Published online Jan 7, 2020. doi: 10.3748/wjg.v26.i1.1
Peer-review started: October 17, 2019
First decision: December 4, 2019
Revised: December 5, 2019
Accepted: December 23, 2019
Article in press: December 23, 2019
Published online: January 7, 2020
Processing time: 82 Days and 7.6 Hours
Coeliac disease (CD) is a complex condition resulting from an interplay between genetic and environmental factors. When diagnosing the condition, serological testing and genotyping are useful in excluding CD, although the gold standard of testing is currently histopathological examination of the small intestine. There are drawbacks associated with this form of testing however and because of this, novel forms of testing are currently under investigation. Before we develop completely novel tests though, it is important to ask whether or not we can simply use the data we gather from coeliac patients more effectively and build a more accurate snapshot of CD through statistical analysis of combined metrics. It is clear that not one single test can accurately diagnose CD and it is also clear that CD patients can no longer be defined by discrete classifications, the continuum of patient presentation needs to be recognised and correctly captured to improve diagnostic accuracy. This review will discuss the current diagnostics for CD and then outline novel diagnostics under investigation for the condition. Finally, improvements to current protocols will be discussed with the need for a holistic “snapshot” of CD using a number of metrics simultaneously.
Core tip: Due to the complexity of the condition, the diagnosis of coeliac disease can pose unique challenges. This review will discuss the current diagnostics for the condition and then outline novel diagnostics currently under investigation. Finally improvements to the current testing protocols will be discussed with the need for a holistic “snapshot” of the condition, using a number of metrics simultaneously.
- Citation: Charlesworth RP. Diagnosing coeliac disease: Out with the old and in with the new? World J Gastroenterol 2020; 26(1): 1-10
- URL: https://www.wjgnet.com/1007-9327/full/v26/i1/1.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i1.1
Coeliac disease (CD) is a chronic autoimmune enteropathy which results from a complex interplay between genetic and environmental factors[1,2]. Inheritance of altered forms of the human leukocyte antigen receptor (HLA-DQ2 or HLA-DQ8) in patients with CD bind epitopes of a dietary protein, gluten, with high affinity. Once cells within the small intestine are sensitised to these epitopes, a destructive autoimmune reaction is triggered which ultimately results in the destruction of the small intestinal wall[3-5]. The prevalence of CD is increasing, with around 1% of the general population now affected by the condition and presentation of the disease in populations not classically affected[1,6,7]. The symptoms of CD are varied; but can include diarrhoea, weight loss, abdominal pain and failure to thrive in children. If left untreated, CD can lead to complications associated with nutrient deficiency; such as anemia, alopecia, fertility issues or increased bone fracture risk[2,8-11]. Long-term untreated CD has been associated with increased risk of enteropathy-associated T-cell lymphoma (EATL) and adenocarcinoma of the small intestine[12,13]. The only current form of treatment is a strict, lifelong gluten-free diet[2,14]. This review will focus on the diagnosis of CD, outlining the current diagnostic guidelines and highlighting the benefits and shortcomings associated with these. Novel diagnostics currently under investigation or improvements to the current diagnostics will then be discussed.
The frontline testing for the presence of CD is serological examination. Currently, serological testing for CD is recommended for patients presenting with chronic/intermittent diarrhoea, unexpected weight loss, recurrent abdominal pain or persistent gastrointestinal symptoms. Serological screening for CD is also offered to patients with associated conditions such as autoimmune thyroid disease, irritable bowel syndrome, or type 1 diabetes[15]. Titres of three main types of antibody are assessed in CD screening; IgA-based antibodies against the enzyme tissue transglutaminase (tTG), IgA and IgG-based antibodies against deamidated gliadin peptides (DGP) and IgA-based antibodies against the endomysium (EMA). Of these tests, high titres of IgA-tTG and IgA-EMA accounts for nearly 95% reliability in serological screening[10,15]. IgG/IgA-anti-deamidated gliadin peptide (IgG/IgA-DGP) is used as a companion test for improved accuracy by very specifically detecting antibodies directed against immunogenic peptides of gluten in patients with suspected CD[10,16,17]. Serological testing can also be used as a less invasive method of monitoring treatment progression and adherence to a gluten-free diet after diagnosis[18].
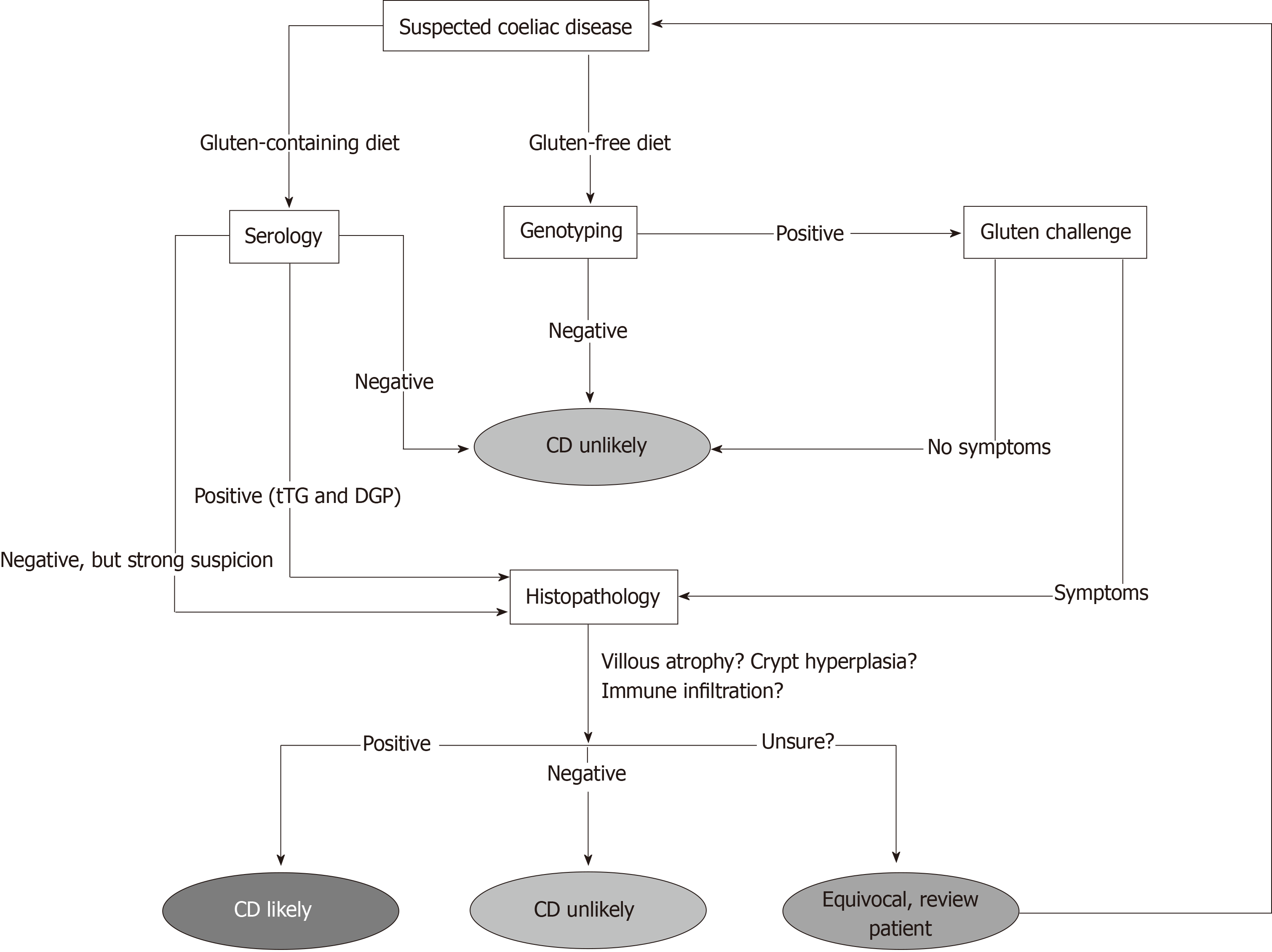
There are shortcomings associated with serological screening however. Firstly, the patient must be on a diet containing gluten for results to be meaningful[10,19]. Secondly, many of the tests rely on IgA-based antibodies and IgA deficiency is far more common in CD patients than in the general population, with a prevalence rate of around 2%-3%[10,19,20]. This can be overcome however by measuring total IgA at the start of testing to ensure sufficient levels and incorporating IgG-based tests into the panel[19]. Furthermore, it has also been shown that the sensitivity of serological tests for CD is far lower than reported when patients with milder pathology are tested[21]. Due to these limitations, patients with negative serology but who are still highly suspected of having CD are often referred for further testing, either by examining biopsy material or genotyping (Figure 1).
Currently, patients who have negative serology (but are suspected of having CD), patients with a family history of CD or patients who are following a gluten-free diet at the time of diagnosis (and unwilling to undergo a gluten challenge) are offered genetic testing for CD in the form of HLA genotyping (Figure 1)[2,19]. Ninety-nine percent of patients with CD express either of the MHC Class II antigens HLA-DQ2 or HLA-DQ8, variants of the Human Leukocyte Antigen class II receptors[3,5]. As MHC II molecules are heterodimers, on a genetic level these variants result from the inheritance of several key alleles. HLA-DQ2 results from the expression of two alleles, HLA-DQA1*0501 and HLA-DQB1*02 whose gene products combine to form the altered MHC II receptor. HLA-DQ8 results from the expression of the variant HLA-DQB1*0302 and HLA-DQB1*03 alleles[22]. These variants are MHC II molecules that favour the binding of negatively charged residues in a 3-anchor point configuration, usually at positions 4, 6 and 7 on the gliadin epitope. HLA-DQ8 is a very similar molecule, although with anchor points usually at positions 1, 4 and 9 on the gliadin epitope[23,24].
HLA-genotyping is therefore useful in excluding a diagnosis of CD in cases where serological or histological results are difficult to interpret or to determine the prevalence of CD amongst relatives[22,25]. The test can be performed either with blood or buccal samples and a negative result effectively rules out the presence of CD entirely. It is also not dependent on gluten intake, so can be administered without the need for the patient to commence a gluten-containing diet[15,19].
Unfortunately, the frequency of these genes has been reported to vary geographically, with the DQ2.5 allele being reported at higher frequency in north-western Europeans, such as those from Ireland[26] and the DQ8 allele showing a higher frequency in Amerindian populations[27], thus creating differences in expression independent of the presence of CD. In Australia specifically, approximately 20% of the population have been estimated to have the DQ2 allele, whilst less than 5% of the population have been estimated to have the DQ8 allele[28-30]. At the same time, the cost of the test excludes its use as a front line diagnostic for CD and it also cannot diagnose CD effectively in its own right, as only around 1 in 30 people with the DQ2 or DQ8 variants will eventually develop the condition[22]. Thus HLA-genotyping only provides a risk profile for developing CD. For this reason, even if the gene test returns a positive result, clinical guidelines state that patients still need to undergo small bowel biopsy to confirm the diagnosis[2,15,19].
Histopathological examination of duodenal biopsy material is currently the most conclusive test for the presence of CD. Using image enhancement on modern endoscopes, it is currently recommended that if villous atrophy is suspected during upper-gastrointestinal assessment that 2-3 biopsies are taken from the duodenal bulb and 4-6 biopsies are taken from along the distal duodenum[10,15,18,19,31]. Biopsies from these regions are used as they are the first point of contact with the digesta[10]. The histopathological findings associated with active CD are well documented and include three main findings (Figure 1); blunted or atrophic villi (including complete destruction of the epithelial surface), crypt hyperplasia and mononuclear/ lymphocytic infiltration into the lamina propria[15,18,32].
Villous atrophy and crypt hyperplasia are usually assessed by calculating the crypt-villous ratio, a measure of the height of the villous when compared to the depth of the adjacent intestinal crypt. Using this method, a normal ratio of villous:crypt height in adults is around 3:1 to 5:1, whilst this figure in children is around 2:1. Values significantly less than these give an indication of the degree of villous atrophy present[33,34]. Lymphocytic infiltration can be assessed by directly examining the numbers of lymphocytes present within the lamina propria in the late stages of the condition (usually T and B lymphocytes) and by assessing the numbers of intraepithelial lymphocytes (IEL)[15,34]. The IELs are a specialised, critical part of the gut-associated lymphoid tissue and do not need priming by other immune cells to release cytokines[34-36]. As the population of these cells is expanded in CD, the current diagnostic cut-off is 25 IELs per 100 enterocytes to demonstrate intraepithelial lymphocytosis in the condition[19,34,37,38].
Morphological changes in CD mucosa can then graded according to the Marsh Score[39,40], with “0” indicating no detectable changes and “3a/3b/3c” indicating severely inflamed tissue affected by autoimmune destruction. It should be noted however that some pathologists will prefer to use descriptive terms instead of the Marsh score in routine assessment of CD rather than the Marsh score[10]. There has been considerable debate as to the accuracy of the Marsh score system however, as it is based on subjective observations of intestinal histological sections which must be made by an experienced pathologist[10,39,40,41]. It has been suggested that subjective interpretation of biopsy material may potentially lead to significant inter-observer disagreement and therefore negative or delayed patient outcomes[41]. Further confounding the histological diagnosis of CD is the patchy presentation of the condition and the fact that the lesions that appear during active CD may not be entirely specific and can often be seen in other enteropathies such as giardiasis or gastroenteritis[37,42-44]. However, at present the Marsh score system in conjunction with serology is currently the gold standard for the assessment of CD and a vast majority of pathologists are able to readily recognise active lesions (Marsh type 3). The difficulty arises when assessment of milder lesions is required. Thus, equivocal patients with subtle changes may be missed by the current histological criteria, leading to ambiguity in diagnosis.
It is clear that the current testing regimen for CD is complicated, as shown in Figure 1, and it is also clear that each diagnostic has significant drawbacks associated with its use. Therefore, it is at this point that we need to ask a critical question. Do we need to develop completely novel tests for coeliac disease or can we use the data we currently generate from patients more effectively? A number of studies have investigated whether or not the application of statistical analysis to existing measurements can increase the diagnostic sensitivity of CD screening. One such technique is linear discriminant analysis (LDA). This technique, when applied to biological data, aims to assign patients to one or more groups on the basis of a series of measurements from which a linear function has been defined[45,46]. Discriminant analysis has been shown to be able to predict patient groupings in conditions such as rheumatoid arthritis[47], Parkinson’s disease[48], diabetes[49], Alzheimer’s disease[50] and coronary disease[51]. In CD, improving diagnosis with discriminant function analysis has been previously investigated with IgA/IgG and absorptive serology[52,53], IEL counts with crypt/villous ratios[54] and immunohistochemistry data[55]. More recently, studies in CD have used this technique with capsule endoscopy images[56], histology data[57] and gene expression data[58]. The use of this technique has further highlighted the inherit difficulties in classifying CD patients using the discrete divisions of the Marsh subclassifications[59]. Clearly, the full spectrum of CD presentation needs to be captured with continuous categories along a scale to be able to accurately diagnose all who present with CD-like symptomology. Improved and more accurate diagnostics could then also be used to separate other inflammatory conditions, such as Crohn’s disease.
As the current technique of sampling from the small intestinal mucosa relies on the patient being on a gluten-containing diet and actively having CD damage to evaluate, previous research has also focused on determining other biopsy-based tests which could be implemented after a patient has commenced treatment. It has been shown that a rectal challenge with gluten can induce CD-like pathology in the rectal mucosa that is specific enough to attain a diagnosis of CD as both a screening and confirmatory test, with reported sensitivities of 90%-100% and specificities of 91%-100%[60,61]. As sensitised gluten-specific T lymphocytes circulate within the gastrointestinal mucosa, they can be rapidly deployed to sites of localised antigen presentation to initiate a localised inflammatory reaction[60,62]. Although experimental, this test involves the introduction of a slurry of gluten to the rectum that the patient is instructed to retain for as long as possible, preferably for at least 2-4 hours. Biopsies are then taken from the rectal mucosa and the intraepithelial lymphocyte numbers are assessed within the tissue[60-63].
More recently, a novel serological test for CD[64] has also been demonstrated to show high sensitivity and specificity for CD patients who possess the DQ2.5 allele and does not require the patient to be on a gluten-containing diet. Using a whole blood cytokine release assay (primarily used for infectious diseases) focused on IFN-γ, these authors took whole blood from treated CD patients after an oral challenge of gluten and cultured it in the presence of gliadin epitopes. They found that there was no test which could detect changes between the treated CD patients and controls before the gluten challenge; but after gluten was consumed by the treated CD patients, significant elevations in IFN-γ and IFN-γ inducible protein 10 (IP-10; CXCL10) resulted in 85% IFN-γ and 94% IP-10 sensitivity and 100% specificity for DQ2.5+ CD patients. These authors concluded that further clinical studies investigating the utility of these tests were required[64]. Similar testing using tetramers of gluten and HLA has recently been shown to be able to specifically detect gluten-reactive T cells in coeliac patients with a high degree of accuracy and regardless of current gluten intake[65].
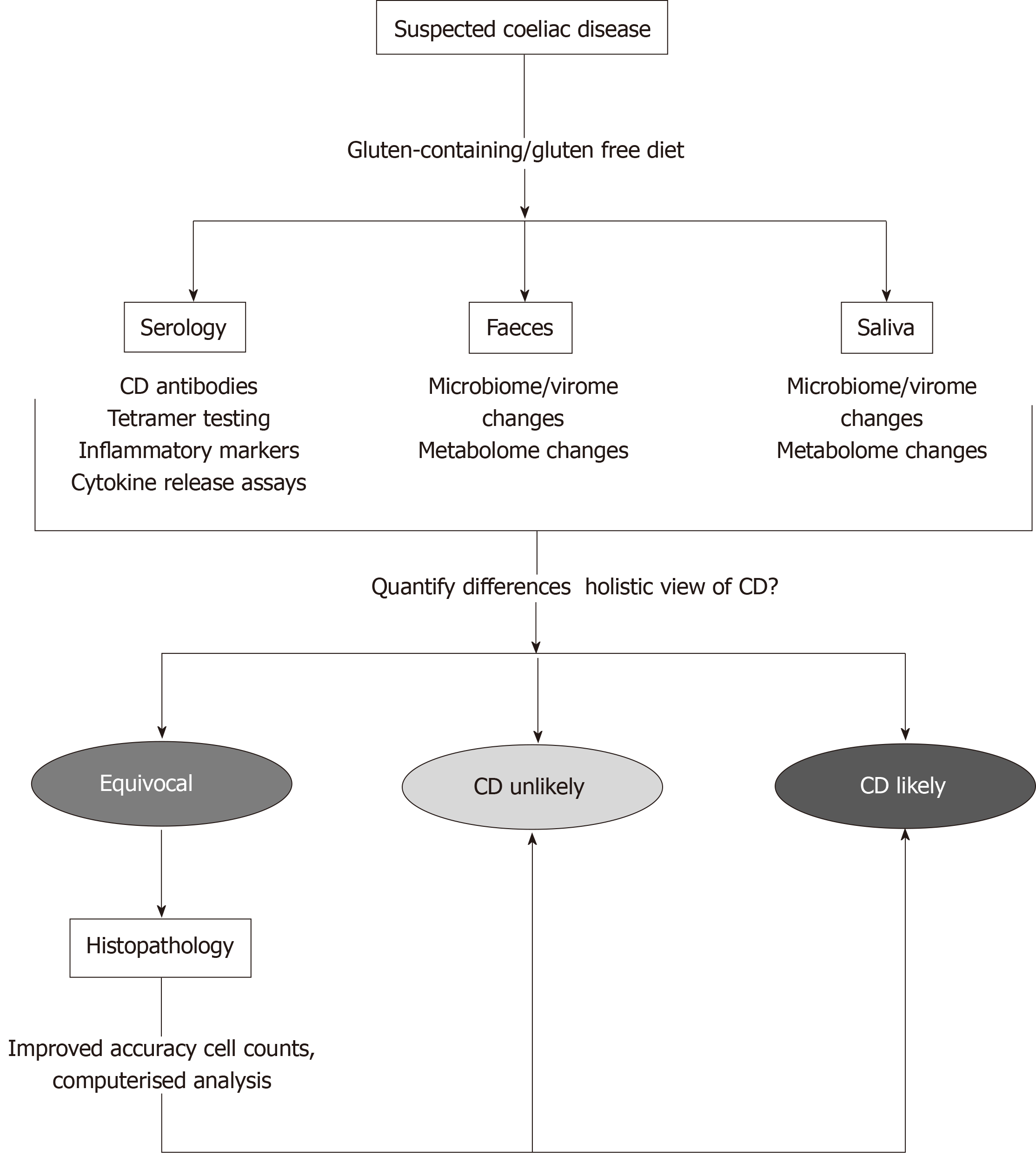
Due to the complexity of the coeliac reaction, it is clear that not one single test can fully capture the coeliac continuum, data from many different parameters would need to be combined for the most accuracy. So where should this data come from and how could it be used as a single diagnostic? Histology of the duodenal mucosa should always a play a part in CD diagnosis, however we must first define what a normal duodenal mucosa is before we can begin to compare pathological specimens. The upper and lower borders of the mucosal surface need to be defined, in particular where the exact border of the crypt and the villus meet, perhaps through the use of mRNA expression[59]. At the same time, the surface of the duodenum is a complex and 3-dimensional environment which is poorly represented on a 2-dimensinal microscope slide. Computerised analysis is needed to fully understand the 3-dimensional structure of the duodenal mucosa and how this relates to our 2-dimensional representations[59,66]. Once we can overcome these shortfalls, we need to take numerical values of histological parameters from slides instead of making subjective assessments or attempting to put patients into discrete categories. These numbers can then be used to improve diagnostics as previously shown[57].
There is a wealth of data which could potentially be collected from CD patients, with the most recent being insights into the microbiome of these patients. In the mouth of CD patients, it has been shown that differences occur in the microbial population and that these organisms display proteolytic activity against gliadin, possibly generating immunogenic peptides in the process[67]. In the small intestine, although it was demonstrated that the microbiome did not differ in children with CD when compared to healthy controls, this same effect has not been well investigated in adults to date[68]. It is hypothesised however that changes in the small intestinal microbiome may be involved in the pathogenesis of CD through immune reactions generated against translocated bacterial proteins, resulting from decreased intestinal barrier integrity[69-71]. Within the large intestine, shifts in microbial populations have been shown in CD with a number of genera, including Lactobacillus, Streptococcus and Clostridium demonstrating proteolytic activity against gluten proteins. Members of these strains may possibly be used in the treatment of the condition by digesting immunogenic fragments of gliadin[72]. Significant changes in the colonic microbiome, including increases in the Veillonellaceae family and other taxa involved in starch metabolism, have also been observed in patients who have started treatment with a gluten-free diet in CD[73]. If it is possible to numerically categorise these changes in CD patients when compared to non-CD patients, this data could then be used in a diagnostic sense. At the same time, the collection of faeces and saliva is more efficient and less invasive than intestinal tissue sampling.
Along a similar vein is the categorisation of the CD metabolome; that is, the complete set of metabolites present in a patient sample at a given time point[74,75]. This holistic assessment of end-products can therefore indirectly take into account a variety of changes which may occur from genotype to phenotype[74]. In CD, the metabolites studied are most commonly from pathways associated with malabsorption, energy metabolism and alterations in intestinal permeability and these can be assessed in a diverse range of fluids, including saliva, CSF, amniotic fluid, breath condensate and faecal extract[74,75]. As there is currently no one particular metabolite which has been shown to have a high predictive value for CD, assessing panels of these potential biomarkers currently holds the most promise for novel diagnostic tests[74-77]. Most recently, this approach has identified a phospholipid signature in HLA at-risk infants which has diagnostic capacity for CD well before antibodies or clinical symptoms appear[78].
“Fingerprinting” of microbial and metabolomic signatures in CD therefore has potential to generate a large amount of data from individuals from a relatively non-invasive test. With the use of LDA, the more variables which can be added to build diagnostic equations, the more accurate the outcome[45]. Combining these new definitions of CD with current diagnostics would allow for a snapshot of CD presentation from all angles. Measuring these parameters (histology, mRNA expression, microbiome change, metabolome change) simultaneously would allow for the most accurate diagnosis and secure the best outcome for patients, as shown in Figure 2.
Having accurate diagnostics for CD is critical moving forward, with increasing prevalence of the condition and the risk of serious effects if treatment is not commenced early enough. Current diagnostics have significant drawbacks however and the accuracy of these tests needs to be improved to successfully detect CD in all patients who present with the condition. This is particularly true for those who lack classical symptomology or those who have very mild histopathology. This is also true for tracking treatment progression and healing in patients once a gluten-free diet has been commenced. We need to move away from the discrete definition of CD and towards a continuous scale to fully capture the complete spectrum of patient presentation. To do this, we need diagnostic tests which are holistic; that is, they can take a range of measures from a patient at once and can then be combined to improve diagnostic accuracy. This is where new diagnostic tests need to be defined which can assess CD less invasively. Of most interest is the changes which appear in the microbiome of CD patients and if these changes can be numerically defined, this could lead to a range of novel tests for the condition, either alone or in combination with the traditional CD diagnostics.
With our original question in mind then, novel diagnostic advances in CD are welcomed, particularly if they can assess the condition less invasively and increase the accuracy and speed of screening. The current diagnostics for the condition need to be revisited for the next generation of CD patients and their accuracy needs to be improved, particularly for equivocal presentation. It is hoped then that a balance can be found between novel tests and traditional methods to provide an accurate snapshot of the condition and improve the outcomes of CD patients.
The author would like to thank Dr. Gal Winter for her editing assistance.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Australia
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good):
Grade C (Good):
Grade D (Fair):
Grade E (Poor):
P-Reviewer: Can G S-Editor: Gong ZM L-Editor: A E-Editor: Ma YJ
| 1. | Catassi C, Gatti S, Fasano A. The new epidemiology of celiac disease. J Pediatr Gastroenterol Nutr. 2014;59 Suppl 1:S7-S9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 2. | Ludvigsson JF, Bai JC, Biagi F, Card TR, Ciacci C, Ciclitira PJ, Green PH, Hadjivassiliou M, Holdoway A, van Heel DA, Kaukinen K, Leffler DA, Leonard JN, Lundin KE, McGough N, Davidson M, Murray JA, Swift GL, Walker MM, Zingone F, Sanders DS; BSG Coeliac Disease Guidelines Development Group; British Society of Gastroenterology. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut. 2014;63:1210-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 689] [Cited by in RCA: 783] [Article Influence: 71.2] [Reference Citation Analysis (3)] |
| 3. | Jabri B, Sollid LM. Mechanisms of disease: immunopathogenesis of celiac disease. Nat Clin Pract Gastroenterol Hepatol. 2006;3:516-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 125] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 4. | Mowat AM. Coeliac disease--a meeting point for genetics, immunology, and protein chemistry. Lancet. 2003;361:1290-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 55] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Sollid LM. Molecular basis of celiac disease. Annu Rev Immunol. 2000;18:53-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 452] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 6. | Lionetti E, Gatti S, Pulvirenti A, Catassi C. Celiac disease from a global perspective. Best Pract Res Clin Gastroenterol. 2015;29:365-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 7. | Mustalahti K, Catassi C, Reunanen A, Fabiani E, Heier M, McMillan S, Murray L, Metzger MH, Gasparin M, Bravi E, Mäki M; Coeliac EU Cluster, Project Epidemiology. The prevalence of celiac disease in Europe: results of a centralized, international mass screening project. Ann Med. 2010;42:587-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 523] [Article Influence: 34.9] [Reference Citation Analysis (1)] |
| 8. | Lebwohl B, Michaëlsson K, Green PH, Ludvigsson JF. Persistent mucosal damage and risk of fracture in celiac disease. J Clin Endocrinol Metab. 2014;99:609-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 9. | Murray JA, Watson T, Clearman B, Mitros F. Effect of a gluten-free diet on gastrointestinal symptoms in celiac disease. Am J Clin Nutr. 2004;79:669-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 212] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 10. | Walker MM, Murray JA. An update in the diagnosis of coeliac disease. Histopathology. 2011;59:166-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Zugna D, Richiardi L, Akre O, Stephansson O, Ludvigsson JF. A nationwide population-based study to determine whether coeliac disease is associated with infertility. Gut. 2010;59:1471-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Catassi C, Bearzi I, Holmes GK. Association of celiac disease and intestinal lymphomas and other cancers. Gastroenterology. 2005;128:S79-S86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 194] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 13. | Silano M, Volta U, Mecchia AM, Dessì M, Di Benedetto R, De Vincenzi M; Collaborating centers of the Italian registry of the complications of coeliac disease. Delayed diagnosis of coeliac disease increases cancer risk. BMC Gastroenterol. 2007;7:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 14. | Newnham ED, Shepherd SJ, Strauss BJ, Hosking P, Gibson PR. Adherence to the gluten-free diet can achieve the therapeutic goals in almost all patients with coeliac disease: A 5-year longitudinal study from diagnosis. J Gastroenterol Hepatol. 2016;31:342-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Centre for Clinical Practice at NICE (UK). Coeliac Disease: Recognition and Assessment of Coeliac Disease [Internet]. 2009;. [PubMed] |
| 16. | Lerner A, Kumar V, Iancu TC. Immunological diagnosis of childhood coeliac disease: comparison between antigliadin, antireticulin and antiendomysial antibodies. Clin Exp Immunol. 1994;95:78-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 109] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 17. | Volta U, Granito A, Parisi C, Fabbri A, Fiorini E, Piscaglia M, Tovoli F, Grasso V, Muratori P, Pappas G, De Giorgio R. Deamidated gliadin peptide antibodies as a routine test for celiac disease: a prospective analysis. J Clin Gastroenterol. 2010;44:186-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 86] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | Iacucci M, Ghosh S. Routine duodenal biopsies to diagnose celiac disease. Can J Gastroenterol. 2013;27:385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | Rubio-Tapia A, Hill ID, Kelly CP, Calderwood AH, Murray JA; American College of Gastroenterology. ACG clinical guidelines: diagnosis and management of celiac disease. Am J Gastroenterol. 2013;108:656-76; quiz 677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 1158] [Article Influence: 96.5] [Reference Citation Analysis (0)] |
| 20. | McGowan KE, Lyon ME, Butzner JD. Celiac disease and IgA deficiency: complications of serological testing approaches encountered in the clinic. Clin Chem. 2008;54:1203-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 21. | Rostom A, Dubé C, Cranney A, Saloojee N, Sy R, Garritty C, Sampson M, Zhang L, Yazdi F, Mamaladze V, Pan I, MacNeil J, Mack D, Patel D, Moher D. The diagnostic accuracy of serologic tests for celiac disease: a systematic review. Gastroenterology. 2005;128:S38-S46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 332] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 22. | Megiorni F, Pizzuti A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J Biomed Sci. 2012;19:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 23. | Broughton SE, Petersen J, Theodossis A, Scally SW, Loh KL, Thompson A, van Bergen J, Kooy-Winkelaar Y, Henderson KN, Beddoe T, Tye-Din JA, Mannering SI, Purcell AW, McCluskey J, Anderson RP, Koning F, Reid HH, Rossjohn J. Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity. 2012;37:611-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 24. | Tjon JM, van Bergen J, Koning F. Celiac disease: how complicated can it get? Immunogenetics. 2010;62:641-651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 25. | Abadie V, Sollid LM, Barreiro LB, Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol. 2011;29:493-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 354] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 26. | Michalski JP, McCombs CC, Arai T, Elston RC, Cao T, McCarthy CF, Stevens FM. HLA-DR, DQ genotypes of celiac disease patients and healthy subjects from the West of Ireland. Tissue Antigens. 1996;47:127-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Layrisse Z, Guedez Y, Domínguez E, Paz N, Montagnani S, Matos M, Herrera F, Ogando V, Balbas O, Rodríguez-Larralde A. Extended HLA haplotypes in a Carib Amerindian population: the Yucpa of the Perija Range. Hum Immunol. 2001;62:992-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Cummins AG, Roberts-Thomson IC. Prevalence of celiac disease in the Asia-Pacific region. J Gastroenterol Hepatol. 2009;24:1347-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 29. | Gujral N, Freeman HJ, Thomson AB. Celiac disease: prevalence, diagnosis, pathogenesis and treatment. World J Gastroenterol. 2012;18:6036-6059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 412] [Cited by in RCA: 415] [Article Influence: 31.9] [Reference Citation Analysis (6)] |
| 30. | Lionetti E, Catassi C. Co-localization of gluten consumption and HLA-DQ2 and -DQ8 genotypes, a clue to the history of celiac disease. Dig Liver Dis. 2014;46:1057-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 31. | Antonioli DA. Celiac disease: a progress report. Mod Pathol. 2003;16:342-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 32. | Ferguson A, Arranz E, O'Mahony S. Clinical and pathological spectrum of coeliac disease--active, silent, latent, potential. Gut. 1993;34:150-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 264] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 33. | Dickson BC, Streutker CJ, Chetty R. Coeliac disease: an update for pathologists. J Clin Pathol. 2006;59:1008-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 202] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 34. | Serra S, Jani PA. An approach to duodenal biopsies. J Clin Pathol. 2006;59:1133-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 35. | Cellier C, Patey N, Mauvieux L, Jabri B, Delabesse E, Cervoni JP, Burtin ML, Guy-Grand D, Bouhnik Y, Modigliani R, Barbier JP, Macintyre E, Brousse N, Cerf-Bensussan N. Abnormal intestinal intraepithelial lymphocytes in refractory sprue. Gastroenterology. 1998;114:471-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 250] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 36. | Sanchez-Muñoz LB, Santón A, Cano A, Lopez A, Almeida J, Orfao A, Escribano L, Roy G. Flow cytometric analysis of intestinal intraepithelial lymphocytes in the diagnosis of refractory celiac sprue. Eur J Gastroenterol Hepatol. 2008;20:478-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Kamboj AK, Oxentenko AS. Clinical and Histologic Mimickers of Celiac Disease. Clin Transl Gastroenterol. 2017;8:e114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 38. | Rostami K, Marsh MN, Johnson MW, Mohaghegh H, Heal C, Holmes G, Ensari A, Aldulaimi D, Bancel B, Bassotti G, Bateman A, Becheanu G, Bozzola A, Carroccio A, Catassi C, Ciacci C, Ciobanu A, Danciu M, Derakhshan MH, Elli L, Ferrero S, Fiorentino M, Fiorino M, Ganji A, Ghaffarzadehgan K, Going JJ, Ishaq S, Mandolesi A, Mathews S, Maxim R, Mulder CJ, Neefjes-Borst A, Robert M, Russo I, Rostami-Nejad M, Sidoni A, Sotoudeh M, Villanacci V, Volta U, Zali MR, Srivastava A. ROC-king onwards: intraepithelial lymphocyte counts, distribution & role in coeliac disease mucosal interpretation. Gut. 2017;66:2080-2086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 39. | Marsh MN. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity ('celiac sprue'). Gastroenterology. 1992;102:330-354. [PubMed] |
| 40. | Oberhuber G, Granditsch G, Vogelsang H. The histopathology of coeliac disease: time for a standardized report scheme for pathologists. Eur J Gastroenterol Hepatol. 1999;11:1185-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1142] [Cited by in RCA: 1205] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 41. | Corazza GR, Villanacci V. Coeliac disease. J Clin Pathol. 2005;58:573-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 189] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 42. | Ravelli A, Bolognini S, Gambarotti M, Villanacci V. Variability of histologic lesions in relation to biopsy site in gluten-sensitive enteropathy. Am J Gastroenterol. 2005;100:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 43. | Wahab PJ, Meijer JW, Mulder CJ. Histologic follow-up of people with celiac disease on a gluten-free diet: slow and incomplete recovery. Am J Clin Pathol. 2002;118:459-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 248] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 44. | Weir DC, Glickman JN, Roiff T, Valim C, Leichtner AM. Variability of histopathological changes in childhood celiac disease. Am J Gastroenterol. 2010;105:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 45. | Fisher RA. The use of multiple measurements in taxonomic problems. Ann Hum Genet. 1936;7:179-188. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8484] [Cited by in RCA: 3648] [Article Influence: 280.6] [Reference Citation Analysis (0)] |
| 46. | Friedman JH. Regularized discriminant analysis. J Am Stat Assoc. 1989;84:165-175. [RCA] [DOI] [Full Text] [Cited by in Crossref: 885] [Cited by in RCA: 563] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 47. | Prevoo ML, van 't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4129] [Cited by in RCA: 4607] [Article Influence: 153.6] [Reference Citation Analysis (0)] |
| 48. | Burn DJ, Sawle GV, Brooks DJ. Differential diagnosis of Parkinson's disease, multiple system atrophy, and Steele-Richardson-Olszewski syndrome: discriminant analysis of striatal 18F-dopa PET data. J Neurol Neurosurg Psychiatry. 1994;57:278-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 106] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 49. | Polat K, Güneş S, Arslan A. A cascade learning system for classification of diabetes disease: Generalized Discriminant Analysis and Least Square Support Vector Machine. Expert Syst Appl. 2008;34:482-487. [DOI] [Full Text] |
| 50. | Charpentier P, Lavenu I, Defebvre L, Duhamel A, Lecouffe P, Pasquier F, Steinling M. Alzheimer's disease and frontotemporal dementia are differentiated by discriminant analysis applied to (99m)Tc HmPAO SPECT data. J Neurol Neurosurg Psychiatry. 2000;69:661-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 51. | Hammermeister KE, DeRouen TA, Dodge HT. Variables predictive of survival in patients with coronary disease. Selection by univariate and multivariate analyses from the clinical, electrocardiographic, exercise, arteriographic, and quantitative angiographic evaluations. Circulation. 1979;59:421-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 483] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 52. | Greco L, Troncone R, De Vizia B, Poggi V, Mayer M, Grimaldi M. Discriminant analysis for the diagnosis of childhood celiac disease. J Pediatr Gastroenterol Nutr. 1987;6:538-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 53. | Mayer M, Greco L, Troncone R, Grimaldi M, Pansa G. Early prediction of relapse during gluten challenge in childhood celiac disease. J Pediatr Gastroenterol Nutr. 1989;8:474-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 54. | Catassi C, Rossini M, Rätsch IM, Bearzi I, Santinelli A, Castagnani R, Pisani E, Coppa GV, Giorgi PL. Dose dependent effects of protracted ingestion of small amounts of gliadin in coeliac disease children: a clinical and jejunal morphometric study. Gut. 1993;34:1515-1519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 55. | Troncone R, Franzese A, Mazzarella G, Paparo F, Auricchio R, Coto I, Mayer M, Greco L. Gluten sensitivity in a subset of children with insulin dependent diabetes mellitus. Am J Gastroenterol. 2003;98:590-595. [PubMed] |
| 56. | Zhou T, Han G, Li BN, Lin Z, Ciaccio EJ, Green PH, Qin J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med. 2017;85:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 57. | Charlesworth RPG, Andronicos NM, Scott DR, McFarlane JR, Agnew LL. Can the sensitivity of the histopathological diagnosis of coeliac disease be increased and can treatment progression be monitored using mathematical modelling of histological sections? - A pilot study. Adv Med Sci. 2017;62:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 58. | Charlesworth RPG, Agnew LL, Scott DR, Andronicos NM. Celiac disease gene expression data can be used to classify biopsies along the Marsh score severity scale. J Gastroenterol Hepatol. 2019;34:169-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 59. | Charlesworth RP, Marsh MN. From 2-dimensional to 3-dimensional: Overcoming dilemmas in intestinal mucosal interpretation. World J Gastroenterol. 2019;25:2402-2415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 60. | Ensari A, Marsh MN, Morgan S, Lobley R, Unsworth DJ, Kounali D, Crowe PT, Paisley J, Moriarty KJ, Lowry J. Diagnosing coeliac disease by rectal gluten challenge: a prospective study based on immunopathology, computerized image analysis and logistic regression analysis. Clin Sci (Lond). 2001;101:199-207. [PubMed] |
| 61. | Loft DE, Marsh MN, Crowe PT. Rectal gluten challenge and diagnosis of coeliac disease. Lancet. 1990;335:1293-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 62. | Green PH, Rostami K, Marsh MN. Diagnosis of coeliac disease. Best Pract Res Clin Gastroenterol. 2005;19:389-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 101] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 63. | Dezi R, Niveloni S, Sugai E, Pedreira S, Smecuol E, Vazquez H, Doldan I, Cabanne A, Boerr L, Valero J, Kogan Z, Mauriño E, Bai JC. Gluten sensitivity in the rectal mucosa of first-degree relatives of celiac disease patients. Am J Gastroenterol. 1997;92:1326-1330. [PubMed] |
| 64. | Ontiveros N, Tye-Din JA, Hardy MY, Anderson RP. Ex-vivo whole blood secretion of interferon (IFN)-γ and IFN-γ-inducible protein-10 measured by enzyme-linked immunosorbent assay are as sensitive as IFN-γ enzyme-linked immunospot for the detection of gluten-reactive T cells in human leucocyte antigen (HLA)-DQ2·5(+) -associated coeliac disease. Clin Exp Immunol. 2014;175:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 65. | Sarna VK, Lundin KEA, Mørkrid L, Qiao SW, Sollid LM, Christophersen A. HLA-DQ-Gluten Tetramer Blood Test Accurately Identifies Patients With and Without Celiac Disease in Absence of Gluten Consumption. Gastroenterology. 2018;154:886-896.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 66. | Marsh MN, Rostami K. What Is A Normal Intestinal Mucosa? Gastroenterology. 2016;151:784-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 67. | Fernandez-Feo M, Wei G, Blumenkranz G, Dewhirst FE, Schuppan D, Oppenheim FG, Helmerhorst EJ. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin Microbiol Infect. 2013;19:E386-E394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 68. | de Meij TG, Budding AE, Grasman ME, Kneepkens CM, Savelkoul PH, Mearin ML. Composition and diversity of the duodenal mucosa-associated microbiome in children with untreated coeliac disease. Scand J Gastroenterol. 2013;48:530-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 69. | Rostami Nejad M, Ishaq S, Al Dulaimi D, Zali MR, Rostami K. The role of infectious mediators and gut microbiome in the pathogenesis of celiac disease. Arch Iran Med. 2015;18:244-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 70. | Sanz Y. Microbiome and Gluten. Ann Nutr Metab. 2015;67 Suppl 2:28-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 71. | Verdu EF, Galipeau HJ, Jabri B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2015;12:497-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 169] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 72. | Caminero A, Herrán AR, Nistal E, Pérez-Andrés J, Vaquero L, Vivas S, Ruiz de Morales JM, Albillos SM, Casqueiro J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol. 2014;88:309-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 73. | Bonder MJ, Tigchelaar EF, Cai X, Trynka G, Cenit MC, Hrdlickova B, Zhong H, Vatanen T, Gevers D, Wijmenga C, Wang Y, Zhernakova A. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016;8:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 179] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 74. | Calabrò A, Gralka E, Luchinat C, Saccenti E, Tenori L. A metabolomic perspective on coeliac disease. Autoimmune Dis. 2014;2014:756138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 75. | Ryan D, Newnham ED, Prenzler PD, Gibson PR. Metabolomics as a tool for diagnosis and monitoring in coeliac disease. Metabolomics. 2015;11:980-990. [DOI] [Full Text] |
| 76. | Singh A, Pramanik A, Acharya P, Makharia GK. Non-Invasive Biomarkers for Celiac Disease. J Clin Med. 2019;8:pii: E885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 77. | Upadhyay D, Sharma U, Makharia GK, Jagannathan NR. Role of NMR metabonomics in Celiac Disease (CeD). Biomed Spectrosco Imag. 2016;5:27-40. [DOI] [Full Text] |
| 78. | Auricchio R, Galatola M, Cielo D, Amoresano A, Caterino M, De Vita E, Illiano A, Troncone R, Greco L, Ruoppolo M. A Phospholipid Profile at 4 Months Predicts the Onset of Celiac Disease in at-Risk Infants. Sci Rep. 2019;9:14303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |