Published online Jan 14, 2020. doi: 10.3748/wjg.v26.i2.154
Peer-review started: October 24, 2019
First decision: December 5, 2019
Revised: December 14, 2019
Accepted: December 21, 2019
Article in press: December 21, 2019
Published online: January 14, 2020
Processing time: 81 Days and 7.5 Hours
It is evident that current clinical criteria are suboptimal to accurately estimate patient prognosis. Studies have identified epigenetic aberrant changes as novel prognostic factors for colorectal cancer (CRC).
To estimate whether a methylation gene panel in different clinical stages can reflect a different prognosis.
We enrolled 120 CRC patients from Tri-Service General Hospital in Taiwan and used the candidate gene approach to select six genes involved in carcinogenesis pathways. Patients were divided into two groups based on the methylation status of the six evaluated genes, namely, the < 3 aberrancy group and ≥ 3 aberrancy group. Various tumor stages were divided into two subgroups (local and advanced stages) on the basis of the pathological type of the following tissues: Tumor and adjacent normal tissues (matched normal). We assessed DNA methylation in tumors and adjacent normal tissues from CRC patients and analyzed the association between DNA methylation with different cancer stages and the prognostic outcome including time to progression (TTP) and overall survival.
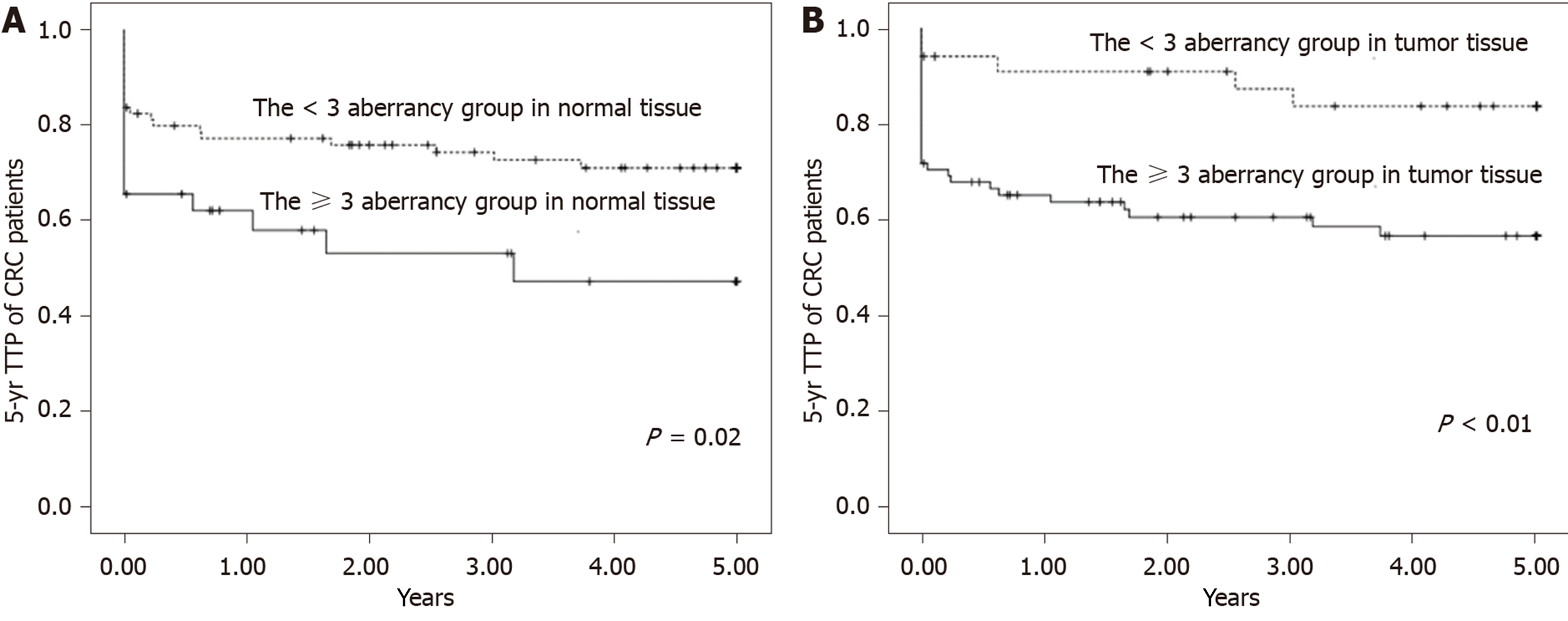
We observed a significantly increasing trend of hazard ratio as the number of hypermethylated genes increased both in normal tissue and tumor tissue. The 5-year TTP survival curves showed a significant difference between the ≥ 3 aberrancy group and the < 3 aberrancy group. Compared with the < 3 aberrancy group, a significantly shorter TTP was observed in the ≥ 3 aberrancy group. We further analyzed the interaction between CRC prognosis and different cancer stages (local and advanced) according to the methylation status of the selected genes in both types of tissues. There was a significantly shorter 5-year TTP for tumors at advanced stages with the promoter methylation status of selected genes than for those with local stages. We found an interaction between cancer stages and the promoter methylation status of selected genes in both types of tissues.
Our data provide a significant association between the methylation markers in normal tissues with advanced stage and prognosis of CRC. We recommend using these novel markers to assist in clinical decision-making.
Core tip: Our data show that a novel methylation gene panel in adjacent normal tissues predicts a poor prognosis of colorectal cancer. We recommend that the matched normal tissues of patients with colorectal cancer could be an alternative source of prognostic markers to assist clinical decision-making.
- Citation: Hsu CH, Hsiao CW, Sun CA, Wu WC, Yang T, Hu JM, Huang CH, Liao YC, Chen CY, Lin FH, Chou YC. Novel methylation gene panel in adjacent normal tissues predicts poor prognosis of colorectal cancer in Taiwan. World J Gastroenterol 2020; 26(2): 154-167
- URL: https://www.wjgnet.com/1007-9327/full/v26/i2/154.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i2.154
Colorectal cancer (CRC) is a leading cause of cancer-related morbidity and mortality worldwide. In 2017, in the United States, approximately 135430 patients were newly diagnosed with CRC, and 50260 deaths from CRC were reported[1]. Survival of patients with CRC is closely linked to the tumor stage at diagnosis, and the 5-year relative survival rates are 64.9% for all stages and 89.9% for local, 71.3% for regional, and 13.9% for distant disease[2]. Cancer staging systems enable reasonable adjuvant treatment, help stratify tumors according to the risk of recurrence, and help establish precise prognoses. Using resection specimens, the pathologic staging of CRC is conducted according to the tumor-node-metastasis (TNM) classification from the Seventh Edition of the American Joint Committee on Cancer (AJCC) Staging Manual. According to the TNM staging system, the survival of patients with CRC is related to the size of the primary tumor (T), nearby affected lymph nodes (N), and distant metastasis (M)[3]. According to this classification, patients with stage II CRC have a low or high risk based on clinical risk factors including tumor size, number of lymph nodes investigated, tumor differentiation, perforation, obstruction, and lymphovascular invasion. Routine adjuvant therapy after surgical resection is recommended for patients with high-risk stage II CRC as well as for those with stage III and IV tumors[4]. However, 10%-20% of patients with stage II CRC and 30%-40% of patients with stage III CRC ultimately develop recurrence after therapeutic intervention. Low-risk patients with stage II CRC show a good prognosis, as only a small proportion of these patients experience relapse[5]. The prognostic factors that define these relapse-prone patients should be identified to optimize treatment selection.
Several studies have been conducted to identify novel prognostic and predictive biomarkers for CRC, including both genetic and epigenetic aberrant changes. Genetic abnormalities include microsatellite instability; chromosomal instability; mutations of cancer driver genes such as KRAS, BRAF, TP53, and PIK3CA; certain proteins; microRNAs; and gene expression signatures[6,7]. Research has demonstrated that the epigenetic mechanism of DNA methylation plays an important role in several essential biological processes, such as development, cell differentiation, and gene silencing[8]. Epigenetic silencing of multiple genes involved in DNA repair, cell cycle, and apoptosis through promoter hypermethylation is a common event in various cancers including CRC[9]. These molecular biomarkers could be used to stratify patients with the same tumor stage according to different molecular factors for optimal adjuvant chemotherapy[10,11].
Previous studies have demonstrated that CDKN2A, hMLH1, and MGMT hypermethylation, which is related to carcinogenesis pathways via gene silencing, could serve as a diagnostic prognostic marker for CRC[12,13]. In the present study, in addition to the aforementioned genes, we selected three other candidate genes, namely, CSF2, DIS3L2, and OAF; CSF2 and DIS3L2, which are involved in inhibitory effects on tumor growth[14,15], were selected from a previous study[16]. OAF (the out at first homolog gene), selected from PRECOG (PREdiction of Clinical Outcomes from Genomic Profiles, https://precog.stanford.edu/) and MethHC (a database of DNA Methylation and gene expression in Human Cancer, http://methhc. mbc.nctu.edu.tw/php/index.php), is located at chromosome 11q23.2 and is ubiquitously expressed in the liver (RPKM 56.8) and colon (RPKM 31.5) (NCBI Gene, https://http://www.ncbi.nlm.nih.gov/gene). To the best of our knowledge, little is known about this gene. According to browser data from genome-wide association studies (FANTOM CAT, http://fantom.gsc.riken.jp/cat/v1/#/), the OAF locus is related to small-cell lung carcinoma[17].
To determine the effect of the methylation status of candidate genes on the relationship between the histological stage and prognosis of CRC, we examined DNA methylation in tumor tissues and adjacent normal tissues (matched normal). In this study, we propose the understanding that the methylation status of a multiple-gene panel, even in matched normal tissues, combined with different clinical stages may predict the prognosis and provide clinical recommendations for optimal treatment for CRC.
The methods applied in this study are described in detail elsewhere[13]. In this hospital-based retrospective cohort study, we analyzed the data of patients diagnosed with CRC between 2006 and 2010 who underwent surgical resection at Tri-Service General Hospital (TSGH), Taiwan, to evaluate their prognosis in 5 years. Written informed consent was obtained from all patients before enrollment into the study to evaluate their prognosis. This study was approved by the TSGH Institutional Review Board (TSGHIRB approval number: 098-05-292 and 2-105-05-129). According to the clinical practice guideline of the Division of Colon and Rectum of TSGH, the enrollees should return for a check-up once every 3 mo in the first year after undergoing surgical resection and once every 3-6 mo thereafter. From the cancer registration database of TSGH, information on registered patients, including their sex, age at surgery (continuous variable), adjuvant chemotherapy, histological grade, and tumor location and their follow-up data on recurrence, metastasis, and survival, was obtained.
Time to progression (TTP) and overall survival (OS) were calculated from the date of surgery to the presentation of disease progression (including cancer recurrence or metastasis), death from any cause, or till the last follow-up date before December 31, 2010. On the basis of the inclusion criteria, 120 tumor tissues and matched normal tissues (240 samples) were obtained from the enrollees.
Cellulose-coated magnetic beads were employed to extract genomic DNA from the samples by using the MagCore Compact Automated Nucleic Acid Extractor (Catalogue No. MCA0801; RBC Bioscience, Taipei, Taiwan) and the Genomic DNA Tissue Kit (Catalogue No. 69504; Qiagen, Taipei, Taiwan), according to the manufacturer’s protocol. Isolated DNA was treated with sodium bisulfite using the EZ DNA Methylation Kit (Zymo Research Corporation, Orange, CA, United States).
The promoter methylation status of CDKN2A, hMLH1, MGMT, CSF2, DIS3L2, and OAF genes was assessed using methylation-specific PCR (MS-PCR), as described in our previous research[13]. The reaction solution (25 µL) contained HotStart Taq Premix (12.5 µL, RBC Bioscience, Taipei, Taiwan), 1.2-μL aliquots of forward and reverse primers, and bisulfite-converted DNA.
For MS-PCR, we used the following oligonucleotide primers: CDKN2A: 5′-TTATTAGAGGGTGGGGCGGATCGC-3′ (forward primer) and 5′-GACCCCGAACCGCGACCGTAA-3′ (reverse primer) to amplify the methylated sequence (PCR annealing at 62 °C, product size: 150 bp) and 5′-TTATTAGAGGGTGGGGTGGATTGT-3′ (forward primer) and 5′-CAACCCCAAACCACAACCATAA-3′ (reverse primer) to amplify the unmethylated sequence (PCR annealing at 62 °C, product size: 151 bp); hMLH1: 5′-ACGTAGACGTTTTATTAGGGTCGC-3′ (forward primer) and 5′-CCTCATCGTAACTACCCGCG-3′ (reverse primer) to amplify the methylated sequence (PCR annealing at 60 °C, product size: 118 bp) and 5′-TTTTGATGTAGATGTTTTATTAGGGTTGT-3′ (forward primer) and 5′-ACCACCTCATCATAACTACCCACA-3′ (reverse primer) to amplify the unmethylated sequence (PCR annealing at 60 °C, product size: 124 bp); MGMT: 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ (forward primer) and 5′-GCACTCTTCCGAAAACGAAACG-3′ (reverse primer) to amplify the methylated sequence (PCR annealing at 53 °C, product size: 81 bp) and 5′-TTTGTGTTTTGATGTTTGTAGGTTTTTGT-3′ (forward primer) and 5′-AACTCCACACTCTTCCAAAAACAAAACA-3′ (reverse primer) to amplify the unmethylated sequence (PCR annealing at 53 °C, product size: 93 bp); CSF2: 5′-TGATTATTTAGGGAAAAGGTTTATC-3′ (forward primer) and 5′-ATAACCACAAAATACCAAAAAAACG-3′ (reverse primer) to amplify the methylated sequence (PCR annealing at 56 °C, product size: 105 bp) and 5′-ATTATTTAGGGAAAAGGTTTATTGT-3′ (forward primer) and 5′-AATAACCACAAAATACCAAAAAAACA-3′ (reverse primer) to amplify the unmethylated sequence (PCR annealing at 60 °C, product size: 104 bp); DIS3L2: 5′-GTCGTAGTTGAATCGTCGATTAC-3′ (forward primer) and 5′-TTACTAAAAAAAATACTCTTCCGAA-3′ (reverse primer) to amplify the methylated sequence (PCR annealing at 54 °C, product size: 134 bp) and 5′-GTTGTAGTTGAATTGTTGATTATGA-3′ (forward primer) and 5′-TTACTAAAAAAAATACTCTTCCAAA-3′ (reverse primer) to amplify the unmethylated sequence (PCR annealing at 55 °C, product size: 134 bp); and OAF: 5′-GTTATTGTCGTGGAGCGTTAGC-3′ (forward primer) and 5′-CCTACCTCCCGTACTTCCCG-3′ (reverse primer) to amplify the methylated sequence (PCR annealing at 59.4 °C, product size: 170 bp) and 5′-TTATTGTTGTGGAGTGTTAGTGTTT-3′ (forward primer) and 5′-CCTACCTCCCATACTTCCCACAT-3′ (reverse primer) to amplify the unmethylated sequence (PCR annealing at 59.4 °C, product size: 169 bp). The PCR cycling conditions were as follows: 10 min at 95 °C; 35 cycles of denaturation for 30 s at 95 °C; 30-s annealing at gene-appropriate temperature; 30-s extension at 72 °C; and final extension for 4 min at 72 °C. After the amplification, PCR products were mixed with a loading buffer, electrophoresed (for 25 min) on a 2% agarose gel by using 0.2 μL of gel-stained dye, and visualized using an ultraviolet transilluminator.
Patients were divided into two groups based on the methylation status of the six evaluated genes, namely, < 3 aberrancy group and ≥ 3 aberrancy group. In addition, various tumor stages were divided into two subgroups (local and advanced stages) on the basis of the pathological type of the following tissues: Tumor and adjacent normal tissues (matched normal). Associations of the number of hypermethylated genes under study and different clinical stages with TTP or OS were assessed using the univariate Cox proportional hazards regression model. The multivariate Cox regression model was then employed to estimate the independent prognostic effect of the number of methylated genes, with adjustment for sex, age at surgery (continuous variable), adjuvant chemotherapy, histological grade, and tumor location, based on a previous study[13]. The positive predictive value (PPV) and negative predictive value (NPV) of the ≥ 3 aberrancy group in predicting the prognosis of CRC were calculated. The area under the receiver operating characteristic (ROC) curve is reported along with its 95% confidence interval (CI). The 5-year TTP and OS curves for the < 3 aberrancy and ≥ 3 aberrancy groups were plotted using the Kaplan–Meier method and were compared using the log-rank test. Statistical analyses were performed using IBM Statistical Package for the Social Sciences (SPSS) V.22 (IBM SPSS Statistics 22). All statistical tests were two-sided, and P values less than 0.05 were considered statistically significant.
In the study, 120 CRC tumor samples and adjacent normal samples from the TSGH tumor bank were analyzed. The relationship between the methylation status of the selected genes and the demographic and clinicopathological features of patients with CRC was evaluated. As shown in Table 1, the progression of CRC in 5 years indicated that 37.5% of the enrollees had cancer recurrence or metastasis, and 19.2% of the enrollees died during the period. Although the six genes were methylated in both tumor and matched normal tissue samples, the percentage of methylation was higher in tumor tissues than in normal tissues (CDKN2A, 67.3% vs 32.7%; MGMT, 76.3% vs 23.7%; MLH1, 51.6% vs 48.4%; CSF2, 51.6% vs 48.4%; DIS3L2, 55.1% vs 44.9%; OAF, 68.1% vs 31.9%). In addition, progression was detected in significantly higher percentages in the normal tissue with OAF hypermethylation and in the ≥ 3 aberrancy group. The PPV and NPV of ≥ 3 aberrancies in predicting the progression of CRC were 51.4% and 68.2% in normal tissues and 43.9% and 76.3% in tumor tissues, respectively. The PPV and NPV of the ≥ 3 aberrancy group in predicting the survival of CRC were 20.0% and 81.2% in normal tissues and 18.3% and 78.9% in tumor tissues, respectively. No other associations were found between the methylation statuses of the six genes and the demographic and clinicopathological characteristics of the study patients.
| Variables | Total | Methylation status | |||
| OAF | ≥ 3 of genes1 | ||||
| Normal | Tumors | Normal | Tumors | ||
| Sex | |||||
| Male | 60 (50.0) | 13 (28.9) | 28 (62.2) | 16 (26.7) | 41 (68.3) |
| Female | 60 (50.0) | 16 (34.8) | 34 (73.9) | 19 (31.7) | 41 (68.3) |
| Age in yr at surgery | |||||
| Mean (SD) | 63.8 (14.8) | 62.6 (12.6) | 63.2 (15.0) | 62.5 (13.5) | 62.5 (15.3) |
| < 65 | 61 (50.8) | 15 (30.0) | 33 (66.0) | 19 (31.1) | 45 (73.8) |
| ≥ 65 | 59 (49.2) | 14 (34.1) | 29 (70.7) | 16 (27.1) | 37 (62.7) |
| Stage | |||||
| 19 (15.8) | 2 (16.7) | 6 (50.0) | 6 (31.6) | 10 (52.6) | |
| 39 (32.5) | 6 (20.7) | 22 (75.9) | 11 (28.2) | 26 (66.7) | |
| 40 (33.3) | 14 (45.2) | 22 (71.0) | 11 (27.5) | 29 (72.5) | |
| 22 (18.3) | 7 (36.8) | 12 (63.2) | 7 (31.8) | 17 (77.3) | |
| Adjuvant chemotherapy | |||||
| No | 32 (28.3) | 5 (22.7) | 13 (59.1) | 13 (40.6) | 23 (71.9) |
| Yes | 81 (71.7) | 24 (36.9) | 46 (70.8) | 22 (27.2) | 55 (67.9) |
| Lymph/vascular invasion | |||||
| No | 44 (97.8) | 12 (35.3) | 21 (61.8) | 18 (40.9) | 28 (63.6) |
| Yes | 1 (2.2) | 0 (0) | 1 (100.0) | 0 (0) | 1 (100.0) |
| Histological grade2 | |||||
| Well or Moderately | 96 (88.9) | 23 (31.9) | 45 (62.5) | 31 (32.3) | 62 (64.6) |
| Poor or undifferentiated | 12 (11.1) | 5 (45.5) | 10 (90.9) | 2 (16.7) | 11 (91.7) |
| Tumor location2 | |||||
| Colon | 89 (78.8) | 19 (28.4) | 46 (68.7) | 27 (30.3) | 59 (66.3) |
| Rectum | 24 (21.2) | 10 (50.0) | 13 (65.0) | 8 (33.3) | 19 (79.2) |
| Progression in 5 yr | |||||
| No | 75 (62.5) | 11 (22.0)a | 33 (66.0) | 17 (22.7)a | 46 (61.3) |
| Yes | 45 (37.5) | 18 (43.9) | 29 (70.7) | 18 (40.0) | 36 (80.0) |
| All-cause death in 5 yr | |||||
| No | 97 (80.8) | 22 (31.0) | 50 (70.4) | 28 (28.9) | 67 (69.1) |
| Yes | 23 (19.2) | 7 (35.0) | 12 (60.0) | 7 (30.4) | 15 (65.2) |
The association between the number of hypermethylated genes and the 5-year TTP and OS of patients with CRC was separately assessed. We observed a significantly increasing trend of hazard ratio (HR) as the number of hypermethylated genes increased both in normal tissue (P = 0.02) and tumor tissue (P = 0.02) (Table 2).
| Normal tissues | Tumor tissues | |||||||||||
| No. of subjects | No. of cases | Crude | Adjusted | No. of subjects | No. of cases | Crude | Adjusted | |||||
| HR | 95%CI | HR | 95%CI | HR | 95%CI | HR | 95%CI | |||||
| NO. of hypermethylated genes | ||||||||||||
| 0 | 9 | 3 (33.3) | 1.00 | Referent | 1.00 | Referent | 3 | 1 (33.3) | 1.00 | Referent | 1.00 | Referent |
| 1 | 34 | 8 (23.5) | 0.76 | 0.15-3.74 | 0.64 | 0.13-3.23 | 7 | 0 (0) | N/A | N/A | N/A | N/A |
| 2 | 42 | 16 (38.1) | 1.58 | 0.36-6.94 | 1.62 | 0.34-7.62 | 28 | 8 (28.6) | 0.47 | 0.05-4.20 | 0.65 | 0.07-5.88 |
| 3 | 21 | 12 (57.1) | 2.66 | 0.58-12.19 | 2.18 | 0.46-10.4 | 39 | 15 (38.5) | 1.02 | 0.13-7.86 | 1.36 | 0.17-10.7 |
| 4 | 11 | 3 (27.3) | 1.28 | 0.21-7.65 | 1.48 | 0.18-12.2 | 23 | 14 (60.9) | 2.12 | 0.28-16.3 | 2.56 | 0.33-20.2 |
| 5 | 3 | 3 (100) | 7.26a | 1.00-52.84 | 4.70 | 0.57-39.0 | 17 | 6 (35.3) | 1.17 | 0.14-9.71 | 1.52 | 0.17-13.5 |
| 6 | 0 | 0 | N/A | N/A | N/A | N/A | 3 | 1 (33.3) | 1.13 | 0.07-18.0 | 2.10 | 0.12-38.3 |
| P value1 | 0.02 | 0.02 | 0.02 | 0.02 | ||||||||
| ≥ 3 of genes2 | 35 | 18 (51.4) | 1.99a | 1.03-3.85 | 2.01a | 1.00-4.01 | 82 | 36(43.9) | 3.26a | 1.27-8.39 | 3.18a | 1.21-8.39 |
The 5-year TTP survival curves showed a significant difference between the ≥ 3 aberrancy group and the < 3 aberrancy group (P = 0.02 for normal tissue; P < 0.01 for tumor tissue) (Figure 1). Compared with the < 3 aberrancy group, a significantly shorter TTP was observed in the ≥ 3 aberrancy group [HR (95%CI) = 1.99 (1.03-3.85) for normal tissue, 3.26 (1.27-8.39) for tumor tissue], even after adjustment for confounders in multivariable analysis [HR (95%CI) = 2.01 (1.00-4.01) for normal tissue, 3.18 (1.21-8.39) for tumor tissue].
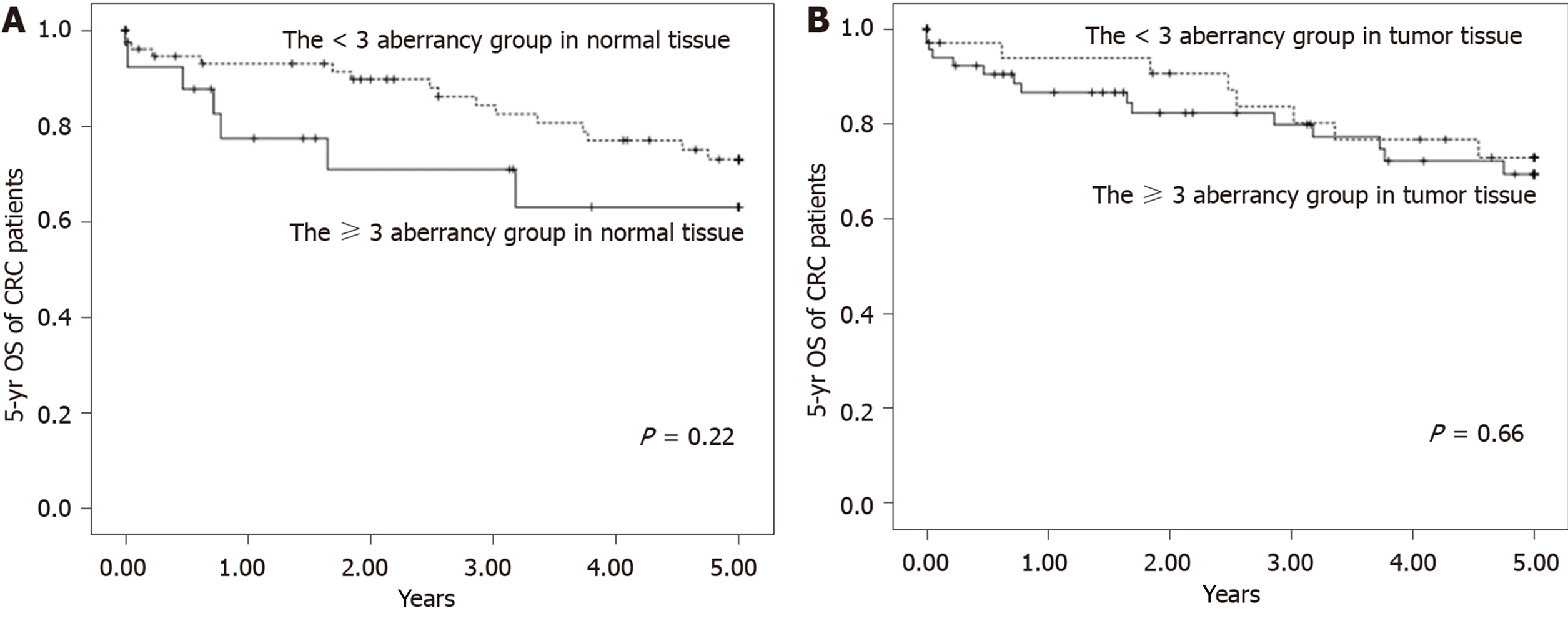
Although there was an increasing trend of HR of the 5-year OS of patients with CRC with the increasing number of hypermethylated genes in normal tissue, this difference did not reach statistical significance (P = 0.33) (Table 3). The Kaplan–Meier curves of 5-year OS of patients with CRC between the ≥ 3 aberrancy group and the < 3 aberrancy group are shown in Figure 2. The log-rank test revealed no significant differences between the groups in both types of tissues over the entire Kaplan–Meier curve. The area under the ROC curve of CRC progression and survival was 0.59 (95%CI: 0.49-0.70, P = 0.09) and 0.48 (95%CI: 0.35-0.61, P = 0.77) in tumor tissue, respectively. The area under the ROC curve of CRC progression and survival was 0.59 (95%CI: 0.48-0.69, P = 0.11) and 0.51 (95%CI: 0.38-0.64, P = 0.91) in normal tissue, respectively.
| Normal tissues | Tumor tissues | |||||||||||
| No. of subjects | No. of cases | Crude | Adjusted | No. of subjects | No. of cases | Crude | Adjusted | |||||
| HR | 95%CI | HR | 95%CI | HR | 95%CI | HR | 95%CI | |||||
| NO. of methylated genes | ||||||||||||
| 0 | 9 | 1 (11.11) | 1.00 | Referent | 1.00 | Referent | 3 | 1 (33.33) | 1.00 | Referent | 1.00 | Referent |
| 1 | 34 | 7 (20.59) | 1.52 | 0.19-12.4 | 1.19 | 0.14-10.1 | 7 | 0 (0) | N/A | N/A | N/A | N/A |
| 2 | 42 | 8 (19.05) | 1.76 | 0.22-14.1 | 0.89 | 0.10-8.40 | 28 | 7 (25) | 0.95 | 0.12-7.72 | 0.73 | 0.08-6.55 |
| 3 | 21 | 4 (19.05) | 1.79 | 0.20-16.0 | 1.41 | 0.15-13.2 | 39 | 6 (15.38) | 0.66 | 0.08-5.47 | 0.59 | 0.07-5.2 |
| 4 | 11 | 2 (18.18) | 2.11 | 0.19-23.3 | 1.40 | 0.10-18.8 | 23 | 6 (26.09) | 1.24 | 0.15-10.3 | 1.16 | 0.13-10.2 |
| 5 | 3 | 1 (33.33) | 4.89 | 0.30-78.8 | 4.91 | 0.29-83.4 | 17 | 3 (17.65) | 0.84 | 0.09-8.06 | 0.60 | 0.05-7.13 |
| 6 | 0 | 0 (0) | N/A | N/A | N/A | N/A | 3 | 0 (0) | N/A | N/A | N/A | N/A |
| P value1 | 0.33 | 0.40 | 0.59 | 0.64 | ||||||||
| ≥ 3 of genes2 | 35 | 7 (20.0) | 1.31 | 0.54-3.18 | 1.50 | 0.59-3.81 | 82 | 15 (18.3) | 1.13 | 0.48-2.66 | 1.21 | 0.47-3.10 |
We further analyzed the interaction between CRC prognosis and different cancer stages (local and advanced) according to the methylation status of the selected genes in both types of tissues. Table 4 reveals a significantly shorter 5-year TTP for tumors at advanced stages with the promoter methylation status of selected genes than for those with local stages. The adjusted HR of OAF methylation in normal tissue in patients with advanced stages was 20.3 (100%CI: 4.12-100). In addition, we found an interaction between cancer stages and the promoter methylation status of selected genes in both types of tissues. In normal tissue, there was a significant joint effect that increased the association of CRC progression with advanced cancer stages in the ≥3 aberrancy group (Me/advanced), with an HR of 33.4 (95%CI: 7.49-149; P < 0.01); after adjusting for confounders, the HR was 28.8 (95%CI: 6.24-133; P < 0.01). A similar result was observed in the tumor tissue, in which the crude and adjusted HRs were 23.1 (95%CI: 3.14-170; P < 0.01) and 20.8 (95%CI: 2.75-157; P < 0.01), respectively. With regard to the effect of the interaction between methylation of the gene promoter region and different cancer stages on the 5-year OS of patients with CRC, no significant joint effect was observed in both types of tissues (Table 5).
| Normal tissues | Tumor tissues | |||||||||||
| No. of subjects | No. of cases | Crude | Adjusted | No. of subjects | No. of cases | Crude | Adjusted | |||||
| HR | 95%CI | HR | 95%CI | HR | 95%CI | HR | 95%CI | |||||
| OAF | ||||||||||||
| UnMe/local (1 and 2)1 | 33 | 3 (9.1) | 1.00 | Referent | 1.00 | Referent | 13 | 0 (0.0) | 1.00 | Referent | 1.00 | Referent |
| UnMe/advanced (3 and 4)2 | 29 | 20 (69.0) | 14.7a | 3.38-63.6 | 16.6a | 3.69-74.6 | 16 | 12 (75.0) | N/A | N/A | N/A | N/A |
| Me/local (1 and 2)3 | 8 | 0 (0.0) | N/A | N/A | N/A | N/A | 28 | 3 (10.7) | N/A | N/A | N/A | N/A |
| Me/advanced (3 and 4)4 | 21 | 18 (85.7) | 19.0a | 4.30-84.6 | 20.3a | 4.12-100 | 34 | 26 (76.5) | N/A | N/A | N/A | N/A |
| P value1 | < 0.01 | < 0.01 | ||||||||||
| ≥ 3 of genes6 | ||||||||||||
| UnMe/local (1 and 2)1 | 41 | 2 (4.88) | 1.00 | Referent | 1.00 | Referent | 22 | 2 (9.09) | 1.00 | Referent | 1.00 | Referent |
| UnMe/advanced (3 and 4)2 | 44 | 25 (56.8) | 13.3a | 3.10-57.0 | 11.9a | 2.72-51.8 | 16 | 7 (43.8) | 7.17 | 0.80-64.2 | 6.56 | 0.72-59.4 |
| Me/local (1 and 2)3 | 17 | 1 (5.88) | N/A | N/A | N/A | N/A | 36 | 1 (2.78) | 0.63 | 0.04-9.99 | 0.59 | 0.04-9.62 |
| Me/advanced (3 and 4)4 | 18 | 17 (94.4) | 33.4a | 7.49-149 | 28.8a | 6.24-133 | 46 | 35 (76.1) | 23.1a | 3.14-170 | 20.8a | 2.75-157 |
| P value5 | < 0.01 | < 0.01 | < 0.01 | < 0.01 | ||||||||
| Normal tissues | Tumor tissues | |||||||||||
| No. of subjects | No. of cases | Crude | Adjusted | No. of subjects | No. of cases | Crude | Adjusted | |||||
| HR | 95%CI | HR | 95%CI | HR | 95%CI | HR | 95%CI | |||||
| OAF | ||||||||||||
| UnMe/local (1 and 2)1 | 33 | 5 (15.2) | 1.00 | Referent | 1.00 | Referent | 13 | 2 (15.4) | 1.00 | Referent | 1.00 | Referent |
| UnMe/advanced (3 and 4)2 | 29 | 8 (27.6) | 2.24 | 0.73-6.86 | 4.84a | 1.22-19.2 | 16 | 6 (37.5) | 3.12 | 0.63-15.5 | 15.7a | 1.98-124 |
| Me/local (1 and 2)3 | 8 | 2 (25.0) | 2.32 | 0.45-12.0 | 7.04 | 0.82-60.4 | 28 | 5 (17.9) | 1.44 | 0.28-7.44 | 3.74 | 0.54-26.0 |
| Me/advanced (3 and 4)4 | 21 | 5 (23.8) | 1.85 | 0.53-6.41 | 5.96a | 1.16-30.6 | 34 | 7 (20.6) | 1.78 | 0.37-8.60 | 6.86 | 0.93-50.5 |
| P value5 | 0.33 | 0.03 | 0.47 | 0.06 | ||||||||
| ≥ 3 of genes6 | ||||||||||||
| UnMe/local (1 and 2)1 | 41 | 5 (12.2) | 1.00 | Referent | 1.00 | Referent | 22 | 3 (13.6) | 1.00 | Referent | 1.00 | Referent |
| UnMe/advanced (3 and 4)2 | 44 | 11 (25.0) | 2.57 | 0.89-7.40 | 5.28a | 1.35-20.6 | 16 | 5 (31.3) | 2.68 | 0.64-11.2 | 5.53 | 1.00-30.6 |
| Me/local (1 and 2)3 | 17 | 4 (23.5) | 2.69 | 0.72-10.0 | 4.46 | 0.93-21.5 | 36 | 6 (16.7) | 1.59 | 0.40-6.35 | 2.04 | 0.38-11.1 |
| Me/advanced (3 and 4)4 | 18 | 3 (16.7) | 1.86 | 0.44-7.82 | 4.27 | 0.76-23.8 | 46 | 9 (19.6) | 2.09 | 0.56-7.76 | 3.93 | 0.78-19.8 |
| P value5 | 0.40 | 0.10 | 0.27 | 0.10 | ||||||||
The TNM staging system, which is based on anatomical information including the size and extent of the primary tumor, is widely used to guide clinical treatment strategy and predict the prognosis of cancer. However, CRC is an etiologically heterogeneous disease involving several distinct biologic pathways, and the survival status of different patients at the same TNM stage varies[3,18].
Aberrant DNA methylation of certain loci is characteristic of certain human cancers, which leads to silencing of tumor-suppressor genes through methylation of the CpG islands in promoters[19,20]. Gene hypermethylation has been reported to be a robust and reliable diagnostic method and, thus, a promising source of biomarkers for cancer[21,22]. Moreover, the gene methylation status can be used as a biomarker for the prognosis of cancer; it can predict the cancer outcome, malignant potential of the tumor, and risk of tumor recurrence regardless of therapy[23]. In the present paper, we provide the results of an analysis of 120 tumor tissues and matched normal tissues from patients with CRC. The promoter methylation status of the selected genes confirmed the presence of DNA methylation: Methylation in CDKN2A, hMLH1, MGMT, CSF2, DIS3L2, and OAF was significantly associated with an increased risk of CRC progression, as revealed by Cox proportional hazards regression analysis and Kaplan–Meier analysis. In addition, the ≥ 3 aberrancy group, defined by the presence of three or more methylated genes, showed an increased risk of 5-year TTP of CRC, with a significant joint effect between DNA methylation and clinical stage, especially in matched normal tissues.
Hence, our findings can be used together with clinical staging to guide the re-evaluation of clinical management of cancer, and they can serve as suitable indicators to identify patients at a higher risk of recurrence and requiring intensive follow-up. Our results revealed that the presence of three or more methylated genes we selected, which was used for marking subgroups of patients with CRC, could be a useful biomarker for cancer prognosis and provide an indication of the need for aggressive surveillance. Our findings showed that predicting prognosis with high accuracy is important, which can be best achieved with a panel of individually well-performing biomarkers rather than any single marker alone[24].
Several studies have shown that the use of gene promoter panels, usually comprising more than four genes, improves the prediction of prognosis, which is consistent with our finding[25-27]. De Sousa e Melo et al[28] showed that the hypermethylation of a panel of WNT target genes, specifically ASCL2, LGR5, APCDD1, DKK1, and AXIN2, was a strong predictor of CRC recurrence. Exner et al[25] found that the CpG island methylator phenotype (CIMP) panel, including CDKN2A, MINT1, MINT2, MINT31, and MLH1, was an independent poor prognostic factor for disease-free survival (DFS) and OS in rectal cancers. Kim et al[27] assessed the methylation status of a panel of genes including p16, p14, MINT1, MINT2, MINT31, hMLH1, DKK3, WNT5A, AXIN2, and TFAP2E in patients with CRC and found that higher number of methylations among the panel genes was related to worse clinical outcomes.
However, some studies have reported no relationship between the methylation status of gene panels and the prognosis of CRC. Recent meta-analyses have also shown no significant effect of the CIMP status on prognosis in 8 of 11 and 13 of 19 previously published studies[29,30]. This inconsistency in results may be attributed to several reasons, including the high heterogeneity of CIMP definitions, the different ethnicities of the study population, and the study method used to detect CpG island methylation.
In addition, genome-wide DNA hypomethylation plays a pivotal role in the development of CRC, as this feature induces genomic instability and global loss of imprinting (LOI); moreover, it facilitates the aberrant expression of proto-oncogenes/oncogenes[31,32]. Rodriguez et al[33] suggested that DNA hypomethylation may induce a cascade effect with direct impact on the progression pathway and hence on patient outcomes. Global hypomethylation usually occurs in repetitive transposable elements, such as long interspersed nucleotide element-1 (LINE-1), which comprises approximately 18% of the human genome. Several studies have demonstrated that LINE-1 hypomethylation is significantly correlated with shorter OS, DFS, and cancer-specific survival[34].
In this study, we found that the methylation status of the gene panel in adjacent normal tissues was significantly associated with a poor prognosis. Recently, an increasing number of studies on carcinogenesis have demonstrated that molecular and microscopic changes in normal tissues surrounding tumors lead to cancer progression. Such changes are generally considered a result of the “field effect”. Field effect theory postulates that repeated exposure to environmental carcinogens could lead to multiple epigenetic and genetic alterations in normal-appearing tissues; the organ site is at an increased risk of developing premalignant lesions and is prone to cancer recurrence. Several studies have shown that the aberrant methylation status of specific genes could be a potential marker of the CRC field effect[35,36], which is in line with our finding that compared with tumor tissues, aberrant DNA methylation in adjacent normal tissues is associated with a poor prognosis after surgical resection.
CDKN2A (p16), which has critical roles in cell cycle progression, cellular senescence, and the development of human cancers, is the most frequently studied methylation biomarker[37]. Many studies, including subgroup evaluations, have demonstrated statistically significant relationships between CDKN2A hypermethylation and poor prognosis[38,39]. The DNA mismatch repair (MMR) defects that result from aberrant hMLH1 methylation are linked to hereditary nonpolyposis CRC[40]. Kuan et al[41] and Iida et al[42] have found a significant association between hMLH1 methylation and a worse prognosis in TNM stages I–IV. Somatic epigenetic inactivation of MGMT has been reported as an early event in CRC, in which it is known to be associated with KRAS and TP53 mutations. The methylation status of MGMT is a key prognostic factor for treatment with alkylating drugs such as temozolomide and carmustine, especially in metastatic CRC[43]. Lee et al[44] showed that CSF2 was the most upregulated gene of significance for tumor development and invasiveness among those associated with positive regulation of tyrosine phosphorylation of STAT5, and it could serve as an important prognosticator of urothelial carcinoma.
DIS3L2 inactivation was associated with mitotic abnormalities and altered the expression of mitotic checkpoint proteins. Overexpression of DIS3L2 inhibited the growth of human cancer cell lines[45]. In this study, we evaluated the methylation status of OAF selected from PRECOG[46] and MethHC[47]. In PRECOG and MethHC, low OAF expression was associated with a worse survival outcome, and the methylation frequency of OAF in tumor tissues was significantly higher than that in normal tissues. Little is known about this gene, and few studies have shown decreased expression of OAF in patients with metastatic breast cancer[48] or ulcerative colitis[49]. Few studies have assessed the association between the methylation status of CSF2, DIS3L2, and OAF and CRC prognosis.
This study has several limitations. The results of this study should be carefully interpreted because of the small sample size. A larger prospective cohort study is warranted to validate these results. Moreover, the enrollees of the present study were from a single population of Taiwan. The utility of the gene panel as a prognostic biomarker for CRC must be validated in other ethnic populations. Finally, data from healthy individuals were unavailable, and the results may not be representative of those in asymptomatic individuals. The development of an acceptable protocol may help study the methylation status of tumor suppressor genes; their distribution in promoter regions; their distribution in the proximal colon, distal colon, and rectum; and their time sequence dependence in healthy individuals, particularly in those who are developing CRC.
It is evident that the current clinical criteria are suboptimal to accurately estimate patient prognosis and outcomes. A new set of methylation markers was identified from our data, particularly in the adjacent normal tissues in patients with advanced stage, providing a basis to apply and investigate the potential of these markers to predict the prognosis of CRC. Future research in large and independent cohorts could define the utility of the new set of markers and address whether the modification of treatment/management decisions based on additional prognostic information from this marker would improve the TTP and OS of patients with CRC. We recommend using these novel markers in adjacent normal tissues of patients with CRC to assist in clinical decision-making.
Cancer staging systems, including tumor-node-metastasis classification, facilitate reasonable adjuvant treatment and help predict the prognoses of tumors. However, part of low-risk colorectal cancer (CRC) patients experience relapse after therapeutic intervention. The prognostic factors that define these relapse-prone patients should be identified to optimize treatment selection. Recently, there were several novel prognostic biomarkers for CRC which involve epigenetic aberrant changes have been reported.
To determine the effect of the methylation status of a novel methylation gene panel on the relationship between the cancer stage and prognosis of CRC.
This study aimed to explore the relationship between the methylation status of candidate genes and prognosis of CRC.
One hundred and twenty CRC patients from Taiwan were enrolled to evaluate the association between hypermethylation of candidate genes and prognosis. The promoter methylation status of CDKN2A, hMLH1, MGMT, CSF2, DIS3L2, and OAF genes in tumor and adjacent normal tissues was assessed using methylation-specific PCR. Associations of the number of hypermethylated genes under study and different clinical stages with time to progression (TTP) or overall survival (OS) were assessed using the Cox proportional hazards regression model. Kaplan-Meier univariate assay was performed to analyze potential prognostic factors including TTP and OS.
The ≥ 3 aberrancy methylation group showed a significantly shorter 5-year TTP than the < 3 aberrancy methylation group. There was a significant interaction between CRC prognosis and different cancer stages (local and advanced) according to the methylation status of the selected genes in both types of tissues. However, the 5-year OS of patients with CRC in the ≥ 3 aberrancy group and the < 3 aberrancy group revealed no significant differences in both types of tissues.
Our data identified these novel methylation markers, particularly in the adjacent normal tissues in patients with advanced stage, and provided a basis to apply and investigate the potential of these markers to predict the prognosis of CRC.
Based on our findings, these novel markers in adjacent normal tissues of patients with CRC are recommended to help in clinical decision-making. Future cohort researches are needed to validate the utility of the new set of markers and address whether the modification of treatment/management decisions based on additional prognostic information from these markers would improve the TTP and OS of patients with CRC.
Manuscript source: Invited Manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Taiwan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Brisinda G, Mohamed SY, Wang K S-Editor: Gong ZM L-Editor: Wang TQ E-Editor: Liu JH
| 1. | Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2526] [Cited by in RCA: 2912] [Article Influence: 364.0] [Reference Citation Analysis (3)] |
| 2. | Howlader N, Noone A, Krapcho M, Miller D, Bishop K, Kosary C, Yu M, Ruhl J, Tatalovich Z, Mariotto A. SEER cancer statistics review, 1975–2014. Bethesda, MD: National Cancer Institute 2017; 2018. Available from: https://seer.cancer.gov/archive/csr/1975_2014/. |
| 3. | Edge SB. AJCC cancer staging manual. 2010;7:97-100 Springer. |
| 4. | DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1980] [Cited by in RCA: 2190] [Article Influence: 199.1] [Reference Citation Analysis (0)] |
| 5. | Broadbridge VT, Karapetis CS, Beeke C, Woodman RJ, Padbury R, Maddern G, Kim SW, Roder D, Hakendorf P, Price TJ. Do metastatic colorectal cancer patients who present with late relapse after curative surgery have a better survival? Br J Cancer. 2013;109:1338-1343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745-1755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 234] [Cited by in RCA: 285] [Article Influence: 31.7] [Reference Citation Analysis (8)] |
| 7. | Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 161] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 8. | Gevensleben H, Holmes EE, Goltz D, Dietrich J, Sailer V, Ellinger J, Dietrich D, Kristiansen G. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget. 2016;7:79943-79955. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 9. | Hashimoto Y, Zumwalt TJ, Goel A. DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer. Epigenomics. 2016;8:685-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Yi JM, Dhir M, Van Neste L, Downing SR, Jeschke J, Glöckner SC, de Freitas Calmon M, Hooker CM, Funes JM, Boshoff C, Smits KM, van Engeland M, Weijenberg MP, Iacobuzio-Donahue CA, Herman JG, Schuebel KE, Baylin SB, Ahuja N. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011;17:1535-1545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 11. | Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 507] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 12. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1241] [Article Influence: 112.8] [Reference Citation Analysis (1)] |
| 13. | Chang HF, Wu CC, Sun CA, Chu CM, Lin FG, Hsieh JF, Hsu CH, Huang CH, Yang T, Tsai YM, Kuan JC, Chou YC. Clinical stage and risk of recurrence and mortality: interaction of DNA methylation factors in patients with colorectal cancer. J Investig Med. 2016;64:1200-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Butterfield LH, Zhao F, Lee S, Tarhini AA, Margolin KA, White RL, Atkins MB, Cohen GI, Whiteside TL, Kirkwood JM, Lawson DH. Immune Correlates of GM-CSF and Melanoma Peptide Vaccination in a Randomized Trial for the Adjuvant Therapy of Resected High-Risk Melanoma (E4697). Clin Cancer Res. 2017;23:5034-5043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Costa P, Romão L, Gama-Carvalho M. Transcriptomic screen for DIS3 and DIS3L1 exosome subunits associated functional networks in colorectal cancer. BioSys PhD Day, Faculdade de Ciências da Universidade de Lisboa, 11 dezembro. 2015; Available from: http://repositorio.insa.pt/bitstream/10400.18/3657/1/Poster_Biosys%20Paulo%20Costa%202015.pdf. |
| 16. | Wu WC, Hsu CH, Kuan JC, Hsieh JF, Sun CA, Yang T, Wu CC, Chou YC. Predicting the progress of colon cancer by DNA methylation markers of the p16 gene in feces - Evidence from an animal model. Genet Mol Biol. 2013;36:323-328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 17. | David D, Anand D, Araújo C, Gloss B, Fino J, Dinger M, Lindahl P, Pöyhönen M, Hannele L, Lavinha J. Identification of OAF and PVRL1 as candidate genes for an ocular anomaly characterized by Peters anomaly type 2 and ectopia lentis. Exp Eye Res. 2018;168:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK. Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol. 2010;28:264-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 438] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 19. | Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2028] [Cited by in RCA: 1998] [Article Influence: 111.0] [Reference Citation Analysis (0)] |
| 20. | Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3811] [Cited by in RCA: 4214] [Article Influence: 324.2] [Reference Citation Analysis (0)] |
| 21. | Jung M, Pützer S, Gevensleben H, Meller S, Kristiansen G, Dietrich D. Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant, and malignant ascites. Clin Epigenetics. 2016;8:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Wick W, Weller M, van den Bent M, Sanson M, Weiler M, von Deimling A, Plass C, Hegi M, Platten M, Reifenberger G. MGMT testing--the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10:372-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 412] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 23. | Ng JM, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci. 2015;16:2472-2496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 24. | Ahmed D, Danielsen SA, Aagesen TH, Bretthauer M, Thiis-Evensen E, Hoff G, Rognum TO, Nesbakken A, Lothe RA, Lind GE. A tissue-based comparative effectiveness analysis of biomarkers for early detection of colorectal tumors. Clin Transl Gastroenterol. 2012;3:e27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Exner R, Pulverer W, Diem M, Spaller L, Woltering L, Schreiber M, Wolf B, Sonntagbauer M, Schröder F, Stift J, Wrba F, Bergmann M, Weinhäusel A, Egger G. Potential of DNA methylation in rectal cancer as diagnostic and prognostic biomarkers. Br J Cancer. 2015;113:1035-1045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 26. | Luo X, Huang R, Sun H, Liu Y, Bi H, Li J, Yu H, Sun J, Lin S, Cui B, Zhao Y. Methylation of a panel of genes in peripheral blood leukocytes is associated with colorectal cancer. Sci Rep. 2016;6:29922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Kim SH, Park KH, Shin SJ, Lee KY, Kim TI, Kim NK, Rha SY, Ahn JB. CpG Island Methylator Phenotype and Methylation of Wnt Pathway Genes Together Predict Survival in Patients with Colorectal Cancer. Yonsei Med J. 2018;59:588-594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | de Sousa E Melo F, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR, Fessler E, van den Bergh SP, Rodermond H, Dekker E, van der Loos CM, Pals ST, van de Vijver MJ, Versteeg R, Richel DJ, Vermeulen L, Medema JP. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9:476-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 269] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 29. | Juo YY, Johnston FM, Zhang DY, Juo HH, Wang H, Pappou EP, Yu T, Easwaran H, Baylin S, van Engeland M, Ahuja N. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann Oncol. 2014;25:2314-2327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 135] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 30. | Freitas M, Ferreira F, Carvalho S, Silva F, Lopes P, Antunes L, Salta S, Diniz F, Santos LL, Videira JF, Henrique R, Jerónimo C. A novel DNA methylation panel accurately detects colorectal cancer independently of molecular pathway. J Transl Med. 2018;16:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 31. | Kushwaha G, Dozmorov M, Wren JD, Qiu J, Shi H, Xu D. Hypomethylation coordinates antagonistically with hypermethylation in cancer development: a case study of leukemia. Hum Genomics. 2016;10 Suppl 2:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Porcellini E, Laprovitera N, Riefolo M, Ravaioli M, Garajova I, Ferracin M. Epigenetic and epitranscriptomic changes in colorectal cancer: Diagnostic, prognostic, and treatment implications. Cancer Lett. 2018;419:84-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 33. | Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capellà G, Ribas M, Peinado MA. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462-9468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 238] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 34. | Ye D, Jiang D, Li Y, Jin M, Chen K. The role of LINE-1 methylation in predicting survival among colorectal cancer patients: a meta-analysis. Int J Clin Oncol. 2017;22:749-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 35. | Luo Y, Yu M, Grady WM. Field cancerization in the colon: a role for aberrant DNA methylation? Gastroenterol Rep (Oxf). 2014;2:16-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Azuara D, Aussó S, Rodriguez-Moranta F, Guardiola J, Sanjuan X, Lobaton T, Boadas J, Piqueras M, Monfort D, Guinó E, Moreno V, Capellá G, de Oca J. New Methylation Biomarker Panel for Early Diagnosis of Dysplasia or Cancer in High-Risk Inflammatory Bowel Disease Patients. Inflamm Bowel Dis. 2018;24:2555-2564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 37. | Li J, Poi MJ, Tsai MD. Regulatory mechanisms of tumor suppressor P16(INK4A) and their relevance to cancer. Biochemistry. 2011;50:5566-5582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 38. | Bihl MP, Foerster A, Lugli A, Zlobec I. Characterization of CDKN2A(p16) methylation and impact in colorectal cancer: systematic analysis using pyrosequencing. J Transl Med. 2012;10:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Kohonen-Corish MR, Tseung J, Chan C, Currey N, Dent OF, Clarke S, Bokey L, Chapuis PH. KRAS mutations and CDKN2A promoter methylation show an interactive adverse effect on survival and predict recurrence of rectal cancer. Int J Cancer. 2014;134:2820-2828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 40. | Lu Y, Wajapeyee N, Turker MS, Glazer PM. Silencing of the DNA mismatch repair gene MLH1 induced by hypoxic stress in a pathway dependent on the histone demethylase LSD1. Cell Rep. 2014;8:501-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 41. | Kuan JC, Wu CC, Sun CA, Chu CM, Lin FG, Hsu CH, Kan PC, Lin SC, Yang T, Chou YC. DNA methylation combinations in adjacent normal colon tissue predict cancer recurrence: evidence from a clinical cohort study. PLoS One. 2015;10:e0123396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 42. | Iida S, Kato S, Ishiguro M, Matsuyama T, Ishikawa T, Kobayashi H, Higuchi T, Uetake H, Enomoto M, Sugihara K. PIK3CA mutation and methylation influences the outcome of colorectal cancer. Oncol Lett. 2012;3:565-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 43. | Belhadj S, Moutinho C, Mur P, Setien F, Llinàs-Arias P, Pérez-Salvia M, Pons T, Pineda M, Brunet J, Navarro M, Capellá G, Esteller M, Valle L. Germline variation in O6-methylguanine-DNA methyltransferase (MGMT) as cause of hereditary colorectal cancer. Cancer Lett. 2019;447:86-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Lee YY, Wu WJ, Huang CN, Li CC, Li WM, Yeh BW, Liang PI, Wu TF, Li CF. CSF2 Overexpression Is Associated with STAT5 Phosphorylation and Poor Prognosis in Patients with Urothelial Carcinoma. J Cancer. 2016;7:711-721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 45. | Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ, Cole T, van Essen AJ, van Lingen RA, Neri G, Opitz JM, Rump P, Stolte-Dijkstra I, Müller F, Pruijn GJ, Latif F, Maher ER. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44:277-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 46. | Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, West RB, Plevritis SK, Alizadeh AA. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2244] [Cited by in RCA: 2348] [Article Influence: 234.8] [Reference Citation Analysis (0)] |
| 47. | Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL, Huang HD. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43:D856-D861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 235] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 48. | Saito T, Mikami Y, Kinugasa M, Mori K, Sugimoto M, Uchida K. Genes for prognosis of cancer. U.S. Patent Application No 12/183,610, 2009. Available from: https://patentimages.storage.googleapis.com/08/9e/de/9cb348d7d14ec5/US20090011423A1.pdf. |
| 49. | Kwon JH, Wu F. Microrna-based diagnostic testing and therapies for inflammatory bowel disease and related diseases. U.S. Patent Application No 12/934,820, 2011. Available from: https://patents.google.com/patent/US20110117111A1/en. |