Published online Feb 14, 2019. doi: 10.3748/wjg.v25.i6.719
Peer-review started: November 5, 2018
First decision: December 28, 2018
Revised: January 11, 2019
Accepted: January 18, 2019
Article in press: January 18, 2019
Published online: February 14, 2019
Processing time: 101 Days and 21.6 Hours
Characteristics of alterations of serum hepatitis B virus (HBV) RNA in different chronic hepatitis B (CHB) patients still cannot be fully explained. Whether HBV RNA can predict HBeAg seroconversion is still controversial.
To investigate whether HBV RNA can predict virological response or HBeAg seroconversion during entecavir (ETV) treatment when HBV DNA is undetectable.
The present study evaluated 61 individuals who were diagnosed and treated with long-term ETV monotherapy at the Department of Infectious Diseases of Peking University First Hospital (China) from September 2006 to December 2007. Finally, 30 treatment-naive individuals were included. Serum HBV RNA were extracted from 140 μL serum samples at two time points. Then they were reverse transcribed to cDNA with the HBV-specific primer. The product was quantified by real-time quantitative PCR (RT-PCR) using TAMARA probes. Statistical analyses were performed with IBM SPSS 20.0.
Level of serum HBV RNA at baseline was 4.15 ± 0.90 log10 copies/mL. HBV RNA levels showed no significant difference between the virological response (VR) and partial VR (PVR) groups at baseline (P = 0.940). Serum HBV RNA significantly decreased among patients who achieved a VR during ETV therapy (P < 0.001). The levels of HBV RNA in both HBeAg-positive patients with seroconversion group and those with no seroconversion increased after 24 wk of treatment. Overall, HBV RNA significantly but mildly correlated to HBsAg (r = 0.265, P = 0.041), and HBV RNA was not correlated to HBV DNA (r = 0.242, P = 0.062). Furthermore, serum HBV RNA was an independent indicator for predicting HBeAg seroconversion and virological response. HBeAg seroconversion was more likely in CHB patients with HBV RNA levels below 4.12 log10 copies/mL before treatment.
The level of serum HBV RNA could predict HBeAg seroconversion and PVR during treatment. In the PVR group, the level of serum HBV RNA tends to be increasing.
Core tip: HBeAg seroconversion was more likely to be achieved for chronic hepatitis B patients with hepatitis B virus (HBV) RNA levels below 4.12 log10 copies/mL before treatment. In the partial virological response group, serum HBV RNA showed an increasing trend.
- Citation: Luo H, Zhang XX, Cao LH, Tan N, Kang Q, Xi HL, Yu M, Xu XY. Serum hepatitis B virus RNA is a predictor of HBeAg seroconversion and virological response with entecavir treatment in chronic hepatitis B patients. World J Gastroenterol 2019; 25(6): 719-728
- URL: https://www.wjgnet.com/1007-9327/full/v25/i6/719.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i6.719
Globally, more than 240 million individuals are infected with hepatitis B virus (HBV)[1]. Nucleos(t)ide analogues (NAs) are effective therapies that are widely used to target reverse transcriptases[2]. However, NAs only suppress HBV DNA replication, resulting in decreased serum HBV DNA, but they do not suppress covalently closed circular DNA (cccDNA), which still resides in the nucleus and leads to long-term infection[3]. Pregenome RNA (pgRNA) is the direct product of cccDNA, and HBV DNA polymerase reverse transcribes pgRNA into rcDNA. Therefore, blocking reverse transcription does not influence the generation of HBV pgRNA. In most studies, HBV pgRNA increased after blocking reverse transcription activity[3,4]. Lu’s studies showed that serum HBV RNA was pgRNA that appeared in virus-like particles.
In 1996, Kock et al[5] first demonstrated that HBV RNA was present in free virus, and a number of studies have shown that the HBV RNA levels in serum were related to virological response (VR) and prognosis[3,4,6-8]. However, characteristics of alterations of serum HBV RNA in different chronic hepatitis B (CHB) patients still cannot be fully explained. Whether HBV RNA can predict HBeAg seroconversion is still controversial.
In this work, we investigated the characteristics of HBV RNA alterations in CHB patients with different treatment effects. The relationships of HBV RNA with other serological markers were also analyzed. Finally, we calculated the predictive value of HBV RNA in anticipating HBeAg seroconversion.
We evaluated 61 individuals from September 2006 to December 2007 at the Department of Infectious Diseases of Peking University First Hospital (China) who had begun long-term entecavir (ETV) monotherapy (Table 1). The criteria for inclusion were: (1) patients who were naive to NA treatment; (2) patients who received monotherapy with ETV, without termination; and (3) patient’s written consent was obtained. The criteria for exclusion were: (1) patients who had severe liver-related complications (including decompensated liver cirrhosis, hepatocellular carcinoma, and liver transplantation); (2) patients with co-infection with human immunodeficiency virus (HIV) or hepatitis C virus (HCV); (3) patients receiving combination therapy with other NAs; (4) an absence of samples at the time points; and (5) patients with poor compliance. Finally, we collected 30 treatment-naive individuals.
| Characteristic | Value |
| Gender (male/female) | 25/5 |
| Age (yr) | 35.47 ± 11.05 (17-58) |
| Body mass index | 24.40 ± 3.50 (18.64-32.27) |
| HBeAg+ (%) | 25 (83%) |
| Alanine aminotransferase (IU/mL) | 159.90 ± 226.70 (40-1308) |
| HBsAg (log10 IU/mL) | 3.97 ± 0.74 (2.29-4.88) |
| HBV DNA (log10 IU/mL) | 7.99 ± 1.32 (4.28-9.52) |
| HBV RNA (log10 copies/mL) | 4.15 ± 0.90 (2.24-5.66) |
| HBV genotype | |
| B (%) | 6 (20.00%) |
| C (%) | 24 (80.00%) |
Patient cohort A included 13 patients who had a VR. Patient cohort B included 17 patients who had a partial VR (PVR). Through 48 weeks of therapy, HBV DNA that was undetected was identified as a VR. After 48 wk of treatment, positive HBV DNA (with a decrease of > 2 log10 copy/mL) was defined as a PVR. At baseline, there were 25 HBeAg positive patients, 10 of whom achieved HBeAg seroconversion during antiviral treatment.
Informed consent was obtained from all enrolled patients and the study protocol conformed to the ethical guidelines of the Declaration of Helsinki and was approved by the Ethic Committee of Shanghai Jing An Central Hospital (Approval No. 090f51e6809a26e1 v1.0).
An automatic biochemical analyzer was used to test the biochemical indicators, including alanine aminotransferase (ALT) and aspartate transaminase (AST). Quantitative HBsAg, HBeAg, and anti-HBe were determined with ELISA kits (Abbott Laboratories, Chicago, IL, United States). COBAS TaqMan assay (Roche Diagnostics, Basel, Switzerland) was used to quantify serum HBV DNA levels, with 100 IU/mL as the lower limit of detection. Identification of HBV genotypes was performed by comparison with the generated preS/S gene sequences in GenBank (NCBI) data[9].
HBV RNA was extracted from 140 μL of serum samples based on the manufacturer’s instructions (QIAamp Viral Mini Kit, Qiagen, Germany). Then, we used a RevertAid First Strand cDNA synthesis Kit (Thermo scientific, MA, United States) to reverse transcribe HBV RNA to cDNA with the HBV-specific primer (5'-ACCACGCTATCGCTACTC-3'). The evaluation of HBV RNA was accomplished by quantitative PCR (qPCR) using the ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, United States) by a Taqman probe method, according to the guidelines provided by the manufacturer. The target section was ligated with the constructed PMD19-T vector. The product was evaluated by real-time quantitative PCR (RT-PCR) using TAMARA probes. The primers and probe utilized to ascertain HBV RNA were as follows: Fw: 5'-ACCACGCTATCGCTACTCAC-3', Rw: 5'-CAACTTTTTCACCTCTGCCTA-3' and probe: (TAMARA) CATGTCCTACTGTTCAA GCCTCCAAG (BHQ2). The qPCR reaction mixture (25 μL) contained 2.5 μL 10 × Buffer, 2 μL dNTPs (2.5 mmol/L), 1 μL cDNA, 1 μL primer each, 0.5 μL probe, 0.25 μL Taq DNA polymerase, and 17.75 μL double-distilled water (ddH2O). The reaction mixture was denatured at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.
Statistical analyses were performed using IBM SPSS statistics, version 18.0.0.1 (SPSS, Chicago, IL, United States). Data are expressed as the mean ± SD or count number. Differences between groups were examined for statistical significance utilizing Student’s t-test. We used the chi-square test of independence to evaluate the distribution of HBeAg positivity in various groups. P < 0.05 was considered statistically significant. The linear relationships between various viral markers were evaluated by Pearson’s correlation coefficient (r) and tested for the significance of the correlation coefficient (P-value reported). Categorical variables were evaluated using Spearman’s correlation coefficient (r). We applied a multivariate regularized linear model to identify the factors closely related to HBV RNA. We used a receiver operating characteristic (ROC) curve to calculate the predictive value.
We studied 30 patients in total, who were divided into two groups depending on whether HBV DNA was undetectable at week 24 of treatment: a VR group (n = 13) and a PVR group (n = 17). The baseline characteristics of the included patients are listed in Table 2. The ratio of males to females was 5:1. The average age was 35 years. Most patients had a body mass index (BMI) of 24.4, within the normal range. The average value of glutamic-pyruvic transaminase was 159.9, two-fold higher than the normal value. There were 25 HBeAg-positive patients (83%). The average value of serum HBV RNA was 4.15 log10 copies/mL. Comparing the baseline characteristics of the VR and PVR groups statistically, the HBV DNA showed a significant difference (P = 0.014), as did HBsAg (P = 0.04). HBV RNA levels showed no significant difference between the VR and PVR groups (P = 0.940).
| Characteristic | Virological response (n = 13) | Partial virological response (n = 17) | P value |
| Gender (male/female) | 11/2 | 14/3 | |
| Age (yr) | 35.69 ± 10.85 | 35.29 ± 11.52 | 0.924 |
| Body mass index | 24.11 ± 3.66 | 24.62 ± 3.46 | 0.704 |
| HBeAg+ (%) | |||
| Week 0 | 9 (69.23%) | 16 (94.12%) | |
| Week 24 | 8 (61.54%) | 16 (94.12%) | |
| Alanine aminotransferase (IU/mL) | |||
| Week 0 | 213.85 ± 72.11 | 118.65 ± 74.25 | 0.262 |
| Week 24 | 30.23 ± 15.76 | 56.71 ± 46.12 | 0.058 |
| HBsAg (log10 IU/mL) | |||
| Week 0 | 3.66 ± 0.66 | 4.21 ± 0.73 | 0.040 |
| Week 24 | 3.49 ± 0.55 | 3.84 ± 0.61 | 0.116 |
| HBV DNA (log10 IU/mL) | |||
| Week 0 | 7.26 ± 1.56 | 8.54 ± 0.75 | 0.014 |
| Week 24 | 2.79 ± 0.27 | 4.20 ± 0.74 | 0.000 |
| HBV RNA (log10 copies/mL) | |||
| Week 0 | 4.17 ± 0.87 | 4.14 ± 0.95 | 0.936 |
| Week 24 | 2.58 ± 0.90 | 5.24 ± 0.51 | 0.000 |
| HBV genotype | |||
| B (%) | 3 (23.08%) | 3 (17.65%) | |
| C (%) | 10 (76.92%) | 14 (82.35%) |
Before NAs treatment, the HBV RNA level had a medium correlation with respect to whether the patient had a VR (r = -0.515, P = 0.004), and the HBV RNA level had no correlation with other clinical characteristics. After 24 wk of treatment, the HBV RNA level had a high correlation with respect to whether the patient had a VR (r = -0.843, P < 0.001), and HBV RNA also had a high correlation with the HBV DNA level (r = 0.758, P < 0.001). The AST and HBsAg levels had medium correlations with the HBV RNA level (r = 0.379, P = 0.039; r = 0.368, P = 0.045).
After 24 wk of treatment, we found a significant variation in HBV RNA between the VR and PVR groups (P = 0.041). In the VR group, the level of HBV RNA decreased, and the changes were significantly different (P < 0.001). In the PVR group, the level of HBV RNA showed an increasing trend, and the changes were significantly different (P < 0.001) (Figure 1A).
Thirty patients were also divided by whether they achieved HBeAg seroconversion. The baseline characteristics of the individuals are listed in Table 3. Comparing the HBeAg-positive patients with seroconversion (group A) with the HBeAg-positive patients with no seroconversion (group B), there were no significant differences in terms of clinical characteristics before treatment except for age: group A was younger than group B (37 ± 6 vs 33 ± 12, P = 0.034). After 24 weeks of treatment, the HBV RNA level showed a difference (3.1 ± 1.4 vs 5.5 ± 1.51, P = 0.032). Comparing group A with the HBeAg-negative group, only the HBV DNA level before treatment showed a significant difference (Figure 1B). The HBV RNA levels in groups A and B increased after 24 wk of treatment.
| Characteristic | (A) HBeAg positive patients with seroconversion (n = 10) | (B) HBeAg positive patients with no seroconversion (n = 15) | P value (A) vs (B) | (C) HBeAg negative patients (n = 5) | P value (A) vs (C) |
| Gender (male/female) | 8/2 | 12/3 | 5/0 | ||
| Age (yR) | 37 ± 6 | 33 ± 12 | 0.034 | 40 ± 150 | 0.505 |
| Body mass index | 25.29 ± 3.86 | 23.82 ± 3.25 | 0.528 | 24.38 ± 3.87 | 0.676 |
| Alanine aminotransferase (IU/mL) | |||||
| Week 0 | 156.10 ± 72.94 | 174.53 ± 317.58 | 0.272 | 123.60 ± 71.81 | 0.428 |
| Week 24 | 46.10 ± 32.61 | 50.00 ± 46.83 | 0.665 | 29.20 ± 8.17 | 0.282 |
| HBsAg (log10 IU/mL) | |||||
| Week 0 | 3.81 ± 0.84 | 4.14 ± 0.57 | 0.068 | 3.79 ± 1.02 | 0.967 |
| Week 24 | 3.51 ± 0.59 | 3.94 ± 0.59 | 0.658 | 3.29 ± 0.32 | 0.474 |
| HBV DNA (log10 IU/mL) | |||||
| Week 0 | 8.05 ± 0.99 | 8.57 ± 0.76 | 0.498 | 6.11 ± 1.64 | 0.013 |
| Week 24 | 3.32 ± 1.11 | 3.68 ± 1.15 | 0.635 | 3.03 ± 0.44 | 0.279 |
| HBV RNA (log10 copies/mL) | |||||
| Week 0 | 3.52 ± 0.84 | 5.59 ± 0.67 | 0.061 | 4.08 ± 0.99 | 0.27 |
| Week 24 | 3.06 ± 1.39 | 5.48 ± 1.51 | 0.032 | 2.93 ± 1.39 | 0.161 |
| HBV genotype | |||||
| B (%) | 3 (30.00%) | 2 (13.33%) | 1 (20%) | ||
| C (%) | 7 (70.00%) | 13 (86.67%) | 4 (80%) |
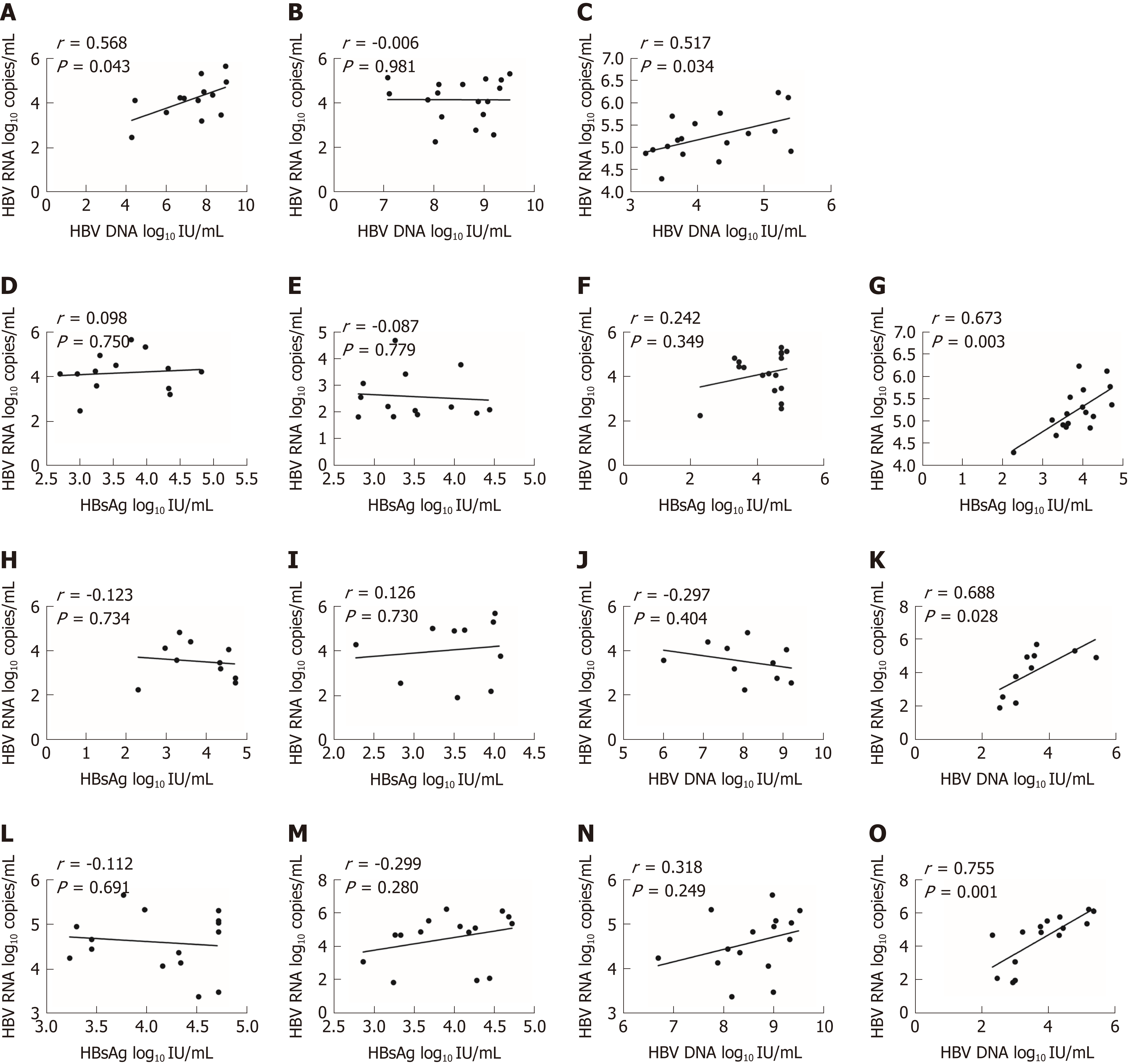
Overall, HBV RNA and HBsAg had a poor correlation (r = 0.265, P = 0.041) (Figure 1C), and HBV RNA had no significant correlation with HBV DNA (r = 0.242, P = 0.062) (Figure 1D).
In the VR group, we only found a moderate correlation between HBV RNA and HBV DNA before treatment (r = 0.568, P = 0.043) (Figure 2A). After 24 wk of treatment, HBV RNA had no correlation with other biomarkers (Figure 2B). In the PVR group, we found that HBV RNA had no correlation with HBV DNA before treatment. After 24 wk of treatment, HBV RNA and HBV DNA had a moderate correlation (r = 0.517, P = 0.034) (Figure 2C). HBV RNA and HBsAg had no correlation before treatment (Figure 2D and E). After 24 wk of treatment, HBV RNA had no correlation with HBsAg in the VR group (Figure 2F). After 24 wk of treatment, HBV RNA had a significant high correlation with HBsAg in the PVR group (r = 0.673, P = 0.003) (Figure 2G).
In group A, HBV RNA had only a moderate correlation with HBV DNA after 24 wk of therapy (r = 0.688, P = 0.028) (Figure 2K). In group B, HBV RNA also had a moderate correlation with HBV DNA after 24 wk of therapy (r = 0.755, P = 0.001) (Figure 2O). HBV RNA had no correlation with HBsAg before or after treatment (Figure 2H, I, L, and M). HBV RNA had no correlation with HBV DNA before treatment (Figure 2J and N).
As HBV patients might have various personal background and their risk factors for prognosis including seroconversion are unknown. It might cause the existence of confounding factors among study subjects which might interfere with the precision of the study. The typical method to adjust confounding factors is multivariate analysis. To evaluate whether HBV RNA was an independent predictor of HBeAg seroconversion and VR, we applied a multivariate regularized linear model. The model was suitable for the following explanatory variables: age, sex, BMI, levels of ALT, HBV DNA, HBsAg, whether there was a VR, and whether there was HBeAg seroconversion.
Before treatment, we identified two models: the first model, whether HBeAg seroconversion was identified as the best indicator associated with HBV RNA level, and the second model, whether HBeAg seroconversion and baseline HBV DNA were the strongest linear factors; however, the P-value for baseline HBV DNA was 0.092, greater than 0.05.
After 24 wk of treatment, we identified one best result model: whether VR was considered the strongest single linear factor related to HBV RNA.
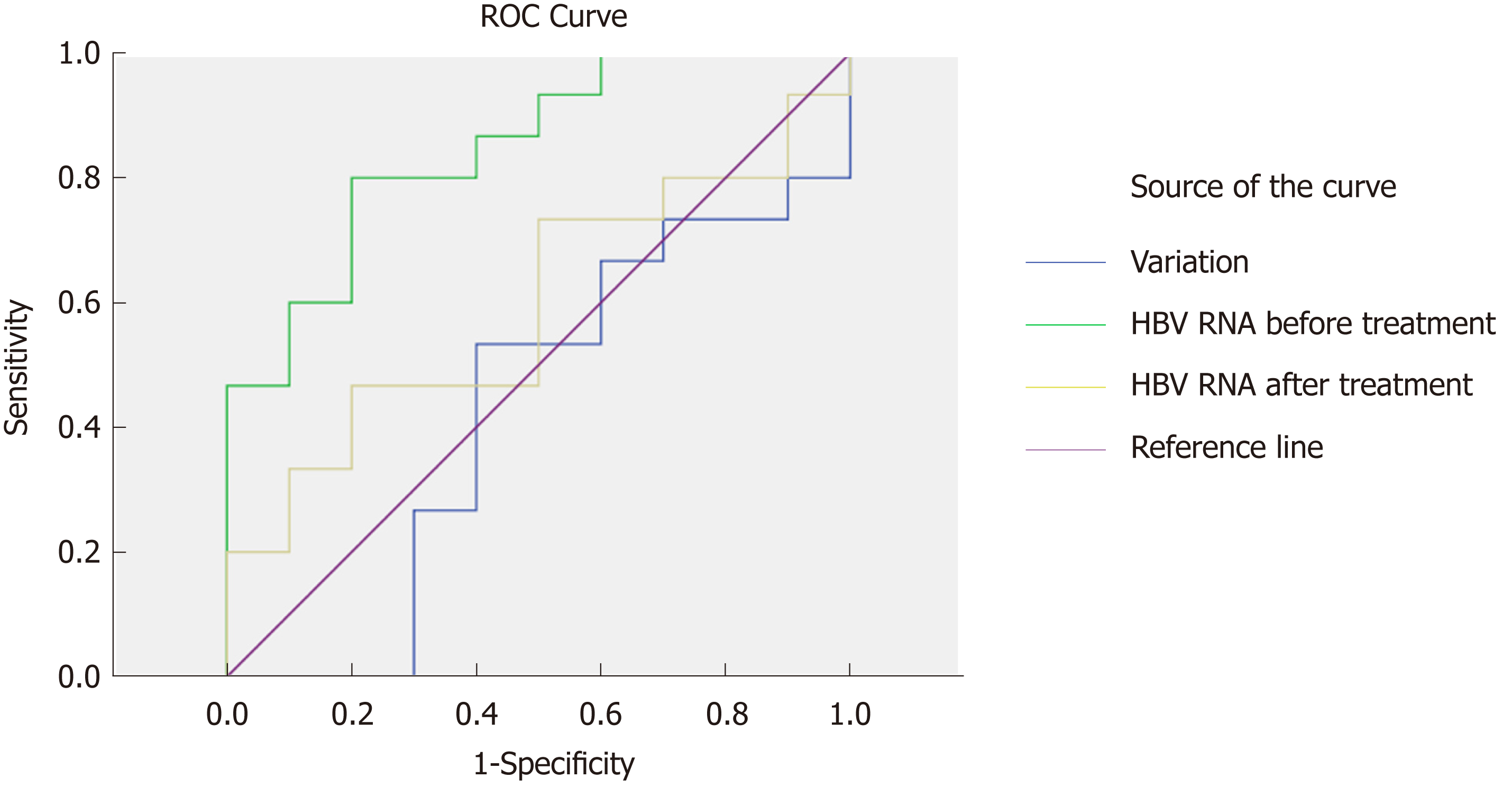
To analyze the predictive value of HBV RNA in HBeAg seroconversion, we generated a ROC curve. We evaluated HBV RNA at baseline and after 24 wk of therapy, measuring the variation in HBV RNA between the time points in terms of three characteristics. The HBV RNA level at baseline had the best predictive value (AUC = 0.847, P = 0.004). The cutoff value was 4.12 (sensitivity = 0.8, specificity = 0.8), suggesting that in CHB patients, an HBV RNA level below 4.12 log10 copies/mL before treatment was more likely to achieve HBeAg seroconversion. The variation and level of HBV RNA at 24 wk did not have predictive value (P = 0.542, P = 0.437) (Figure 3).
Studies have shown that NAs treatment can help more than 60% patients achieve undetectable HBV DNA after 1 year of therapy. However, several patients can achieve serological responses (HBeAg and HBsAg loss, with or without detection of corresponding antibodies)[10]. As a result, if HBV DNA is maintained at undetectable levels, it is difficult to predict whether a patient will have a serological response. To address this question, we evaluated whether HBV RNA had a relationship with VR and HBeAg antigen seroconversion, how HBV RNA was related to other indicators, whether HBV RNA was an independent indicator of HBeAg seroconversion, and finally, the diagnostic value of HBV RNA.
By comparing the dynamic characteristics of HBV RNA in the VR and PVR groups, we found that HBV RNA showed a strong decrease in the VR group. By contrast, HBV RNA increased in the PVR group (Figure 1A). Thus, effectively suppressed viral replication not only suppressed HBV DNA but also led to a decrease in HBV RNA, probably because NAs depleted the cccDNA that is the transcription template of HBV RNA. The serum HBV RNA level increased in the HBeAg no-seroconversion group compared with the HBeAg seroconversion group (Figure 1B). Wang’s study found that blocking HBV polymerase led to HBV pgRNA accumulation in vitro and in transgenic mice, but they also found that in 11 CHB patients, the HBV RNA levels declined after NA treatment[3]. Another study reported that for patients with YMDD mutations, serum HBV RNA was significantly higher than that in others[8]. A study in 2015 reported that during NA therapy, HBV RNA showed remarkable drops in patients undergoing HBeAg seroconversion compared to other patients[7]. The findings of these studies were consistent with our results. They all suggested that HBV RNA reflected the antiviral treatment effect and HBeAg seroconversion.
We also evaluated the relationship of HBV RNA to other biomarkers. Overall, HBsAg had a poor correlation with HBV RNA (r = 0.265, P = 0.041), and HBV DNA and HBV RNA did not show a correlation (r = 0.242, P = 0.062) (Figure 2A and B). Other studies showed that HBV RNA was remarkably related to both HBV DNA and HBsAg before treatment. Nevertheless, those molecules did not show correlations during therapy[7,11]. We hypothesize that HBV RNA may be an independent predictor that is less reflected by the polymerase inhibitors than by HBV DNA. HBsAg can be produced either from intrahepatic cccDNA or from integrated HBV DNA. However, HBV RNA was only produced from cccDNA. As a result, HBV RNA was an independent indicator.
The multivariate regularized linear model was analyzed with the following variables: age, sex, BMI, ALT, HBV DNA, HBsAg, whether there was a VR, and whether there was HBeAg seroconversion. Before treatment, whether there was HBeAg seroconversion was the best single linear indicator associated with HBV RNA level. After 24 wk of treatment, we found one best result model: whether VR was the best single linear indicator related with HBV RNA. The results demonstrated that HBV RNA was an independent molecule predicting HBeAg seroconversion and VR. Despite the fact that VR can be detected by HBV DNA level, we found that when patients had a PVR, the level of HBV RNA increased. Because both serum HBV DNA and HBV RNA can only be generated from cccDNA, the partial effect of blocking reverse transcription activity led to an increase in HBV RNA. A potential explanation may be that the detectable rcDNA translocated into the nucleus. There, it is converted into cccDNA. Because viral mRNA is directly transcribed from cccDNA, an increase in cccDNA will lead to an increase in HBV RNA.
Our results suggested that serum HBV RNA levels predicted HBeAg seroconversion during ETV therapy. The result of ROC curve analysis generated the claim that in patients with subsequent HBeAg seroconversion, serum HBV RNA levels were slightly lower before treatment. A previous study appeared to validate such view[7]. Because most patients (67%-80%) achieved HBeAg seroconversion during therapy[10], patients who had high levels of HBV RNA (over 4.12 log10 copies/mL) before treatment should be given more attention during treatment.
However, the present study had a limitation of a relative small number of enrolled patients. As a result, the conclusion of present study needs to be confirmed by using much larger sample size.
In conclusion, we found that serum HBV RNA predicted both VR and HBeAg seroconversion. The data appeared to suggest that serum HBV RNA decreased in patients who achieved a VR during ETV therapy, and vice versa. Overall, HBsAg and HBV RNA had a poor correlation (r = 0.265, P = 0.041), and HBV DNA had no correlation to HBV RNA (r = 0.242, P = 0.062). Furthermore, serum HBV RNA was an independent predictor of HBeAg seroconversion and VR. HBeAg seroconversion was more likely to be achieved for CHB patients with HBV RNA levels below 4.12 log10 copies/mL before treatment.
Nucleos(t)ide analogues (NAs) only suppress hepatitis B virus (HBV) DNA replication, resulting in decreased serum HBV DNA, but they do not suppress covalently closed circular DNA (cccDNA). Pregenome RNA (pgRNA) is the direct product of cccDNA. A number of studies have shown that HBV RNA levels in serum were related to virological response (VR) and prognosis. However, characteristics of alterations of serum HBV RNA in different chronic hepatitis B (CHB) patients still cannot be fully explained. Whether HBV RNA can predict HBeAg seroconversion is still controversial.
In this work, we investigated the characteristics of HBV RNA alterations in CHB patients with different treatment effects. The relationships of HBV RNA with other serological markers were also analyzed. Finally, we calculated the predictive value of HBV RNA in anticipating HBeAg seroconversion. Solving these problems helps to investigate the predictive value of HBV RNA.
If HBV DNA is maintained at undetectable levels, it is difficult to predict whether a patient will have a serological response. To address this question, we evaluated whether HBV RNA had a relationship with VR and HBeAg antigen seroconversion, how HBV RNA was related to other indicators, whether HBV RNA was an independent indicator of HBeAg seroconversion, and finally, the diagnostic value of HBV RNA.
The present study evaluated 61 CHB patients from September 2006 to December 2007 at the Department of Infectious Diseases of Peking University First Hospital (China) who had begun long-term entecavir (ETV) monotherapy. Finally, we collected 30 treatment-naive individuals. Serum HBV RNA was extracted from 140 μL serum samples at two time points. Then they were reverse transcribed to cDNA with the HBV-specific primer. The product was quantified by real-time quantitative PCR (RT-PCR) using TAMARA probes. Statistical analyses were performed with IBM SPSS 20.0.
By comparing the dynamic characteristics of HBV RNA in the VR and partial VR groups, we found that HBV RNA showed a strong decrease in the VR group. By contrast, HBV RNA increased in the partial VR group. The serum HBV RNA level increased in the HBeAg no-seroconversion group compared with the HBeAg seroconversion group. Overall, HBsAg had a poor correlation with HBV RNA (r = 0.265, P = 0.041), and HBV DNA and HBV RNA did not show a correlation (r = 0.242, P = 0.062). Furthermore, serum HBV RNA was an independent predictor of HBeAg seroconversion and VR. HBeAg seroconversion was more likely to be achieved for CHB patients with HBV RNA levels below 4.12 log10 copies/mL before treatment.
In conclusion, we found that serum HBV RNA predicted both VR and HBeAg seroconversion. The data appeared to suggest that serum HBV RNA decreased in patients who achieved a VR during ETV therapy, and vice versa. Overall, HBsAg and HBV RNA had a poor correlation (r = 0.265, P = 0.041), and HBV DNA had no correlation to HBV RNA (r = 0.242, P = 0.062). Furthermore, serum HBV RNA was an independent predictor of HBeAg seroconversion and VR. HBeAg seroconversion was more likely to be achieved for CHB patients with HBV RNA levels below 4.12 log10 copies/mL before treatment.
The present study suggested that serum HBV RNA decreased in patients who achieved a VR during ETV therapy, and vice versa. HBeAg seroconversion was more likely to be achieved for CHB patients with HBV RNA levels below 4.12 log10 copies/mL before treatment. In the future, the study could focus on the application value of HBV RNA in CHB patients with disease progression.
The authors would like to sincerely thank Professor Ding-Fang Bu (Peking University First Hospital) for his guidance with the statistics.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Dogan UB, Tabll AA, Taylor J S- Editor: Ma RY L- Editor: Wang TQ E- Editor: Yin SY
| 1. | Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1806] [Cited by in RCA: 1995] [Article Influence: 199.5] [Reference Citation Analysis (4)] |
| 2. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 692] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 3. | Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, Zhang R, Chen R, Li T, Zhang T, Yuan Q, Li PC, Huang Q, Colonno R, Jia J, Hou J, McCrae MA, Gao Z, Ren H, Xia N, Zhuang H, Lu F. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016;65:700-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 343] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 4. | Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. Hepatitis B Virus Pregenomic RNA Is Present in Virions in Plasma and Is Associated With a Response to Pegylated Interferon Alfa-2a and Nucleos(t)ide Analogues. J Infect Dis. 2016;213:224-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 5. | Köck J, Theilmann L, Galle P, Schlicht HJ. Hepatitis B virus nucleic acids associated with human peripheral blood mononuclear cells do not originate from replicating virus. Hepatology. 1996;23:405-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 33] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Tsuge M, Murakami E, Imamura M, Abe H, Miki D, Hiraga N, Takahashi S, Ochi H, Nelson Hayes C, Ginba H, Matsuyama K, Kawakami H, Chayama K. Serum HBV RNA and HBeAg are useful markers for the safe discontinuation of nucleotide analogue treatments in chronic hepatitis B patients. J Gastroenterol. 2013;48:1188-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 7. | van Bömmel F, Bartens A, Mysickova A, Hofmann J, Krüger DH, Berg T, Edelmann A. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 198] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 8. | Hatakeyama T, Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Kawakami Y, Fujimoto Y, Ochi H, Abe H, Maekawa T, Kawakami H, Yatsuji H, Aisaka Y, Kohno H, Aimitsu S, Chayama K. Serum HBV RNA is a predictor of early emergence of the YMDD mutant in patients treated with lamivudine. Hepatology. 2007;45:1179-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Ward H, Tang L, Poonia B, Kottilil S. Treatment of hepatitis B virus: an update. Future Microbiol. 2016;11:1581-1597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Tang LSY, Covert E, Wilson E, Kottilil S. Chronic Hepatitis B Infection: A Review. JAMA. 2018;319:1802-1813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 487] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 11. | Lai CL, Wong D, Ip P, Kopaniszen M, Seto WK, Fung J, Huang FY, Lee B, Cullaro G, Chong CK, Wu R, Cheng C, Yuen J, Ngai V, Yuen MF. Reduction of covalently closed circular DNA with long-term nucleos(t)ide analogue treatment in chronic hepatitis B. J Hepatol. 2017;66:275-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 135] [Article Influence: 16.9] [Reference Citation Analysis (0)] |