify conserved regions in viral genes or proteins that may serve as targets for new therapeutic and diagnostic strategies. Moreover, it may help identify prognostic markers of liver disease progression. Here, we detected hyper-conserved nucleotide and amino acid regions regardless of the clinical stage. Moreover, we observed several group-specific conserved and variable regions and an amino acid substitution that could be indicative of different disease progression.
- Citation: Yll M, Cortese MF, Guerrero-Murillo M, Orriols G, Gregori J, Casillas R, González C, Sopena S, Godoy C, Vila M, Tabernero D, Quer J, Rando A, Lopez-Martinez R, Esteban R, Riveiro-Barciela M, Buti M, Rodríguez-Frías F. Conservation and variability of hepatitis B core at different chronic hepatitis stages. World J Gastroenterol 2020; 26(20): 2584-2598
- URL: https://www.wjgnet.com/1007-9327/full/v26/i20/2584.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i20.2584
Hepatitis B virus (HBV) is a small virus with a specific tropism for the liver. It belongs to the Hepadnaviridae family. Despite the existence of effective preventive vaccines, an estimated 257 million people worldwide live with chronic HBV infection and more than 880000 people die every year of HBV-related complications such as liver cirrhosis (LC) and hepatocellular carcinoma (HCC)[1].
HBV is an enveloped virus equipped with 3.2 kb of partially double-stranded circular DNA produced by the reverse transcription of an RNA intermediate known as pregenomic RNA[2]. This ribonucleic intermediate is produced from a viral DNA molecule that interacts with cellular (histone and non-histone) and viral proteins, forming a “mini-chromosome” known as covalently closed circular DNA (cccDNA) that remains in hepatocyte nuclei for the rest of the cell’s life[3]. Although current antiviral therapy can control viral replication, it is not capable of interfering with the formation or persistence of cccDNA, rendering HBV infection eradication impossible. This mini-chromosome could even be a source of HBV reactivation after clinical resolution and HBsAg seroclearance[4]. Due to persistent infection, up to 1% of Caucasian patients with noncirrhotic chronic HBV infection have been found to develop HCC[5].
Gene therapy has emerged as one of the most promising strategies for blocking disease progression, and results from studies investigating the potential of small interfering RNA (siRNA) systems as adjuvant therapy are encouraging[6]. SiRNA is a double-stranded noncoding RNA [with an optimal length of 21 nucleotides (nt)] that interacts with target messenger RNA, promoting its degradation and silencing of the gene[7].
HBV reverse transcriptase lacks 3' to 5' proofreading activity, which leads to viral genome variability comparable to that observed in an RNA virus[8]. This genetic variability is further increased by inter- and intra-genotype recombination events[9]. In short, HBV circulates as a complex mixture of closely related genetic variants (haplotypes) known as quasispecies[10].
The HBV core protein (HBc) [encoded by the HBV core gene (HBC) from the PreCore/Core open reading frame (ORF)] is essential for viral replication. It is a structural 21-kDa protein that self-assembles to create dimers that assemble in hexamers forming the icosahedral viral capsid[11,12]. It has 183 amino acids (aa) (185 for genotype A) with a N-terminal domain and a C-terminal domain (CTD) connected through a linker region. The N-terminal domain ranges from aa position 1 to 149 (including the linker region aa 140 to 149) and constitutes the α helix-rich assembly domain[13]. The CTD is shorter (aa 150 to 183, or 185 for genotype A) and constitutes the functional domain[13]. The CTD allows HBc to intervene in a multitude of processes such as subcellular traffic, viral genome release, capsid assembly and transport, RNA metabolism, and viral pregenomic RNA reverse transcription[14]. Considering just how essential this protein is for viral replication, it could be an optimal target for gene therapy. Moreover, mutations in HBc may have different roles in liver disease progression, positioning them as potentially useful prognostic genetic markers.
Next-generation sequencing (NGS) is a highly sensitive technique for studying viral quasispecies; it is capable of detecting highly conserved regions of the HBV genome, regardless of genome or clinical stage[9]. Moreover, it supports the identification and quantitative determination[15] of specific variants that could be used as markers to predict prognosis and treatment response in patients with HBV infection.
The aim of this study was to apply NGS to analyse HBc conservation and variability at the nt and aa levels in patients with different stages of chronic HBV infection in order to identify hyper-conserved regions of the HBC gene that could be a target for gene therapy and to determine possible prognostic factors of disease progression
The study was reviewed and approved by the Clinical Research Ethics Committee of Hospital Universitari Vall d’Hebron (PR(AG)146/2020). No animals were used.
Forty-five patients with chronic HBV infection were recruited from members of the general population seen at the outpatient clinic at Vall d’Hebron University Hospital in Barcelona, Spain. They tested negative for hepatitis D virus, hepatitis C virus, and human immunodeficiency virus, and had a viral load > 3 log IU/mL, which is the limit of polymerase chain reaction (PCR) amplification sensitivity. HBV serological markers such as the surface antigen (HBsAg), the e antigen (HBeAg), and anti-HBe antibodies were tested using commercial chemiluminescent assays on a COBAS 8000 analyzer (Roche Diagnostics, Rotkreuz, Switzerland). HBV DNA was quantified by real-time PCR with a detection limit of 10 IU/mL (COBAS 6800, Roche Diagnostics). Patients were divided into 3 clinical groups according to liver disease stage determined by biopsy or diagnostic imaging in line with the EASL guidelines[16]: Chronic HBV infection without liver damage (CHB group), chronic HBV infection with liver cirrhosis (LC group), and chronic HBV infection with hepatocellular carcinoma (HCC group).
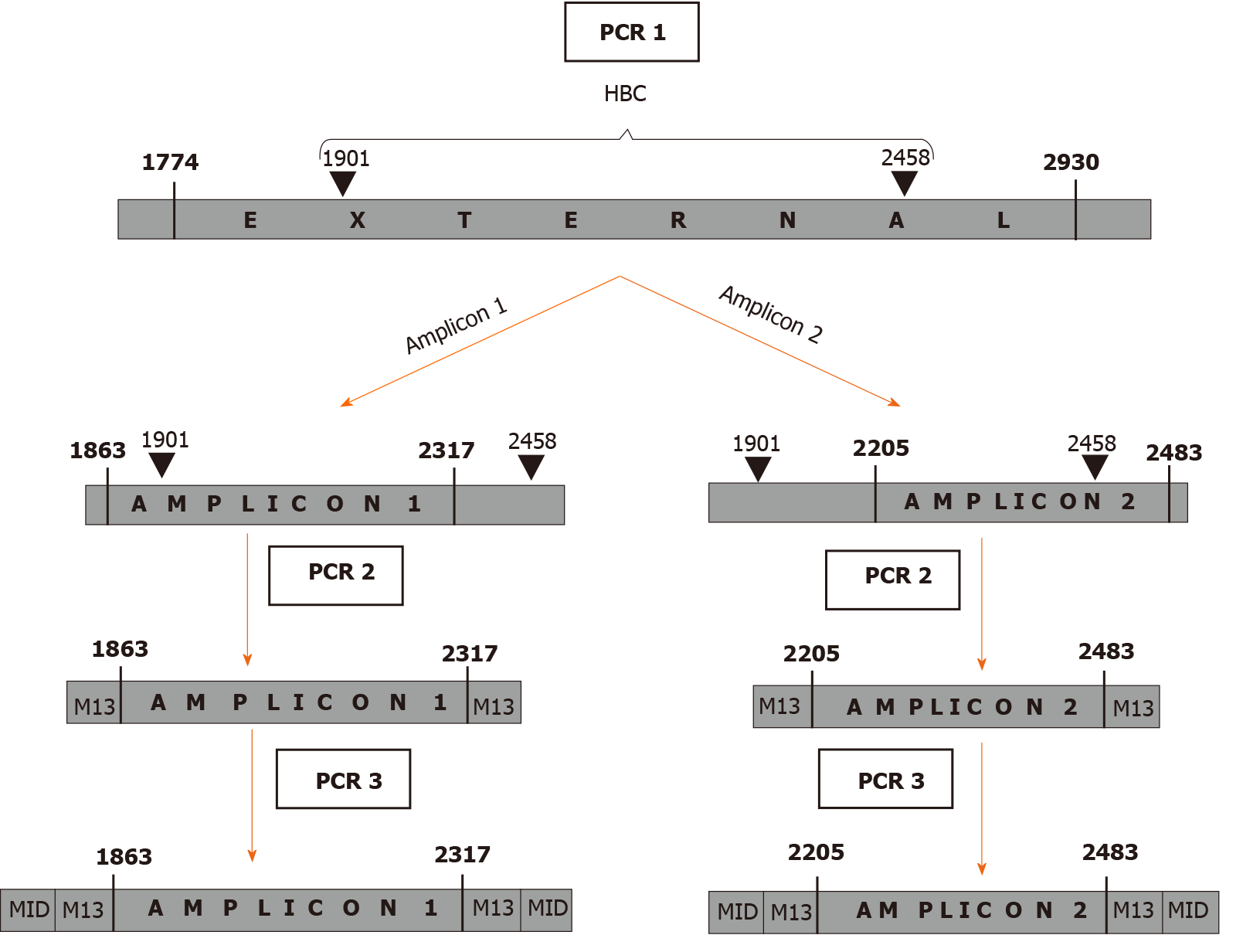
HBV DNA was extracted from 200 µL of serum using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The region of interest was amplified through a 3-step nested PCR protocol (Figure 1). The first step (PCR1) covered a large region between nt 1774-2930 that includes the HBC gene (nt 1901-2464 for genotype A and 1901-2458 for other genotypes). As the Illumina MiSeq platform (Illumina, San Diego, CA, United States) allows read lengths of up to 600 bp, the following amplification steps were performed by dividing HBC into 2 amplicons (amplicon 1 = nt 1863-2317 and amplicon 2 = nt 2205-2483), which overlapped in a 112 nt-long portion (PCR2). The M13-tail, added in step 2, was used for the last step (PCR3), which introduced a 10 nt-long sample-specific multiplex identifier. All the PCR steps were performed using high-fidelity Pfu Ultra II DNA polymerase (Stratagene, Agilent Technologies, Santa Clara, CA, United States). The primers and protocols are reported in Table 1.
| PCR | Primer | Primer sequence (5'- > 3') | Amplified region | Protocol | |
| 1st step | PCR1 | Forward | TAGGAGGCTGTAGGCATA | 1774-2930 | 95 ºC 5 min; (95ºC 20 s, 49 ºC 20 s, 72 ºC 15 s) × 35 cycles; 72 ºC 3 min |
| Reverse | GGAAAGAATCCCAGAGG | ||||
| 2nd step | PCR2 A.1 | Forward A.1 | GTTGTAAAACGACGGCCAGTTTCAAGCCTCCAAGCTGT | 1863-2317 | 95 ºC 2 min; (95 ºC 20 s, 58 ºC 20 s, 72 ºC 15s ) × 35 cycles; 72 ºC 3 min |
| Reverse A.1 | CACAGGAAACAGCTATGACCGATAGGGGCATTTGGTGGTCT | ||||
| PCR2 A.2 | Forward1 A.2 | GTTGTAAAACGACGGCCAGTGGTTTCATATTTCTTGCC | 2205-2483 | 95 ºC 2 min; (95 ºC 20 s, 50 ºC 20 s, 72 ºC 15 s) × 35 cycles; 72 ºC 3 min | |
| Forward2 A.2 | GTTGTAAAACGACGGCCAGTGGTTTCACATTTCCTGTC | ||||
| Forward3 A.2 | GTTGTAAAACGACGGCCAGTGGTTTCACATTTCTTGTC | ||||
| Forward4 A.2 | GTTGTAAAACGACGGCCAGTGGTTTCACATTTCCTGCC | ||||
| Reverse A.2 | CACAGGAAACAGCTATGACCTCCCACCTTATGAGTCCAAG | ||||
| 3rd step | PCR3 | Forward (specific per sample) | GTTGTAAAACGACGGCCAGT+specific 10 nt MID | 95 ºC 2 min; (95 ºC 20 s, 60 ºC 20 s, 72 ºC 15 s) × 20 cycles; 72 ºC 3 min | |
| Reverse (specific per sample) | CACAGGAAACAGCTATGACC+specific 10 nt MID |
The final PCR products were purified with Agencourt AMPure XP magnetic beads (Beckman Coulter, Beverly, LA, United States) and their quality verified using the Agilent 2200 TapeStation System and D1000 ScreenTape kit (Agilent Technologies, Waldbronn, Germany).
Purified amplicons were quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific-Life Technologies, Austin, TX, United States) and pooled to guarantee that the 2 amplicons for each patient were adequately represented in the analysis (2.5x for amplicon 1 and 1x for amplicon 2, due to their different lengths). The amplicon pool was sequenced by NGS on the Illumina MiSeq platform.
The reads obtained underwent an in-house bioinformatics filtering procedure based on R scripts[17], as previously described by our group[18]. For each amplicon, a group of unique sequences (haplotypes) forming the viral quasispecies was obtained. All sequences that did not match in the overlapping 112-nt region between amplicon 1 and 2 were discarded.
The bioinformatics methods used in this study were reviewed by Mercedes Guerrero-Murillo from the Microbiology Department at Vall d’Hebron Hospital (Barcelona, Spain) and by Dr. Josep Gregori from the Liver Disease Viral Hepatitis Laboratory at Vall d’Hebron Hospital (Barcelona, Spain), CIBERehd research group, and Roche Diagnostics SL.
The amplicons from each patient were aligned with the same region of the respective amplicons extracted from 106 full-length HBV genome sequences representative of genotypes A to J obtained from the NCBI GenBank (Supplementary Table 1). Genotyping was conducted by applying distance-based discriminant analysis (DB rule)[19,20], which considers the inter- and intra-class variability of all genotypes. Genetic distances were computed according to the Kimura-80 model[21].
Sequence conservation at nt and aa levels was determined by calculating the information content (IC) of each position in a multiple alignment of all haplotypes detected with a frequency > 0.25.
This analysis calculates the mean IC for windows of 25 nt (or 10 aa), starting from the first position in the multiple alignment and moving forward in steps of 1[9]. The hyper-conserved regions were detected by aligning all haplotypes, regardless of clinical stage. Differences in sequence conservation between the groups were determined by comparing IC values.
To identify specific nt insertions and deletions (indels) and aa substitutions that could discriminate between the groups, haplotype sequences were aligned with their genotype-specific consensus sequence. Consensus was obtained by aligning the sequences of the subgenotypes of interest extracted from the 106 full-length HBV genome sequences. Polymorphisms were identified by aligning haplotype sequences with a population consensus sequence and discarded.
Sequence conservation differences between the groups in the sliding windows were analysed using the Wilcoxon–Mann–Whitney test. Frequencies of aa changes detected were compared with the Kruskal-Wallis test and described as median and interquartile range (IQR). All analyses were performed in R version 3.2.3. P < 0.05 was considered significant.
Of the 45 patients with chronic hepatitis initially included in the study, 38 passed the sequencing quality filters and had correctly overlapping amplicons 1 and 2. After application of the quality filters, a median (IQR) of 133156.5 (85961.25-605212) and 66571 (25958.5-2301225) sequences per patient were obtained respectively for amplicon 1 and amplicon 2. NGS data were submitted to the GenBank SRA database (BioProject accession number PRJNA625435; BioSample accession numbers are reported in Supplementary Table 2). In the clinical groups, there were 16 patients with CHB, 5 with LC, and 17 with HCC. The clinical and viral characteristics (including genotypes) are reported in Table 2.
| Median [IQR] | CHB (n = 16) | HCC (n = 17) | LC (n = 5) | P value |
| Age | 38.5 [33.5-46.5] | 67 [58-69] | 56 [48-66] | 0.002 |
| Viral load (log IU/mL) | 6.8 [5.7-8.0] | 5.5 [4.7-6.7] | 5.7 [4.8-6.2] | NS |
| ALT | 56.5 [41.25-180.5] | 70 [47-212] | 46 [43-79] | NS |
| AST | 56 [34.75-124] | 120.5 [59-163.5] | 66.45 [48.675-84.225] | NS |
| Platelets (109/L) | 183 [161.5-226] | 136 [98.5-255] | 81.5 [61.25-101.75] | NS |
| Proportion | ||||
| Gender (male) | 11/16 | 15/17 | 3/5 | |
| HBeAg (positive) | 8/16 | 3/17 | 0/5 | |
| Genotype, % (n) | ||||
| A | 18.8 (3) | 5.9 (1) | 20.0 (1) | |
| C | 37.5 (6) | 5.9 (1) | 20.0 (1) | |
| D | 25.0 (4) | 64.7 (11) | 40.0 (2) | |
| D/A | (0) | 5.9 (1) | 0.0 (0) | |
| D/E | 6.3 (1) | 11.8 (2) | 0.0 (0) | |
| E | 6.3 (1) | 0.0 (0) | 20.0 (1) | |
| F | 6.3 (1) | 5.9 (1) | 0.0 (0) |
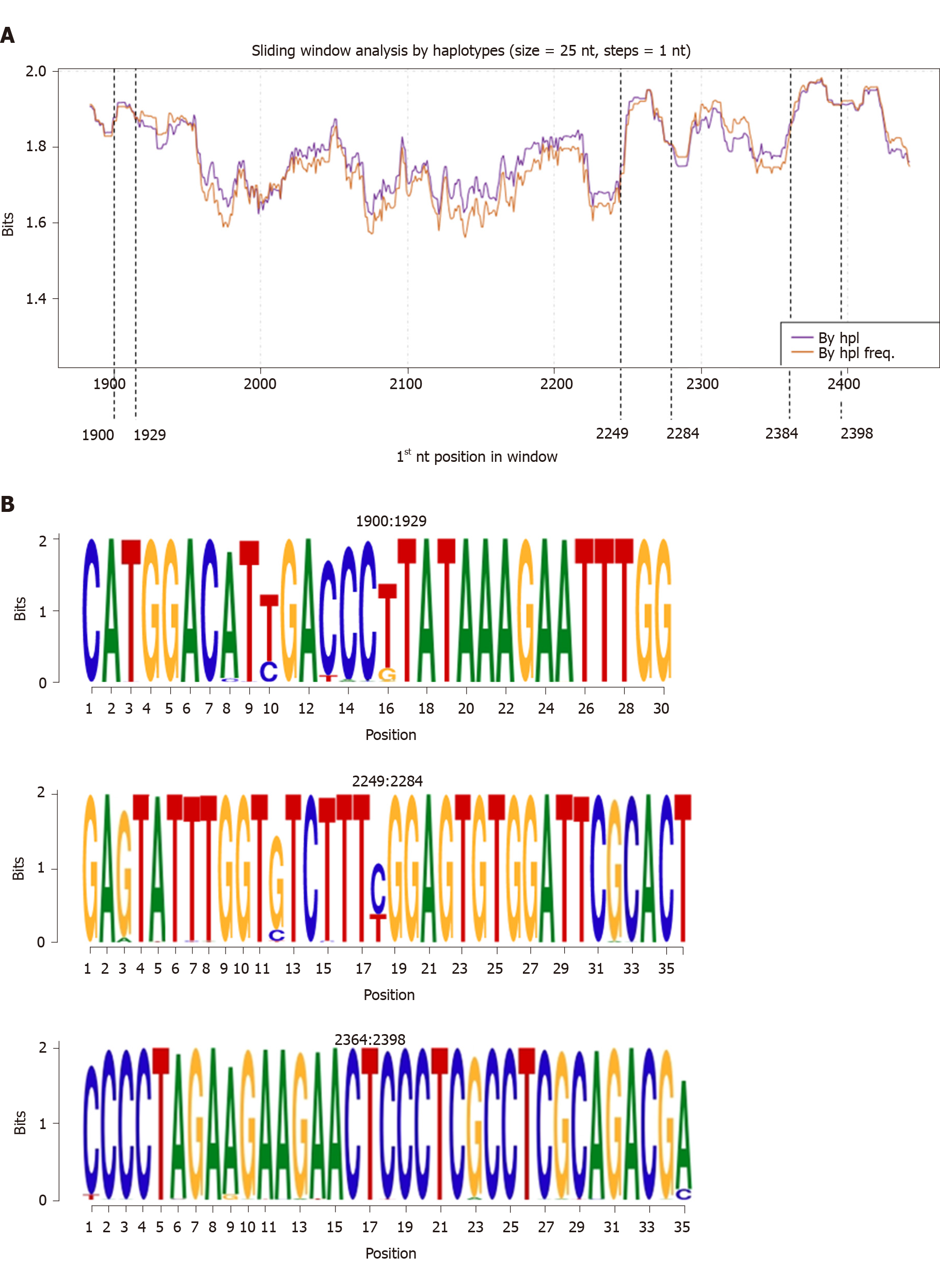
Sequence conservation was studied by applying a sliding window analysis to the entire HBC sequence overlapping the 2 amplicons at the common 112 nt-long portion. No differences in IC were observed on analyzing the sequences by haplotype considering or not their relative frequency (Figure 2A). Considering the IC of all the nt-sequence haplotypes obtained (regardless of clinical group), we identified 3 hyper-conserved regions (nt 1900-1929, 2249-2284, and 2364-2398, Figure 2B). Most of the nt positions within these regions yielded the maximum IC value of 2 bits (100% conservation).
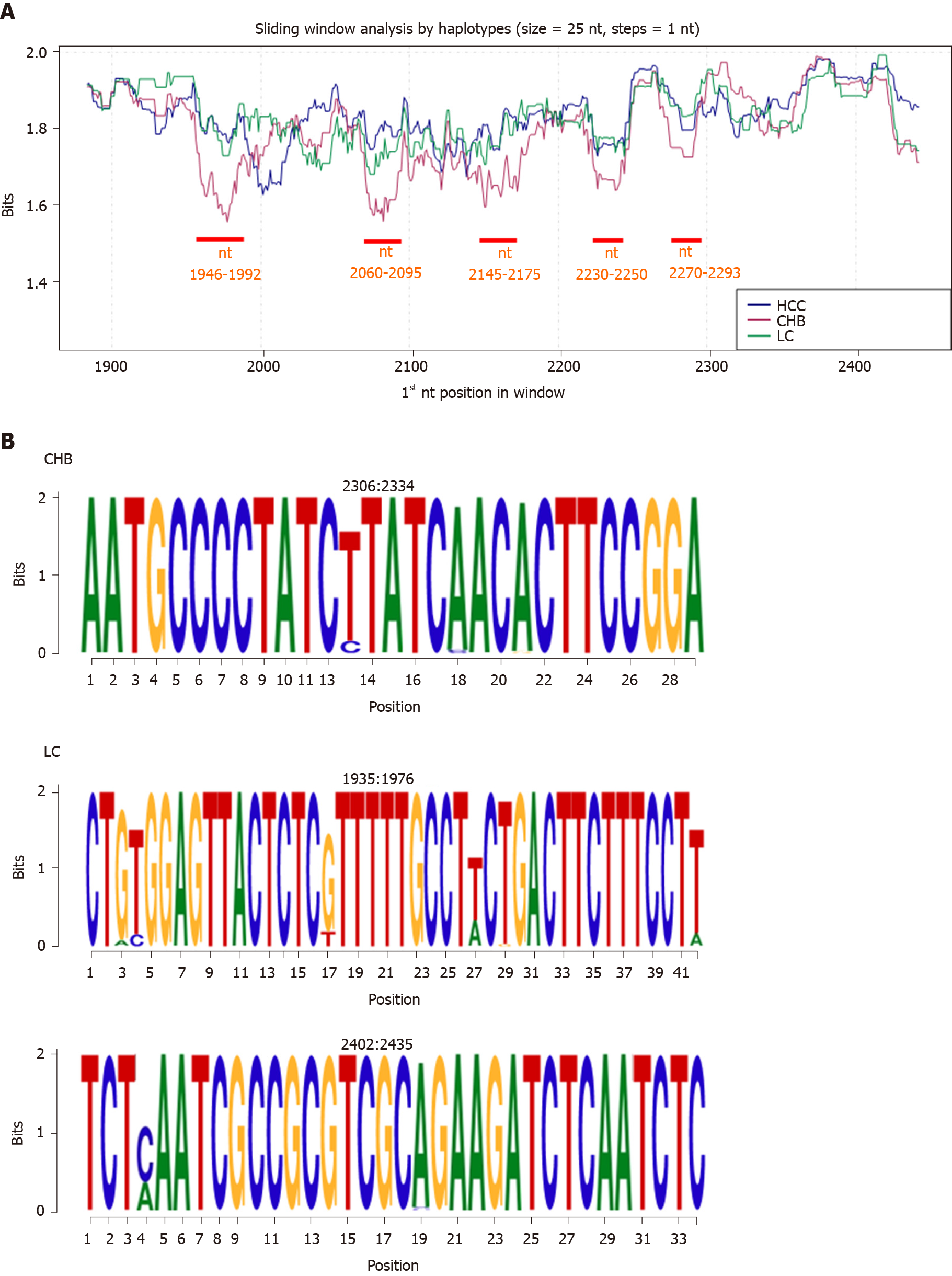
On comparing the IC of each clinical group by haplotype, the HCC and LC groups showed similar conservation patterns; CHB was notably associated with the lowest level of conservation, mainly evident in 5 regions: nt 1946-1992, 2060-2095, 2145-2175, 2230-2250, and 2270-2293 (P < 0.05, Figure 3A). Three group-specific conserved regions were detected: 1 in the CHB group (nt 2306-2334) and 2 in the LC group (nt 1935-1976 and 2402-2435; Figure 3B). Most of the nt positions within these regions yielded the maximum IC value of 2 bits (100% conservation).
The aa sequences of the haplotypes were translated from their respective nt sequences using the HBC reading frame.
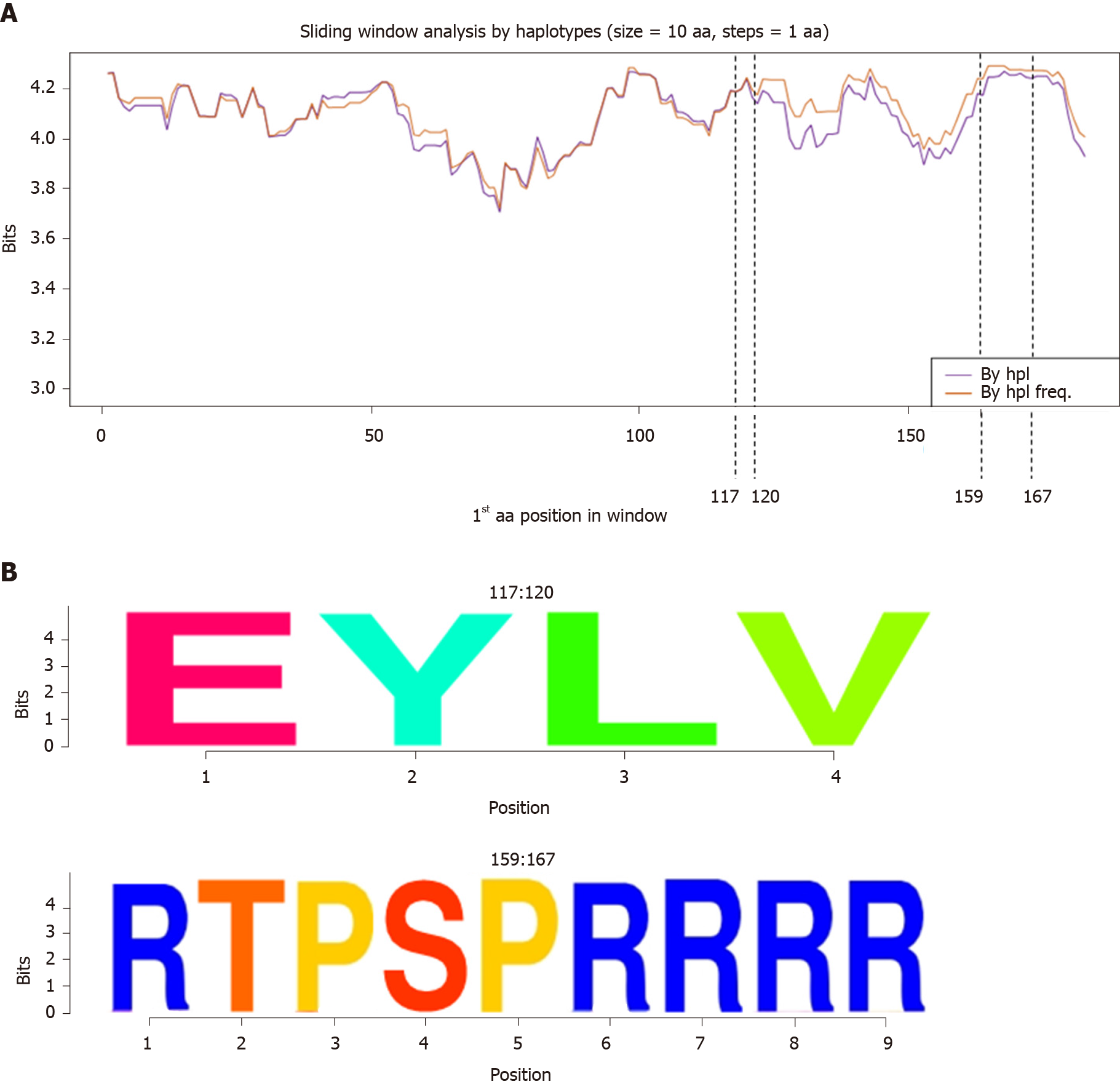
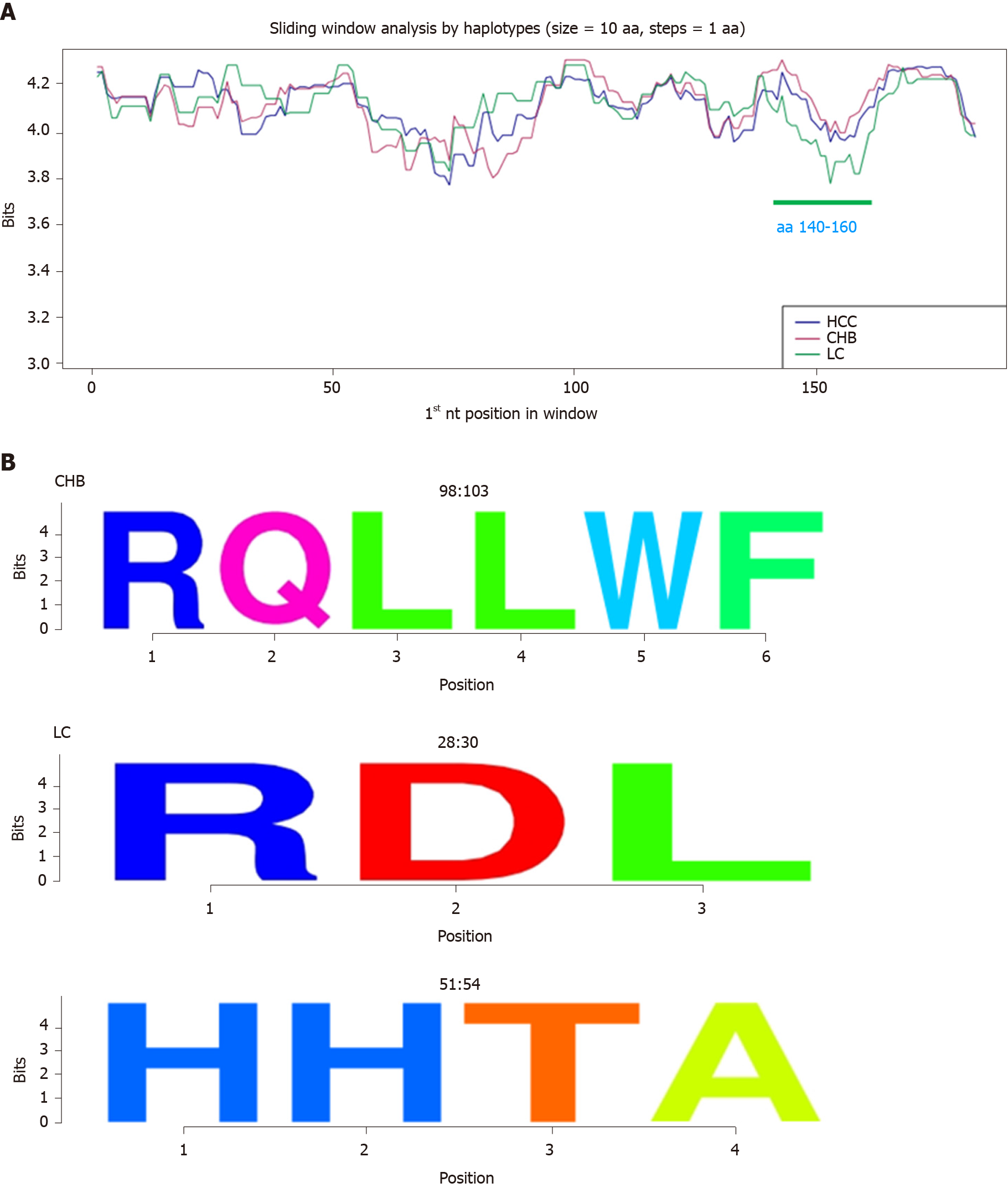
Sliding window analysis of the aa haplotypes of the 38 patients by haplotype and haplotype frequency (Figure 4A) showed that the HBc protein was highly conserved throughout its sequence except for the central region (between aa 50 and 100), where conservation was slightly decreased. Two common hyper-conserved regions were detected: 1 between aa 117-120 and 1 between aa 159-167 (Figure 4B). All the aa in these regions had a conservation of around 100% (4.32 bits).
On analyzing aa conservation by haplotype in relation to clinical stage, the 3 groups showed a similar pattern, except for a region between aa 140 and 160, which was less conserved in the LC group compared with the CHB and HCC groups (P < 0.05, Figure 5A). Again, 3 group-specific conserved aa regions were detected: 1 in the CHB group (aa 98-103) and 2 in the LC group (aa 28-30 and 51-54, Figure 5B). All the aa in these regions had a conservation of around 100% (4.32 bits).
nt indels and aa changes were identified by aligning the patients’ haplotypes with their genotype-specific consensus sequence.
In the CHB group, 8/16 patients had indels in HBC, vs 2/17 in the HCC group and 1/5 in the LC group. The indels consisted of the insertion or deletion of one nt at positions 1951 or 2085 (a thymine in 1951 and a guanine in 2085; Table 3). In all cases, a truncated HBc protein was produced. However, due to the limited number of patients, no statistical differences were observed on comparing the frequencies between the groups.
| Clinical stage (n/total) | Patient | Relative frequency (% of mutated haplotypes) | |
| 1951 (1 nt: T) | 2085 (1 nt: G) | ||
| CHB (8/16) | 1 | 8.36 (8.7) | |
| 2 | 17.12 (40) | ||
| 3 | 3.19 (5) | ||
| 4 | 0.37 (5.9) | ||
| 9 | 2.02 (8.82) | ||
| 10 | 1.34 (50) | ||
| 12 | 1.04 (10) | ||
| 13 | 2.79 (22.22) | ||
| HCC (2/17) | 28 | 0.78 (4) | |
| 33 | 2.42 (4.8) | ||
| LC (1/5) | 34 | 17.42 (19.2) | |
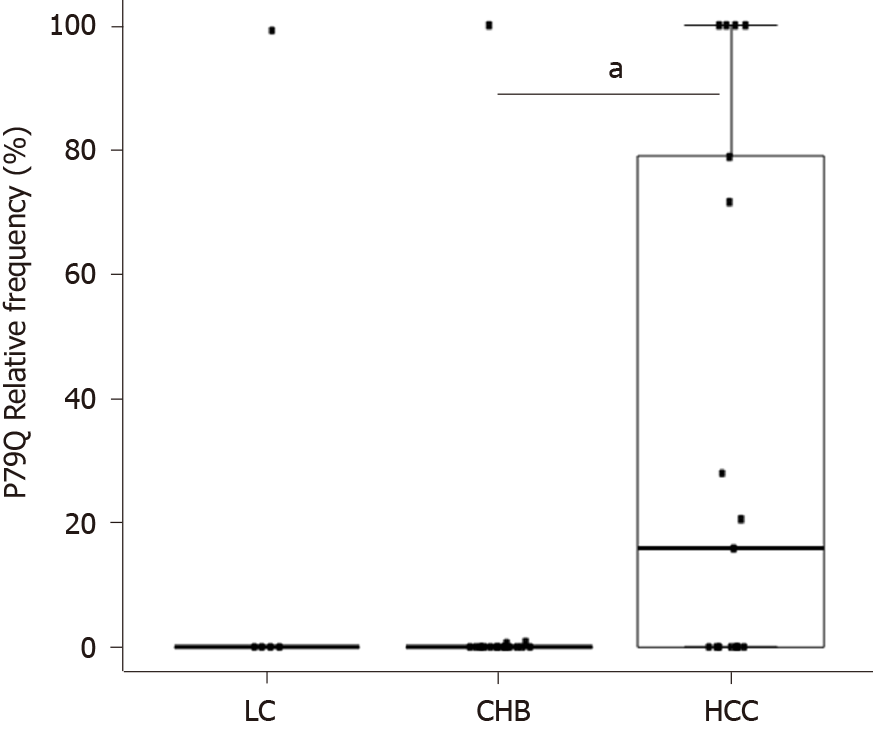
On analysing the presence of aa changes, we identified the aa substitution P79Q (proline to glutamine) in the HCC group with a median (IQR) frequency of 15.82 (0-78.9) vs (0-0) in the CHB group (P < 0.05) and 0 (0-0) in the LC group (Figure 6).
The HBc protein, encoded by the HBC gene, is a key element in viral replication and disease progression and is involved in both structural and functional processes. Studying gene and protein sequences in patients with different clinical stages of HBV infection could provide important information on the pathogenic role of this protein. Moreover, the identification of hyper-conserved regions at both nt and aa levels could help develop new therapeutic approaches, including gene therapy. In this study, we used NGS to analyse HBC quasispecies in a group of patients with chronic HBV infection stratified by liver disease stage.
First, we studied quasispecies conservation to search for hyper-conserved nt and aa regions regardless of clinical stage or viral genotype. Current treatment based on nucleos(t)ide inhibitors does not affect cccDNA levels or transcriptional activity and therefore cannot eliminate HBV infection. This viral mini-chromosome supports the continuous expression of viral antigens that possibly contribute to disease progression, even in the presence of drug-induced viral suppression[22].
New therapeutic approaches are thus required to control HBV expression, and the targeted delivery of siRNA is one of the most promising approaches under investigation[23]. Several siRNAs are currently being tested against X and S ORFs. A study conducted in chimpanzees showed that multiple injections of ARB-1467 (a mixture of 3 interfering RNAs targeting both X and S ORFs[24]) led to a 90% reduction in HBsAg levels and a 50% reduction in cccDNA within 28 d of treatment[25]. None of the molecules currently available, however, target HBC, which considering its role in viral replication could be a valuable target for siRNA-based therapies.
In this study, we analysed quasispecies conservation of the entire HBC gene in patients infected by different HBV genotypes and with different clinical stages of disease in order to identify hyper-conserved regions that might be useful for pangenotypic and panclinical RNA silencing strategies. On analyzing nt conservation for the group of 38 patients, we detected 3 shared hyper-conserved regions, namely the start codon of HBC expression (nt 1900-1929), a portion with 2 CD8 epitopes (HLA-A24 and A3303) (nt 2249-2284)[26], and an arginine-rich portion of the CTD (nt 2364-2398). All 3 sequences could be valuables targets for a new gene silencing strategy.
At the aa level, we observed 2 common hyper-conserved regions (aa 117-120 and 159-167), which fell into the second and third hyper-conserved nt portions (nt 2249-2284 and 2364-2398 respectively). The CTD plays a key role in HBc function. It contains the 4 arginine-rich domains (RRR aa 150-152, RRR aa 157-159, RRRR aa 164-167, and RRRR aa 172-175) that guarantee adequate protein subcellular localization acting as nuclear or cytoplasmic localization signals[27]. The second hyper-conserved aa region (aa 159-167) included one of these arginine-rich domains.
The high degree of sequence conservation observed in HBc may be indicative of its importance in protein function, positioning it as a possible target for diagnostic and therapeutic strategies. Recent studies have defined HBV core-related antigen (HBcrAg, which consists of HBc, HBeAg, and HBV p22 protein) as a promising serological viral marker, particularly for patients with low viral loads, such as treated patients[28] and patients with chronic HBeAg-negative infection[29]. This potential marker, however, has some limitations related to its high limits of detection (2 log IU/mL) and quantification (3-7 log IU/mL). The hyper-conserved regions observed in our study could be used as targets to improve HBc detection technology.
Aptamers are emerging as a promising diagnostic and therapeutic option for different diseases[30]. These molecules consist of single-strand DNA or RNA with high affinity and specificity and no toxicity or immunogenicity[31]. In vitro testing of an aptamer generated using the matrix domain of HBV (located in the large surface protein L and related to the nucleocapsid envelope) resulted in a 50% decrease in HBV titre in treated cell supernatants[32]. In another study, an aptamer targeting HBC resulted in a reduction in extracellular HBV DNA by interfering with nucleocapsid assembly[31]. Again, the hyper-conserved regions detected in our study could be novel targets for aptamer-based strategies that might work independently of clinical stage or HBV genotype. They could be also used to elaborate a new HBV detection system, as has been done with hepatitis C virus[33] and syncytial viruses[34].
On analyzing nt and aa conservation in relation to clinical stage of HBV infection, all 3 groups showed similar patterns at the aa level, although the HBV quasispecies in the LC group was slightly less conserved (mainly between aa 140-160). At the nt level, conservation was lower in the CHB group than in the other 2 groups, largely in the 5 regions between nt 1946-1992, 2060-2095, 2145-2175, 2230-2250, and 2270-2293. This finding could be consistent with the high replication rate of HBV during this clinical stage. Moreover, the first variable region (nt 1946-1992) includes three CD8 HLA epitopes (epitopes B5101, B3501, and B0702 at nt positions 1958-1982)[26], suggesting an attempt at immune evasion. Although the CHB group had the lowest levels of sequence conservation, we detected 2 group-specific conserved regions: aa 98-103 and nt 2306-2334. The nt region included the first 5 aa of the linker region, suggesting thus an important role for this region, which is involved in capsid assembly[35,36] and viral DNA synthesis[36]. In the LC group we detected 2 exclusively conserved nt regions (nt 1935-1976 and 2402-2435, which would translate respectively to aa 11-25 and 167-178) and 2 exclusively conserved aa regions (aa 28-30 and 51-54). The first related regions (nt 1935-1976 and aa 28-30) included portions of HBc (aa 14-18 and aa 23-39 respectively) that are involved in capsid assembly and envelopment and virion production[37], highlighting the importance of these functions in LC. The second LC-specific nt region (nt 2402-2435) contained an arginine-rich domain of the CTD when translated.
The identification of group-specific conserved regions suggests different evolutionary histories that may have different effects on disease progression. Further studies, however, are needed to prove the association between these regions and different clinical stages and to investigate their role in liver disease progression.
Considering the risk and severity of disease progression, identification of prognostic factors would be of great help. A number of studies have focused on detecting aa changes possibly related to different clinical stages. The mutations T1753C and A1762T/G1764A (K130M/V131I in HBx) of basal core promoter, for example, were identified as possible prognostic markers for HCC[38,39], while HBc aa mutations F24Y, E64D, E77Q, A80I/T/V, L116I, and E180A were linked to the development of cirrhosis and HCC[40]. In our study, one of the aa changes detected, P79Q, was exclusively observed in the HCC group. Mutations at this position have been found to be slightly associated with tumour relapse after resection[41]. More in vitro studies are required to investigate the role of the P79Q mutation in liver disease progression.
One limitation of our study is that we were not able to include large numbers of patients with different stages of liver disease due to the limits of PCR detection. This was particularly evident in the LC group, which was very small. Larger samples are needed to confirm our results. Moreover, although the Illumina MiSeq platform offers long read lengths, they are not sufficient to cover the entire HBC gene, making it necessary to divide it into 2 partially overlapping amplicons. Nonetheless, these 2 fragments were treated as independent samples during sequencing and subsequently analysed as such.
In summary, we have identified a number of nt and aa hyper-conserved regions that could be valuable targets for new therapeutic and diagnostic strategies. The role of group-specific conserved regions in liver disease progression requires further analysis. The P79Q substitution could be a possible prognostic factor for HCC. In vitro studies, however, are required to determine whether this change might affect viral replication and to investigate associations between cellular damage and onset of HCC.
Despite the existence of effective preventive vaccines, an estimated 257 million people worldwide live with chronic hepatitus B virus (HBV) infection and more than 880000 people die due to the development of liver cirrhosis and/or hepatocellular carcinoma. Although infection can be controlled with existing treatment, eradication is currently impossible due to the persistence of covalently closed circular DNA in hepatocyte nuclei that acts as a template for viral expression. New therapeutic approaches are needed, and gene therapy has been proposed as one of the most promising options. HBV core protein [encoded by the HBV core gene (HBC)] is a structural protein with functional activity that has a key role in viral replication and disease progression. Accordingly, it could be a potential target for new therapeutic and diagnostic strategies, and its variability could be a valuable prognostic factor for disease progression.
As eradication of HBV infection is currently unachievable, new therapeutic strategies are necessary. Moreover, current treatments cannot interfere with the expression of viral proteins that can favor disease progression. Gene therapy based on silencing RNA is one of the most promising therapeutic approaches currently under investigation. The identification of hyper-conserved regions in key viral genes and proteins (such as HBC) is essential to orchestrate an effective strategy regardless of clinical stage or viral genotype.
This study aimed to identify, by next-generation sequencing, hyper-conserved regions in HBC quasispecies of patients with different clinical stages of chronic HBV infection that could be a valuable target for gene therapy. Considering the essential role of the HBC gene and its encoded protein HBV core protein in HBV infection, changes in gene and protein conservation in specific clinical groups could be determining factors in disease progression and hence serve as prognostic factors for clinical follow-up.
The HBC gene was amplified by a 3-nested PCR protocol and later sequenced by next-generation sequencing (MiSeq, Illumina, United States) in 38 HBV-monoinfected chronic patients [16 with chronic hepatitis B infection without liver damages (CHB group), 5 with liver cirrhosis (LC group) and 17 with hepatocellular carcinoma (HCC group)]. Quasispecies sequences were genotyped by distance-based discriminant analysis, and general and intergroup nucleotide (nt) and amino acid (aa) conservation was determined by sliding window analysis. The presence of nt insertion and deletions and/or aa substitutions in the different groups was determined by aligning the sequences with a genotype-specific consensus sequence.
Three nt (nt 1900-1929, 2249-2284, 2364-2398) and two aa (aa 117-120, 159-167) hyper-conserved regions shared by all the clinical groups were identified. By comparing gene and protein conservation between the different clinical groups, a similar pattern of conservation was observed, although CHB showed five nt less conserved regions (nt 1946-1992, 2060-2095, 2145-2175, 2230-2250, 2270-2293) and LC one aa less conserved region (between aa 140 and 160). Moreover, some group-specific conserved regions were detected at both nt (nt 2306-2334 in CHB and 1935-1976 and 2402-2435 in LC) and aa (aa 98-103 in CHB and 28-30 and 51-54 in LC) levels. No differences in indel frequency were observed between the clinical groups. Contrarily, we identified an aa substitution (P79Q) that was more frequent in HCC [median (interquartile range) frequency of 15.82 (0-78.9) vs 0 (0-0) for the other groups; P < 0.05 vs the CHB group].
We have identified a number of nt and aa regions that were highly conserved in the presence of different viral genotypes and clinical stages. These could be valuable targets for future pangenotypic and panclinical therapeutic and diagnostic strategies. The different clinically related conserved regions and the P79Q aa substitution could potentially be used as prognostic factors for disease progression.
Our findings could guide the creation of a new gene therapy strategy based on RNA silencing. In-depth analysis of group-specific conserved or variable regions and their role in disease progression is needed. Further in vitro studies are required to determine whether the P79Q aa substitution might affect viral replication and to investigate associations between cell damage and onset of HCC.
The statistical and bioinformatics methods used in this study were reviewed by Mercedes Guerrero-Murillo from the Microbiology Department at Vall d’Hebron Hospital (Barcelona, Spain) and by Dr. Josep Gregori from the Liver Disease Viral Hepatitis Laboratory of Vall d’Hebron Hospital (Barcelona, Spain), CIBERehd research group, and Roche Diagnostics SL.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: European Association for the Study of the Liver (14916).
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Said ZNA S-Editor: Zhang H L-Editor: A E-Editor: Ma YJ
| 1. | World Health Organization. Global Hepatitis Report, 2017. World Heal Organ [Internet] 2017; 62. Available from: http://www.who.int/hepatitis. |
| 2. | Yuen MF, Chen DS, Dusheiko GM, Janssen HLA, Lau DTY, Locarnini SA, Peters MG, Lai CL. Hepatitis B virus infection. Nat Rev Dis Primers. 2018;4:18035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 538] [Article Influence: 76.9] [Reference Citation Analysis (1)] |
| 3. | Allweiss L, Dandri M. The Role of cccDNA in HBV Maintenance. Viruses. 2017;9:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 4. | Bowden S, Locarnini S, Chang TT, Chao YC, Han KH, Gish RG, de Man RA, Yu M, Llamoso C, Tang H. Covalently closed-circular hepatitis B virus DNA reduction with entecavir or lamivudine. World J Gastroenterol. 2015;21:4644-4651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Papatheodoridis GV, Chan HL, Hansen BE, Janssen HL, Lampertico P. Risk of hepatocellular carcinoma in chronic hepatitis B: assessment and modification with current antiviral therapy. J Hepatol. 2015;62:956-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 408] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 6. | Ren GL, Huang GY, Zheng H, Fang Y, Ma HH, Xu MC, Zhang HB, Zhang WY, Zhao YG, Sun DY, Hu WK, Liu J. Changes in innate and permissive immune responses after hbv transgenic mouse vaccination and long-term-siRNA treatment [corrected]. PLoS One. 2013;8:e57525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Gish RG, Yuen MF, Chan HL, Given BD, Lai CL, Locarnini SA, Lau JY, Wooddell CI, Schluep T, Lewis DL. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antiviral Res. 2015;121:97-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 8. | Lin YY, Liu C, Chien WH, Wu LL, Tao Y, Wu D, Lu X, Hsieh CH, Chen PJ, Wang HY, Kao JH, Chen DS. New insights into the evolutionary rate of hepatitis B virus at different biological scales. J Virol. 2015;89:3512-3522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | González C, Tabernero D, Cortese MF, Gregori J, Casillas R, Riveiro-Barciela M, Godoy C, Sopena S, Rando A, Yll M, Lopez-Martinez R, Quer J, Esteban R, Buti M, Rodríguez-Frías F. Detection of hyper-conserved regions in hepatitis B virus X gene potentially useful for gene therapy. World J Gastroenterol. 2018;24:2095-2107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Locarnini S, Zoulim F. Molecular genetics of HBV infection. Antivir Ther. 2010;15 Suppl 3:3-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 11. | Nassal M, Rieger A, Steinau O. Topological analysis of the hepatitis B virus core particle by cysteine-cysteine cross-linking. J Mol Biol. 1992;225:1013-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Diab A, Foca A, Zoulim F, Durantel D, Andrisani O. The diverse functions of the hepatitis B core/capsid protein (HBc) in the viral life cycle: Implications for the development of HBc-targeting antivirals. Antiviral Res. 2018;149:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 90] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 13. | Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. Core protein: A pleiotropic keystone in the HBV lifecycle. Antiviral Res. 2015;121:82-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 14. | Ning X, Luckenbaugh L, Liu K, Bruss V, Sureau C, Hu J. Common and Distinct Capsid and Surface Protein Requirements for Secretion of Complete and Genome-Free Hepatitis B Virions. J Virol. 2018;92:e00272-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (1)] |
| 15. | Teng CF, Huang HY, Li TC, Shyu WC, Wu HC, Lin CY, Su IJ, Jeng LB. A Next-Generation Sequencing-Based Platform for Quantitative Detection of Hepatitis B Virus Pre-S Mutants in Plasma of Hepatocellular Carcinoma Patients. Sci Rep. 2018;8:14816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3745] [Cited by in RCA: 3800] [Article Influence: 475.0] [Reference Citation Analysis (1)] |
| 17. | Team RC. A language and environment for statistical computing. R Found. Stat. Comput. Vienna, Austria. 2016; Available from: https://www.r-project.org/. |
| 18. | Ramírez C, Gregori J, Buti M, Tabernero D, Camós S, Casillas R, Quer J, Esteban R, Homs M, Rodriguez-Frías F. A comparative study of ultra-deep pyrosequencing and cloning to quantitatively analyze the viral quasispecies using hepatitis B virus infection as a model. Antiviral Res. 2013;98:273-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Cuadras C. A distance approach to discriminant analysis and its properties. In: Mathematics preprint series. Barcelona: 1991. |
| 20. | Cuadras C. Distance analysis in discrimination and classification using both continuous and categorical variables. In: Statistical AData analysis and Interference. Amsterdam, 1989: 459-473. |
| 21. | Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20000] [Cited by in RCA: 18608] [Article Influence: 413.5] [Reference Citation Analysis (0)] |
| 22. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 694] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 23. | Soriano V, Barreiro P, Benitez L, Peña JM, de Mendoza C. New antivirals for the treatment of chronic hepatitis B. Expert Opin Investig Drugs. 2017;26:843-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | Flisiak R, Jaroszewicz J, Łucejko M. siRNA drug development against hepatitis B virus infection. Expert Opin Biol Ther. 2018;18:609-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Durantel D. New treatments to reach functional cure: Virological approaches. Best Pract Res Clin Gastroenterol. 2017;31:329-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Zhang Y, Wu Y, Deng M, Xu D, Li X, Xu Z, Hu J, Zhang H, Liu K, Zhao Y, Gao F, Bi S, Gao GF, Zhao J, Liu WJ, Meng S. CD8+ T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress. J Virol. 2018;92:e02120-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Li HC, Huang EY, Su PY, Wu SY, Yang CC, Lin YS, Chang WC, Shih C. Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathog. 2010;6:e1001162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 117] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 28. | Hosaka T, Suzuki F, Kobayashi M, Fujiyama S, Kawamura Y, Sezaki H, Akuta N, Suzuki Y, Saitoh S, Arase Y, Ikeda K, Kobayashi M, Kumada H. Impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos(t)ide analogues. Aliment Pharmacol Ther. 2019;49:457-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 29. | Riveiro-Barciela M, Bes M, Rodríguez-Frías F, Tabernero D, Ruiz A, Casillas R, Vidal-González J, Homs M, Nieto L, Sauleda S, Esteban R, Buti M. Serum hepatitis B core-related antigen is more accurate than hepatitis B surface antigen to identify inactive carriers, regardless of hepatitis B virus genotype. Clin Microbiol Infect. 2017;23:860-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | Kaur H, Bruno JG, Kumar A, Sharma TK. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics. 2018;8:4016-4032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 261] [Cited by in RCA: 251] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 31. | Zhang Z, Zhang J, Pei X, Zhang Q, Lu B, Zhang X, Liu J. An aptamer targets HBV core protein and suppresses HBV replication in HepG2.2.15 cells. Int J Mol Med. 2014;34:1423-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 32. | Orabi A, Bieringer M, Geerlof A, Bruss V. An Aptamer against the Matrix Binding Domain on the Hepatitis B Virus Capsid Impairs Virion Formation. J Virol. 2015;89:9281-9287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 33. | Pleshakova TO, Kaysheva AL, Shumov ID, Ziborov VS, Bayzyanova JM, Konev VA, Uchaikin VF, Archakov AI, Ivanov YD. Detection of Hepatitis C Virus Core Protein in Serum Using Aptamer-Functionalized AFM Chips. Micromachines (Basel). 2019;10:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 34. | Percze K, Szakács Z, Scholz É, András J, Szeitner Z, Kieboom CH, Ferwerda G, Jonge MI, Gyurcsányi RE, Mészáros T. Aptamers for respiratory syncytial virus detection. Sci Rep. 2017;7:42794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 35. | Ludgate L, Liu K, Luckenbaugh L, Streck N, Eng S, Voitenleitner C, Delaney WE 4th, Hu J. Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation. J Virol. 2016;90:5830-5844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 36. | Liu K, Luckenbaugh L, Ning X, Xi J, Hu J. Multiple roles of core protein linker in hepatitis B virus replication. PLoS Pathog. 2018;14:e1007085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 37. | Zheng CL, Fu YM, Xu ZX, Zou Y, Deng K. Hepatitis B virus core protein dimerdimer interface is critical for viral replication. Mol Med Rep. 2019;19:262-270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 38. | Ge Z, Tian T, Meng L, Song C, Yu C, Xu X, Liu J, Dai J, Hu Z. HBV mutations in EnhII/BCP/PC region contribute to the prognosis of hepatocellular carcinoma. Cancer Med. 2019;8:3086-3093. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 39. | Chiu AP, Tschida BR, Sham TT, Lo LH, Moriarity BS, Li XX, Lo RC, Hinton DE, Rowlands DK, Chan CO, Mok DKW, Largaespada DA, Warner N, Keng VW. HBx-K130M/V131I Promotes Liver Cancer in Transgenic Mice via AKT/FOXO1 Signaling Pathway and Arachidonic Acid Metabolism. Mol Cancer Res. 2019;17:1582-1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 40. | Al-Qahtani AA, Al-Anazi MR, Nazir N, Abdo AA, Sanai FM, Al-Hamoudi WK, Alswat KA, Al-Ashgar HI, Khan MQ, Albenmousa A, El-Shamy A, Alanazi SK, Dela Cruz D, Bohol MFF, Al-Ahdal MN. The Correlation Between Hepatitis B Virus Precore/Core Mutations and the Progression of Severe Liver Disease. Front Cell Infect Microbiol. 2018;8:355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 41. | Jia J, Li H, Wang H, Chen S, Wang M, Feng H, Gao Y, Wang Y, Fang M, Gao C. Hepatitis B virus core antigen mutations predict post-operative prognosis of patients with primary hepatocellular carcinoma. J Gen Virol. 2017;98:1399-1409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |