Published online Oct 21, 2019. doi: 10.3748/wjg.v25.i39.5973
Peer-review started: May 27, 2019
First decision: July 21, 2019
Revised: September 4, 2019
Accepted: September 13, 2019
Article in press: September 13, 2019
Published online: October 21, 2019
Processing time: 148 Days and 0.6 Hours
Long noncoding RNAs (lncRNAs) have been identified to play important roles in the development and progression of various tumors, including gastric cancer (GC). However, the molecular role of lncRNAs in GC progression remains unclear.
To investigate the differential expression of lncRNAs in human GC and elucidate the function and regulatory mechanism of LINC02407.
The Cancer Genome Atlas database was used to investigate the involvement of lncRNAs in GC. Quantitative real-time polymerase chain reaction was used to estimate the relative expression level of LINC02407 in GC tissues and cells. Functional experiments including CCK8 assay, apoptosis assay, wound healing assay, and transwell assay were used to investigate the effect of LINC02407 on GC cells. Some microRNAs were predicted and verified via bioinformatics analysis and the luciferase reporter system. Predictive analysis and Western blot assay were used to analyze the expression of related proteins.
Many differentially expressed lncRNAs were identified in GC, and some of them including LINC02407 can affect the survival. LINC02407 was upregulated in tumor tissues compared with adjacent tissues. HGC-27 cells showed the highest LINC02407 expression and HaCaT cells exhibited the lowest expression. Different experiment groups were constructed using LINC02407 overexpressing plasmids and related siRNAs. The results of functional experiments showed that LINC02407 can promote the proliferation, migration, and invasion of GC cells but inhibit apoptosis. Luciferase reporter assay showed that hsa-miR-6845-5p and hsa-miR-4455 was downstream regulated by LINC02407. Western blot analysis showed that adhesion G protein-coupled receptor D1 (ADGRD1) was regulated by the LINC02407-miR-6845-5p/miR-4455-ADGRD1 pathways.
LINC02407 plays a role in GC through the LINC02407-miR-6845-5p/miR-4455-ADGRD1 pathways, and thus, it may be an important oncogene and has potential value in GC diagnosis and treatment.
Core tip: Many long noncoding RNAs were differentially expressed in gastric cancer (GC), some of which, including LINC02407, had significant impacts on the prognosis of patients. In vitro experiments showed that LINC02407 was significantly upregulated in GC cell lines and tissue samples, and could promote the proliferation and migration and inhibit apoptosis of GC cells. Mechanistic research found that LINC02407 can regulate the expression of adhesion G protein-coupled receptor D1 by targeting miR-6845-5p and miR-4455 and increased the malignancy of GC cells eventually. Our study provides evidence that LINC02407 may be an important oncogene and has potential value in GC diagnosis and treatment.
- Citation: Zhou LL, Jiao Y, Chen HM, Kang LH, Yang Q, Li J, Guan M, Zhu G, Liu FQ, Wang S, Bai X, Song YQ. Differentially expressed long noncoding RNAs and regulatory mechanism of LINC02407 in human gastric adenocarcinoma. World J Gastroenterol 2019; 25(39): 5973-5990
- URL: https://www.wjgnet.com/1007-9327/full/v25/i39/5973.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i39.5973
Gastric cancer (GC) is one of the most common malignancies and is associated with high morbidity and mortality rates worldwide[1]. In spite of the huge progress that has been made in the field of surgical resection and chemotherapy, the 5-year overall survival (OS) rate for patients is incredibly low due to the distant metastasis of primary GC[2]. Recent studies have shown that long noncoding RNAs (lncRNAs) play key roles in the development of GC, metastasis, and disease prognosis[3]. The rapid development of high-throughput sequencing represents a huge breakthrough, and transcriptional studies of short noncoding RNAs and lncRNAs in the human genome are gradually being carried out[4]. Therefore, a deep investigation of the molecular pathophysiological pathways underlying GC could pave the road for the development of an effective therapeutic strategy.
In general, when the length of an lncRNA exceeds 200 nt, its protein coding ability is limited[5]. For example, the lncRNA SNHG7 promotes the proliferation of GC cells and inhibits apoptosis via repressing P15 and P16 expression[6]. The lncRNA DANCR positively promotes the migration and invasion of GC cells by suppressing lncRNA-LET[7]. Studies have shown that the lncRNA SNHG6 is closely related to a poor prognosis of GC, via the epigenetic silencing of p27 and by modulating cellulite miR-101-3p to promote cell proliferation[8]. Here, we performed preliminary research on the effect of LINC02407 in GC. LINC02407 has a transcript size of 966 bp and is located on chromosome 12: 76252275-76297735[9].
Based on existing research, this study applied a luciferase reporter system to verify whether LINC02407 could directly target hsa-miR-6845-5p and hsa-miR-4455, and investigated the function and molecular mechanism of lncRNA LINC02407 in the proliferation, apoptosis, migration, and invasion of GC cells.
Information of 343 tumor samples and 30 normal samples was obtained from the Cancer Genome Atlas (TCGA) database. For in vitro experiments, the samples in this study were obtained from patients who underwent pathological diagnosis and surgical treatment of gastric diseases at the First Hospital of Jilin University. In total, 20 tumor samples and 20 paired normal samples were obtained from dissected tumors and adjacent normal gastric mucosa tissues, respectively. All patients provided written informed consent. The study was approved by the Medical Ethics Committee of the First Hospital of Jilin University and was implemented in strict accordance with the Helsinki Declaration.
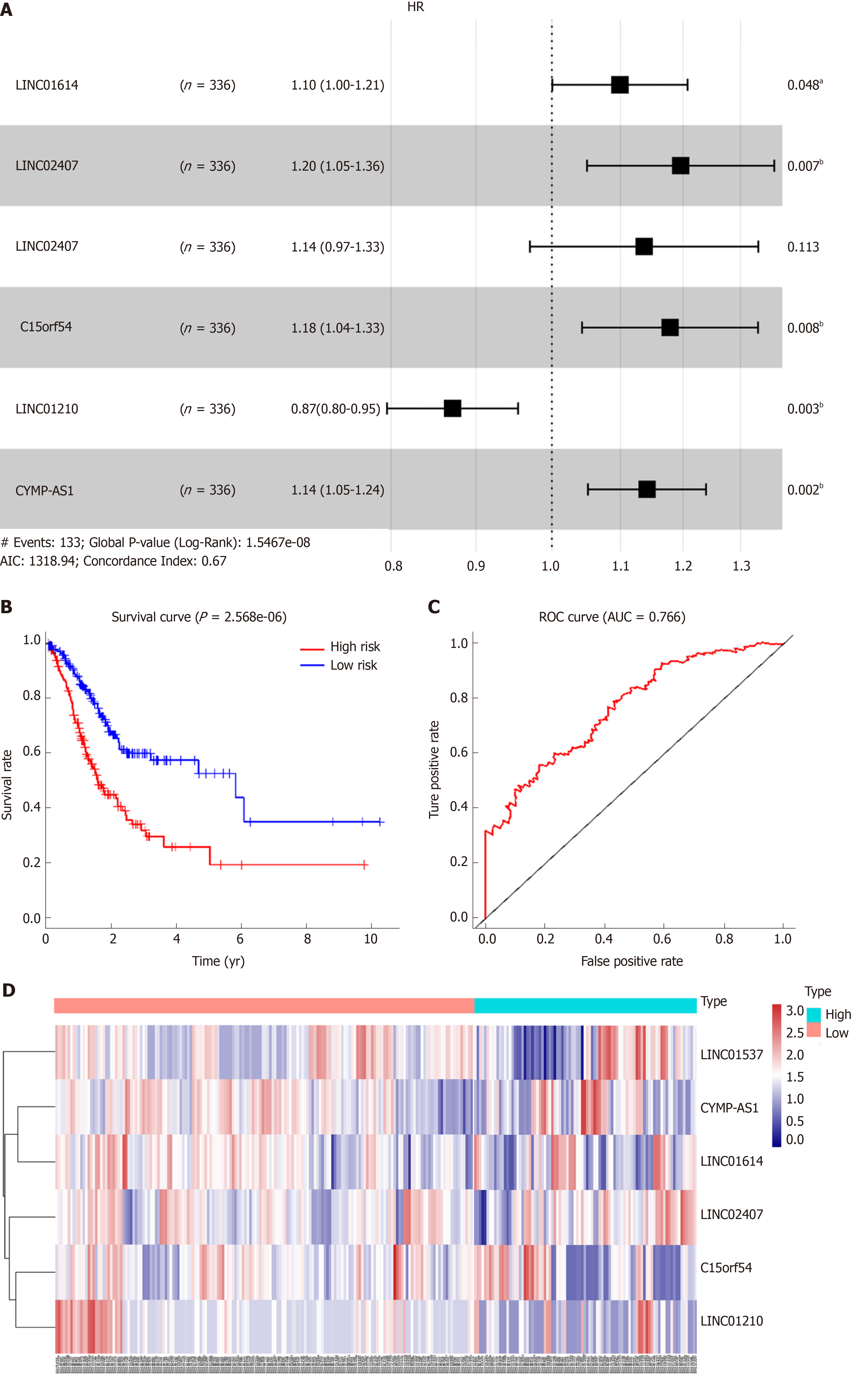
LncRNA expression levels and clinical information were obtained from the TCGA website, and differentially expressed lncRNAs between tumor tissue samples and adjacent nontumor tissue samples were analyzed with edegR, and the filtering condition was log fold change (FC) = 1 and padj = 0.05. Using the clinical information of differentially expressed lncRNAs in GC, univariate analysis of survival was performed to screen a series of potential lncRNAs which have impacts on survival. These lncRNAs were then sorted by hazard ratio (HR) and P value to pick out top 6 lncRNAs. Then we drew forest plots, Kaplan-Meier curves, receiver operating characteristic (ROC) curves, and heat maps for the top 6 lncRNAs based on the Cox proportional hazards model using R-3.5.2 software to pick the most significant lncRNA eventually.
Human GC cell lines, including MGC-803, MKN45, SGC-7901, and HGC-27, were purchased from the China Center for Type Culture Collection. The human normal gastric mucosal cell line GES-1 was purchased from the American Type Culture Collection. The culture medium for MGC-803, MKN45, SGC-7901, and HGC-27 cells was DMEM supplemented with 10% fetal bovine serum (FBS), while GES-1 cells were cultured in DMEM supplemented with 15% FBS. The cells were cultured in a 5% CO2 incubator at 37 °C. LINC02407-overexpressing and overexpression-control lentiviruses were purchased from Hanbio (Shanghai, China), and the LINC02407-overexpressing lentivirus was used to upregulate LINC02407 activity. Cells were infected with lentiviral particles in a specific medium for 48 h until they reached 50% confluence. To strengthen the infection efficiency, cells were cotreated with the cationic polymer polybrene. The vectors si-LINC02407 and si-NC are chemically modified small RNAs constructed by Biosyntech (Suzhou, China). Vector si-LINC02407 was applied to inhibit endogenous si-LINC02407 activity by silencing LINC02407. si-LINC02407 at 10 nmol/L was used for transfection with lipofectamine 3000, and the same amount of si-NC was used for transfection.
Total RNA was extracted with TRIzol reagent (Invitrogen, China), and oligo(dT)18 primers and superscript reverse transcriptase were used for reverse transcription with a miRNeasy Mini Kit (QIAGEN 217004). qRT-PCR experiments were performed using the QuantiNova SYBR Green RT-PCR Kit (QIAGEN 208152). The standardized internal reference was glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers in this study were from Takara Bio Co., Ltd. The sequences of the primers for qRT-PCR are shown in Supplemental Table 1.
CCK8 assays were performed to investigate the influence of LINC02407 overexpression and knockdown on HGC-27 proliferation. In brief, after culturing and transfecting HGC-27 cells as mentioned ahead, 3 × 104 cells were seeded per well in a 96-well plate. CCK8 reagent was added at 0, 12, 24, 36, 48, 60, 72, and 84 h after transfection, and cell viability was analyzed with a microplate reader (K3; Thermo Fisher Scientific, Inc). Each value represents the average of three replicates in a representative experiment from at least two independent experiments.
Hoechst staining assay was performed to analyze cell apoptosis. Briefly, HGC-27 cells were cultured in a 6-well plate and transfected according to the aforementioned method, and then cells were cultured on glass coverslips. Thereafter, the cells were fixed and stained with 10 μg/mL Hoechst 33258 (Beyotime Institute of Biotechnology) for 10 min in the dark. The samples were observed under an inverted fluorescence microscope, and the coverslips were washed with PBS. The nuclei of apoptotic cells have significant morphological changes, and such cells can be identified by their dense nuclei or densely stained fragments. HGC-27 cells were harvested 48 h after transfection and stained with annexin V-FITC and propidium iodide (PI), and then the relative amount of annexin V-FITC positive-PI negative cells was calculated by flow cytometry analysis. Furthermore, the apoptosis rate of si-LINC02407 and si-NC-transfected HGC-27 cells was evaluated.
In brief, HGC-27 cells were cultured in a 6-well plate and transfected according to the aforementioned method. The transfection ratio reached 90% at 24 h post-transfection, and then a straight line was scratched on the 6-well plate with a 200 μL pipette tip. The cell culture plate was washed three times with PBS, and the cells were cultured in DMEM (serum-free) for 24 h in 5% CO2 at 37 °C. The width of the scratch was recorded at several time points from 0-24 h under a light microscope (Olympus IM2), and the assay was run for three independent experiments.
MiRanda software was used to predict the target microRNAs (miRNAs) of LINC02407 and list top of them. Then, several miRNAs were selected after literature review for further research. The luciferase reporter system was purchased from Biosyntech (Suzhou, China). In brief, site-directed mutations were introduced to the LINC02407 binding site of hsa-miR-6845-5p, hsa-miR-4455, hsa-miR-4316, and hsa-miR-1258. PCR was used to subclone the 3’ untranslated region fragment of LINC02407 into the pGL3 luciferase vector (Invitrogen, United States). The hsa-miR-6845-5p-mimic, hsa-miR-4455-mimic, hsa-miR-4316-mimic, and hsa-miR-1258-mimic were transfected with the vector into 293T cells for 12 h in 96-well plates in the presence of 5 μg/mL polybrene (Biosyntech, Suzhou, China). The 293T cells were cultured for 24 h and lysed to determinate the luciferase activity. Renilla (Promega, United States) activity was employed as the internal control[1].
Migration assays were performed using a 24-well Boyden chamber (BD Falcon, Corning-Costar, New York, NY, United States) with an uncoated 8-mm pore size filter. The cells were collected from the culture dishes and washed twice with PBS. Then, the cells were suspended in smooth muscle cell (SMCM) medium (without FBS), and 3 × 104 cells were seeded into the insertion chamber. Subsequently, the cells were cultured in the bottom chamber (containing 0.6 mL of SMCM medium with 10% FBS) at 37 °C (5% CO2) for 24 h. Cells were stained with 4,6-diamidino-2-phenylindole and counted under a vertical microscope.
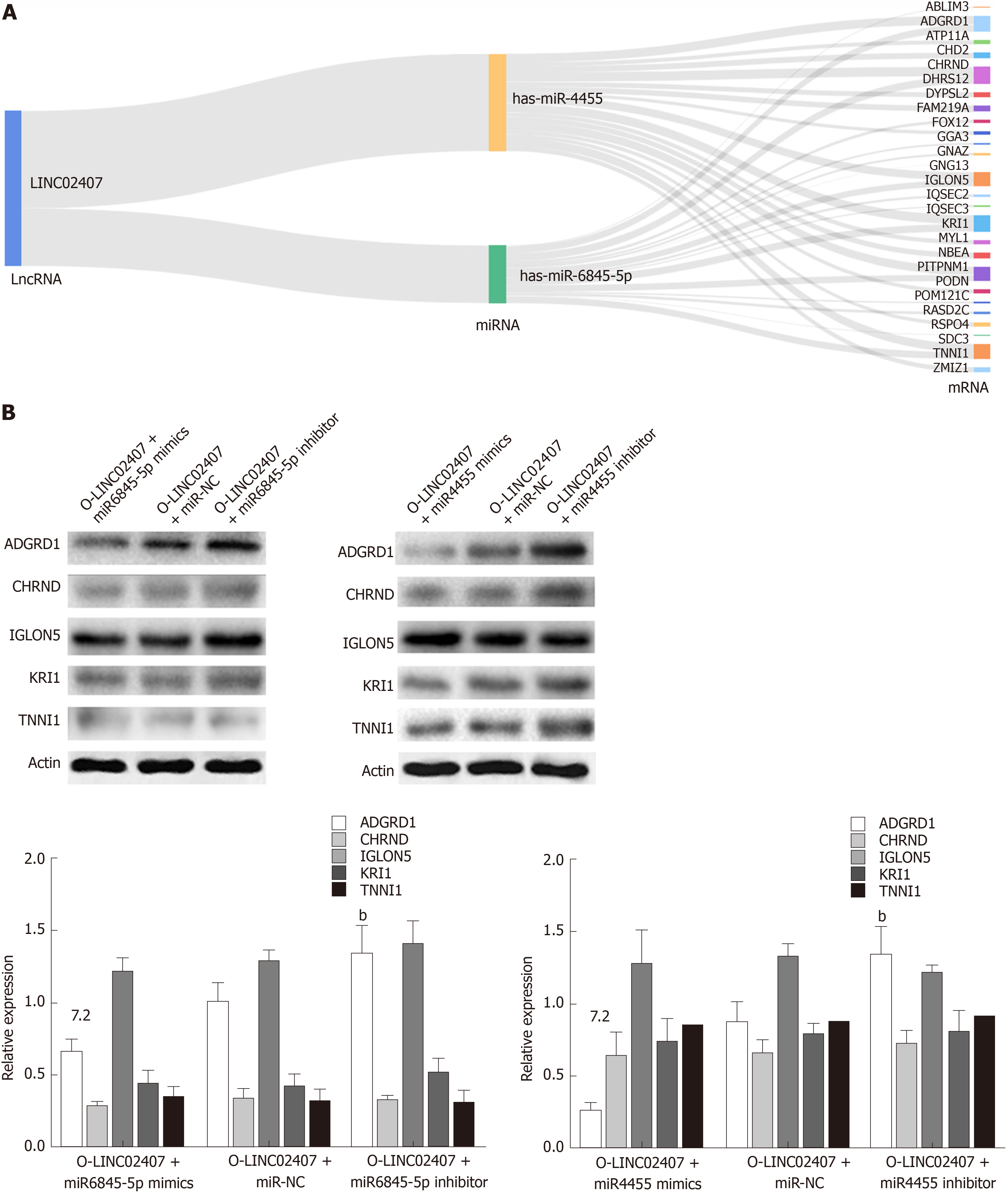
Sankey plot, also known as Sankey diagram, was used to depict the quantity of evidence between LINC02407, miR-6845-5p, miR-4455, and mRNAs. And the data that were used to draw Sankey plot were collected using miRanda software. MiRanda was applied to predict the potential mRNA targets of hsa-miR-6845-5p and hsa-miR4455, the target mRNAs were collected in a list, and then the R-River plot was used to draw a Sankey diagram.
Cells were lysed in an ice bath for 30 min in NP40 lysis buffer (50 mmol/L Tris-HCI pH 7.4, 150 mmol/L NaCl, 1% NP-40, 1 mmol/L EDTA) containing protease and phosphatase inhibitors (5 mmol/L PMSF, 3 mmol/L NaF, 1 mmol/L DTT, 1 mmol/L NaVO4). Proteins in 20 μg of protein cell lysate were separated by SDS-PAGE and electrophoretically transferred onto a polyvinylidene fluoride membrane. The membrane was then washed in PBS, blocked for 1 h (5% milk-TBS-0.05% Tween 20), and incubated with primary antibody overnight at 4 °C. The following primary antibodies were used: anti-GAPDH antibody (Abcam.ab181602, 1:1000), anti-ETV7 antibody (Abcam.ab229832, 1:1000), anti-FOXO3 antibody (Abcam.ab47285, 1:1000), anti-FOXO4 antibody (Abcam.ab128908, 1:1000), and anti-c-MAF antibody (Abcam.ab77071, 1:1000). The membrane was washed with PBS and incubated with an appropriate amount of secondary antibody for 1 h at room temperature (Li-Cor, IRDye 600LT; IRDye 800CW, 1:10000). The membrane was visually analyzed using an imaging system (ChemiDocXRS+, BIO-RAD). Uvitec Alliance software (Eppendorf, Hamburg, Germany) was used to quantify the data. All data were from three independent biological repeats.
The data were processed, and statistical analyses were performed using R Studio (R version 3.5.2) and GraphPad Prism 8.0 software (GraphPad Software Inc., La Jolla, CA, United States). EdgeR was used to identify the differential expression of RNAs. The role of related differential genes was explored by Cox regression analysis. P values < 0.05 were considered statistically significant.
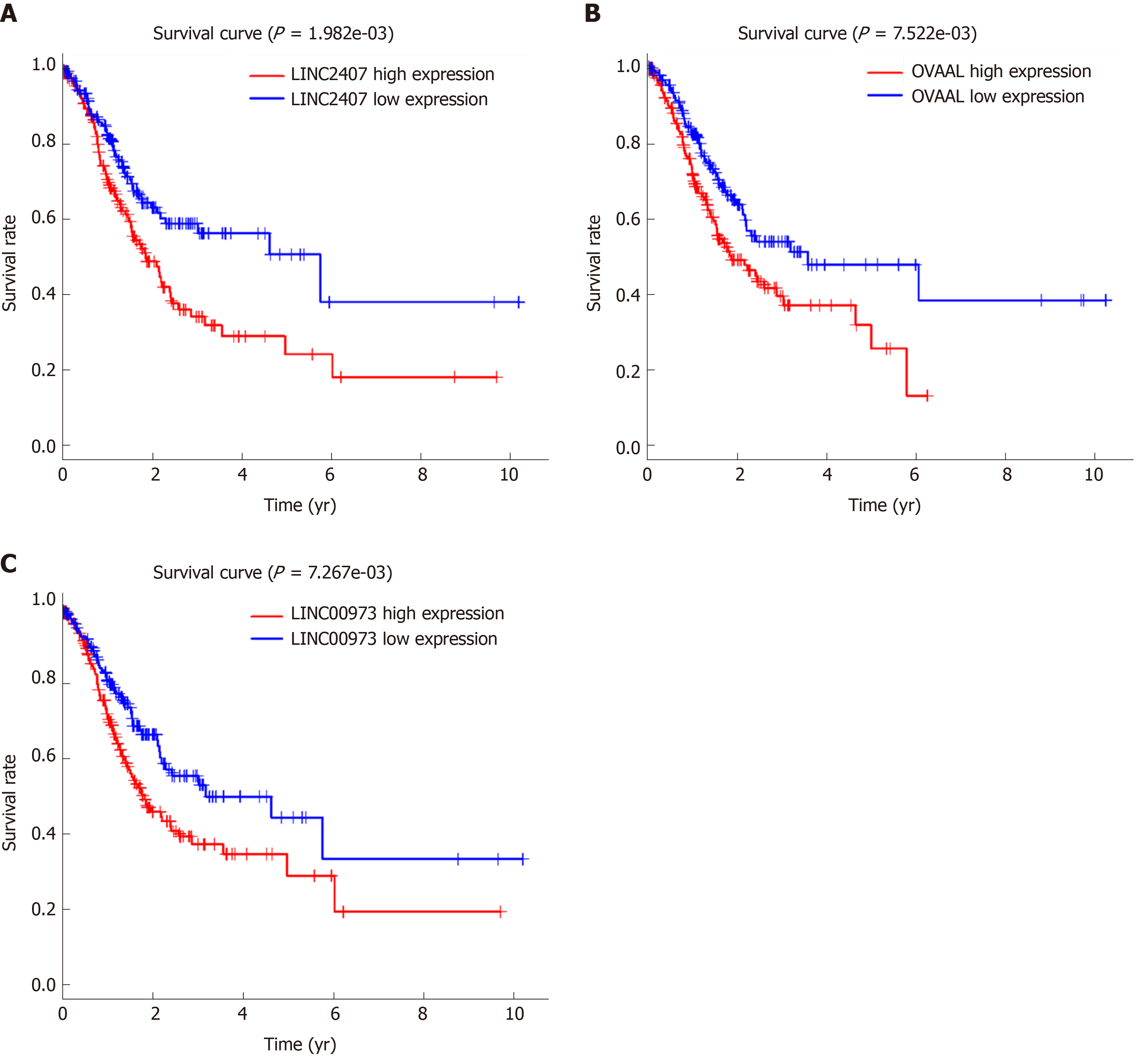
The volcano map (Figure 1A) indicates that there were differences in the expression of lncRNAs between tumor tissues (log FC = 1, padj = 0.05) and adjacent nontumor tissues from TCGA. Among all differentially expressed lncRNAs, there were 16 lncRNAs with P < 0.01. Subsequently, a significant difference was shown by plotting the cluster heat map (Figure 1B). For specific lncRNAs, we depicted the top 20 upregulated and top 20 downregulated lncRNAs involved in GC. Table 1 shows their log FC and -log false discovery rate (log FDR) data based on the results from integrated tumor tissues. The Kaplan-Meier survival curve of LINC02407 (Figure 2A) is shown, and the association of 16 key lncRNAs with OS was analyzed based on TCGA data to study the main features of prognostic outcomes. Ovarian adenocarcinoma amplified long non-coding RNA (Figure 2B) and LINC00973 (Figure 2C) were positively associated with OS (log-rank P < 0.01).
| LncRNA (up-regulated) | Log FC | -LogFDR | LncRNA (down-regulated) | Log FC | -LogFDR |
| MAGEA4-AS1 | 8.20 | 6.39 | HCG22 | -4.59 | 30.13 |
| LINC02582 | 7.29 | 7.04 | LINC00955 | -3.87 | 16.19 |
| LINC00392 | 6.98 | 5.23 | PGM5-AS1 | -3.75 | 25.23 |
| HOXC-AS3 | 6.91 | 17.67 | IL12A-AS1 | -3.31 | 38.15 |
| DSCR8 | 6.78 | 6.14 | C5orf66-AS1 | -3.19 | 22.45 |
| LINC02474 | 6.76 | 9.65 | PCAT18 | -3.15 | 17.08 |
| TMEM132D-AS1 | 6.67 | 6.35 | LINC02404 | -3.11 | 10.98 |
| LINC01234 | 6.55 | 14.68 | UBXN10-AS1 | -3.07 | 30.68 |
| LINC01980 | 6.49 | 7.77 | LINC02560 | -3.06 | 13.89 |
| BANCR | 6.21 | 7.50 | LRRC3-DT | -3.01 | 22.82 |
| ERVMER61-1 | 6.17 | 5.87 | LINC01697 | -2.99 | 24.83 |
| LINC02163 | 5.90 | 17.22 | HAND2-AS1 | -2.96 | 14.27 |
| DSCR4 | 5.75 | 5.33 | LINC00330 | -2.91 | 13.05 |
| LINC01050 | 5.75 | 16.19 | MIR205HG | -2.86 | 4.34 |
| LINC00973 | 5.72 | 4.97 | CERS3-AS1 | -2.82 | 15.20 |
| LINC01419 | 5.72 | 6.63 | LINC02487 | -2.81 | 16.19 |
| HOTAIR | 5.55 | 17.76 | LINC02106 | -2.70 | 16.69 |
| LINC01287 | 5.48 | 6.53 | LINC00671 | -2.69 | 21.70 |
| OVAAL | 5.44 | 4.37 | ADAMTS9-AS1 | -2.66 | 29.85 |
| LINC01194 | 5.36 | 5.12 | MIR1-1HG-AS1 | -2.64 | 12.84 |
Using univariate analysis of survival, the lncRNAs were ranked by HR and P value, and the list of lncRNA correlations is shown in Table 2. For the polygenic risk-score analysis and clinical information data from TCGA, R-3.5.2 software was applied to construct the forest plots based on the Cox proportional hazards model for LIN01614, LIN01537, LIN02407, C15orf54, LIN01210, and CYMP-AS1 lncRNA (Figure 3A), the survival curves (Figure 3B), and the ROC curves (Figure 3C). LIN01614, LIN01537, LIN02407, C15orf54, and CYMP-AS1 positively correlated with the survival of patients. We could infer that these lncRNAs could increase the survival of patients based on the ROC curves (Figure 3C). A HR heatmap representing the associations of LIN01614, LIN01537, LIN02407, C15orf54, and CYMP-AS1 expression with OS in patients with different clinicopathological characteristics (Figure 3D) was acquired by applying R-3.5.2 software.
| Gene | HR | Z value | P value |
| FLJ16779 | 1.16 | 3.48 | 4.93E-04 |
| LINC01537 | 1.25 | 3.46 | 5.34E-04 |
| LINC02407 | 1.28 | 3.41 | 6.49E-04 |
| LINC00565 | 1.21 | 3.18 | 1.46E-03 |
| ADAMTS9-AS1 | 1.18 | 2.99 | 2.81E-03 |
| LINC02408 | 1.20 | 2.98 | 2.90E-03 |
| LINC01614 | 1.13 | 2.93 | 3.35E-03 |
| RNF144A-AS1 | 1.21 | 2.93 | 3.40E-03 |
| MAGI2-AS3 | 1.23 | 2.87 | 4.09E-03 |
| CYMP-AS1 | 1.13 | 2.86 | 4.18E-03 |
| PTPRD-AS1 | 1.16 | 2.86 | 4.27E-03 |
| APCDD1L-DT | 1.18 | 2.84 | 4.56E-03 |
| C15orf54 | 1.18 | 2.83 | 4.68E-03 |
| LINC00973 | 1.12 | 2.72 | 6.49E-03 |
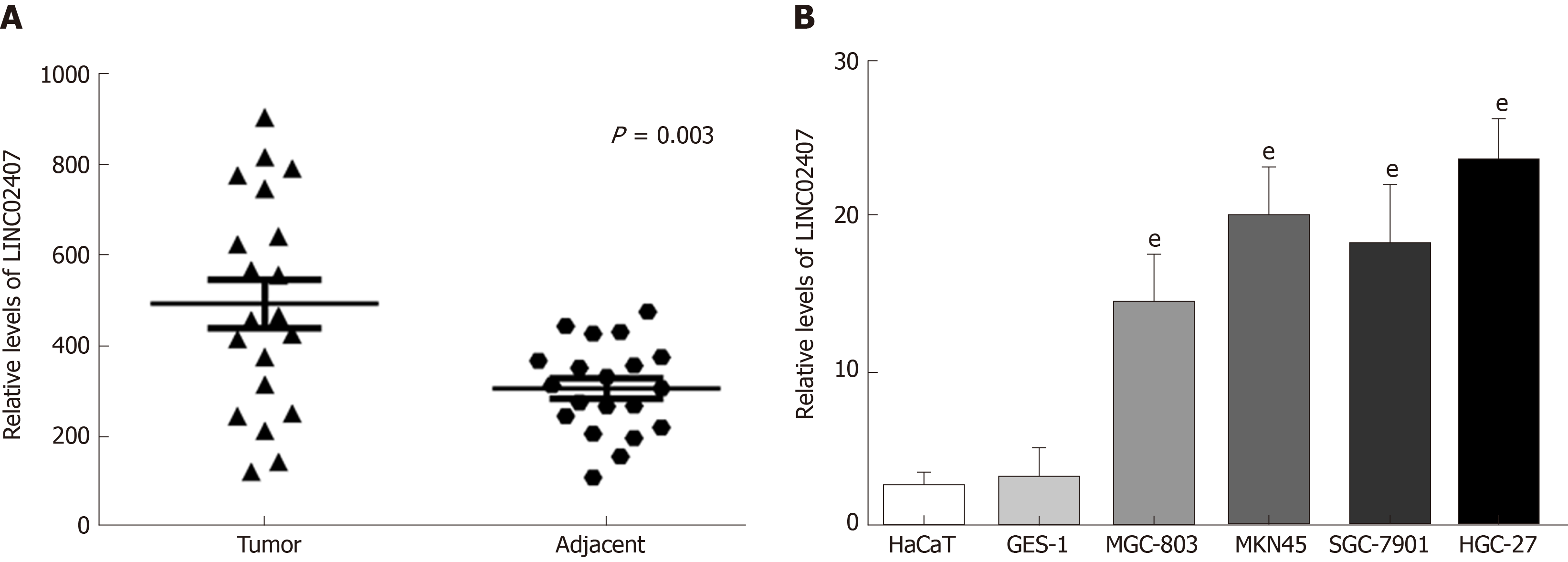
The expression levels of LINC02407 in 20 pairs of GC tissues and adjacent normal tissues were measured by qRT-PCR. The results showed that LINC02407 expression was upregulated in GC samples relative to adjacent normal mucosal tissues (Figure 4A). In addition, the expression levels of LINC02407 in a gastric mucosal normal cell line, GES-1, and five GC cell lines (HaCaT, MGC-803, MKN45, SGC-7901, and HGC-27) were analyzed. The results showed that LINC01354 expression was the highest in HGC-27 cells and the lowest in HaCaT cells (Figure 4B).
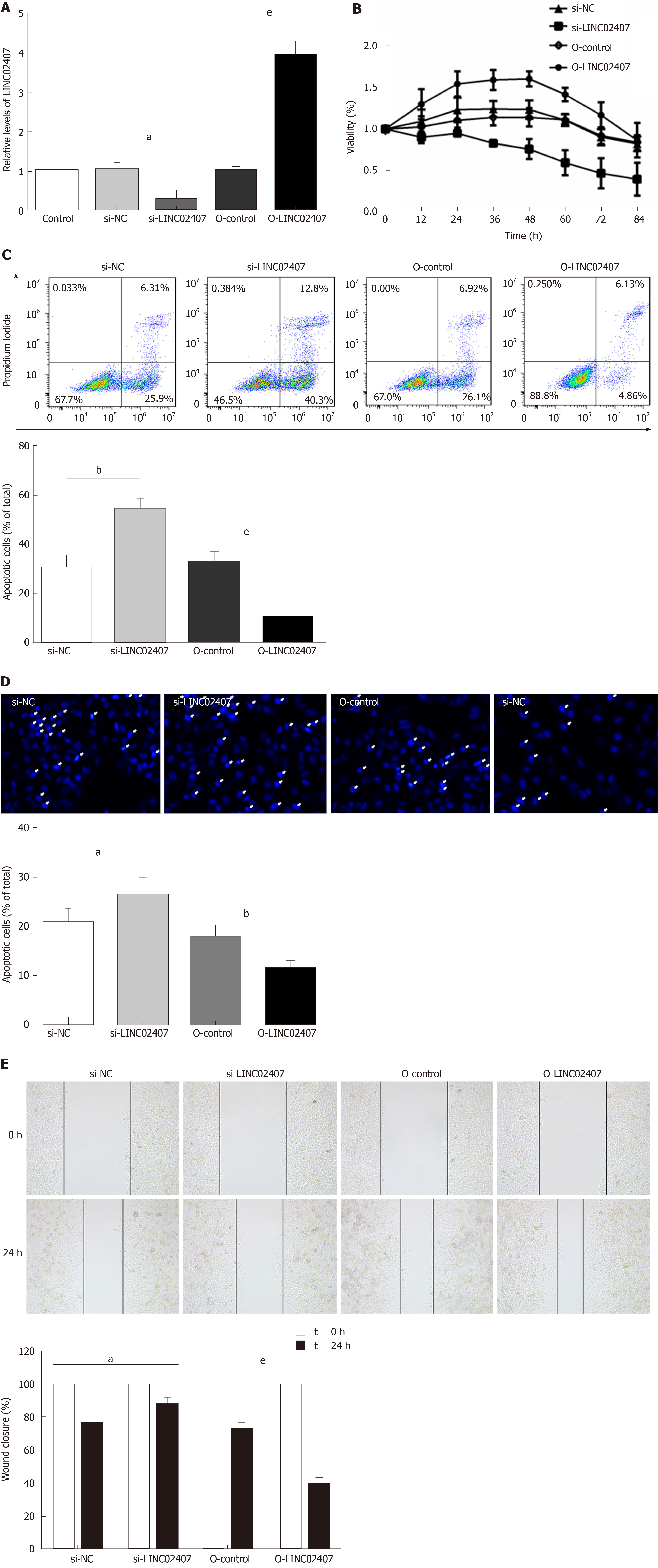
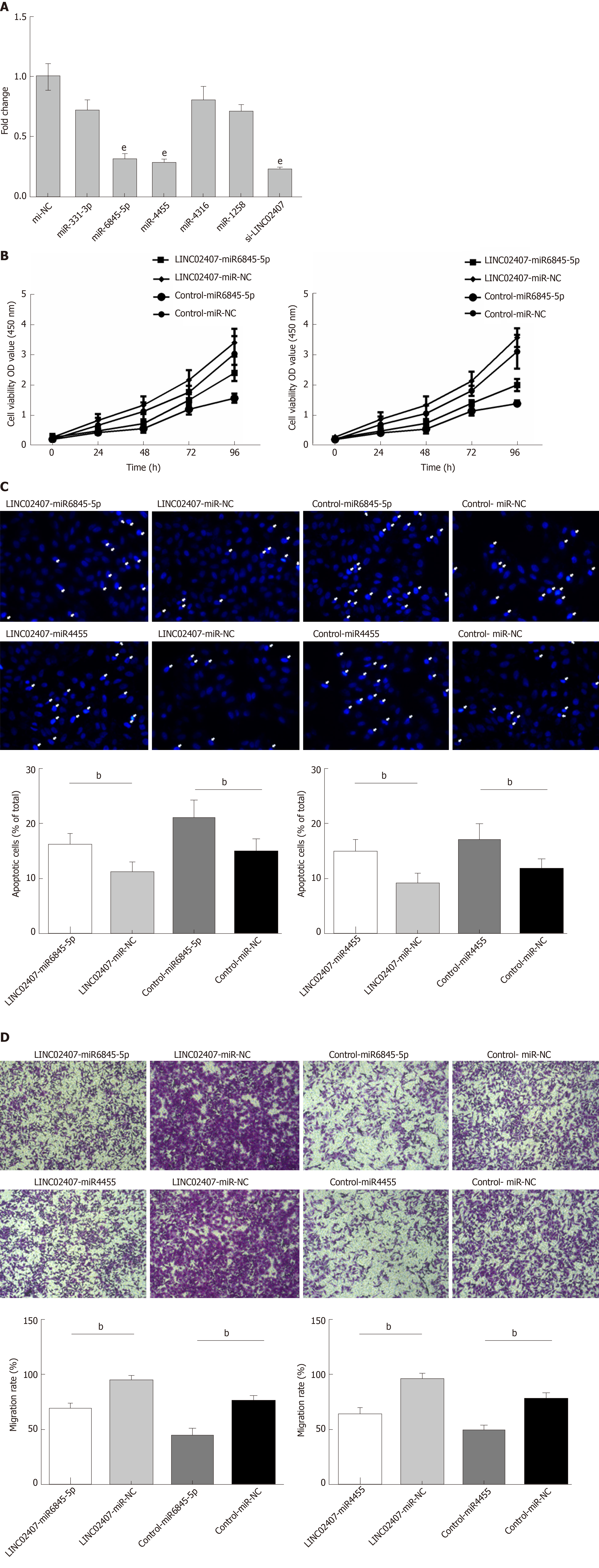
Specially designed primers were applied to detect LINC02407 expression in transfected HGC-27 cells by qRT-PCR. Chemically synthetic si-LINC02407 and si-NC were successfully transfected into HGC-27 cells, and si-LINC02407 successfully downregulated the LINC02407 expression level (Figure 5A). LINC02407-overexpressing lentivirus was successfully transfected into HGC-27 cells, and LINC02407 was upregulated. Cell viability was strengthened by the LINC02407-overexpressing lentivirus relative to the overexpression-control lentivirus as shown in Figure 5B. Apoptosis analysis was performed as previously mentioned, and the results revealed that the LINC02407-overexpressing lentivirus could inhibit apoptosis by 67% in HGC-27 cells, while on the contrary, si-LINC02407 could increase apoptosis by 83% in HGC-27 cells (Figure 5C and Figure 5D). A wound healing assay indicated that the LINC02407-overexpressing lentivirus could enhance the migration and invasion rate by 48% in HGC-27 cells relative to the overexpression-control lentivirus, and si-LINC02407 could decrease the migration and invasion rate by 15% in HGC-27 cells compared with si-NC.
The predicted results of miRanda software were considered to be miRNAs corresponding to mRNA. LncRNA and its candidate miRNAs are shown in Supplemental Table 2. We selected 5 of the 37 miRNAs reported to be closely related to the progression of GC, including hsa-miR-331-3p, hsa-miR-6845-5p, hsa-miR-4455, hsa-miR-4316, and hsa-miR-1258. A luciferase reporter assay was used to verify that LINC02407 could directly target hsa-miR-6845-5p and hsa-miR-4455. The results showed that luciferase activity was reduced in wild-type LINC02407 3’-UTR- and hsa-miR-6845-5p mimic co-transfected 293T cells in comparison to that in wild-type LINC02407 3’-UTR- and hsa-miR-6845-5p mimic NC co-transfected 293T cells (P < 0.05); the same result was found with hsa-miR-4455. Next, the effect of hsa-miR-6845-5p and hsa-miR-4455 on a GC cell line was evaluated. The hsa-miR-6845-5p-mimic could inhibit cell viability in hsa-miR-6845-5p-mimic and LINC02407-overexpressing vector-transfected HGC-27 cells (Figure 6B). Additionally, the hsa-miR-6845-5p-mimic and hsa-miR-4455-mimic had negative effects on the apoptosis in hsa-miR-6845-5p-mimic- and LINC02407-Overexpressing vector-transfected HGC-27 cells (Figure 6C). The hsa-miR-6845-5p-mimic and hsa-miR-4455-mimic could negatively impact the transwell invasion of HGC-27 cells (Figure 6D).
The potential target mRNAs of hsa-miR-6845-5p were analyzed using miRanda software, and the results are showed in Supplemental Table 3. Figure 7A shows the number of studies linking the potential target mRNAs of miR-6845-5p and the number of studies of potential target mRNAs linked to miR-4455 under the regulation of LINC02407, resulting in a Sankey diagram generated with R-3.5.2. Five mRNAs with high scores (ADGRD1, cholinergic receptor nicotinic delta, IgLON family member 5, KRI1, and troponin I1) were identified in a LINC02407 regulated pathway. A Western blot assay revealed that ADGRD1 was regulated by the LINC02407-miR-6845-5p and miR-4455- ADGRD1 pathways (Figure 7B).
LncRNAs refer to noncoding RNAs of more than 200 nt in length, and current studies indicate that lncRNAs are associated with various types of cancer[10,11]. Moreover, lncRNAs with abnormal expression in cancer are often found to be potential diagnostic biomarkers and therapeutic targets. However, tumor cells exhibit different cell morphologies, metastatic potential, and gene expression, and thus one of the major challenges in developing cancer biomarkers is tumor heterogeneity[12,13]. Based on lncRNA expression and its interrelationships with genomic location and miRNA, both the functional roles of common lncRNAs and the identification of tissue-specific mRNAs and lncRNAs can be inferred to indicate their specific roles in biogenesis and different organ functions[13,14]. In summary, lncRNAs may provide common or specific new biomarkers for different types of cancer[15,16].
Numerous studies have revealed that lncRNAs are key regulators of many biological processes, and abnormal lncRNA expression plays an important role in tumor genesis and progression. However, the molecular mechanisms of lncRNAs in GC remain unclear[17,18]. Studying the expression of lncRNAs in different diseases and its possible mechanism of action can contribute to the continuous development of cancer gene therapy. So far, there have been few studies on the relationship between lncRNAs and GC[19]. Therefore, it is necessary to study their role in GC.
The results of this study indicated that the expression of LINC02407 was significantly upregulated in GC cell lines and tissue samples. Therefore, it can be inferred that LINC02407 may be an important lncRNA that regulates the heterogeneity of GC cells. Numerous studies have shown that lncRNAs can promote cancer progression by affecting the proliferation, migration, and invasion of cancer cells[20,21,22]. We speculate that LINC02407 may be involved in the progression of GC. As expected, in vitro experiments have shown that LINC02407 can increase the malignancy of GC cells, promote the invasion of GC cells, and decrease apoptosis.
Numerous studies showed that lncRNAs can act through cavernous miRNAs as competing endogenous RNAs (ceRNAs)[23]. For example, the lncRNA SNHG7 sponge miR-216b upregulates GALNT1, which ultimately promotes CRC cell proliferation and liver metastasis[24,25]. There also have been studies of the abnormal expression of miRNAs in several malignancies. These include breast cancer, ovarian cancer, glioma, and GC[26,27,28]. Our study suggests that LINC02407 may promote GC progression by targeting miRNAs. The current investigation showed that LINC02407 can affect GC through the LINC02407-miR-6845-5p/miR-4455-ADGRD1 pathways. Moreover, hsa-miR-6845-5p and hsa-miR-4455 were negatively regulated by LINC02407. These results indicate that hsa-miR-6845-5p and hsa-miR-4455 may exert tumor suppressive effects. With the deepening of research on cancer, studies found that malignant tumors are driven by their almost unlimited ability to proliferate and metastasize[29-31]. In our study, we observed that LINC02407 acts as a ceRNA and controls the availability of miRNAs that can be acted upon by LINC02407. Importantly, LINC02407 is closely related to CASC19 and cancer cell survival, and our results strongly suggest that LINC02407 can affect GC via the LINC02407-miR-6845-5p/miR-4455-ADGRD1 pathways. In further research, we could collect more GC patients’ tissues and complete clinical information, like survival information and pathological information, to study more about the influence of LINC02407 on GC patients’ survival and the underlying mechanism.
In conclusion, overexpression of LINC02407 enhances the proliferation and inhibits the apoptosis of GC cells. Our study reports the mechanism of action of LINC02407 in GC. It was demonstrated that LINC02407 can affect GC through the LINC02407-miR-6845-5p/miR-4455-ADGRD1 pathways. Studying the LINC02407/miR-6845-5p/miR-4455-ADGRD1 pathways can provide a deeper understanding of the pathogenesis of GC and identify potential essential targets for GC diagnosis and treatment.
The study of the role and mechanism of long non-coding RNAs (lncRNAs) in tumor progression has been gradually developed and validated. The expression of lncRNAs in gastric cancer (GC) may be related to the progression of GC. However, there is little experimental evidence about their mechanism of action in GC.
The study of lncRNA-related genes may suggest its carcinogenic effects and potential molecular mechanisms in GC, and may further provide a new direction for the diagnosis and treatment of GC.
The main objectives of our study were to investigate the differential expression of lncRNAs in human GC and elucidate the function and regulatory mechanism of LINC02407.
Quantitative real-time PCR was used to detect lncRNA gene expression in GC tissues and matched adjacent non-tumor tissues and used the Cancer Genome Atlas database to verify the role of lncRNAs in GC. In a further molecular mechanism study, we confirmed the possible molecular mechanisms and regulatory pathways by which LINC02407 exerts its role by overexpressing and knocking down the expression of downstream molecules of LINC02407.
LncRNA LINC02407 was up-regulated in GC tissues and cell line and promoted proliferation and metastasis and inhibited apoptosis of GC cells. LINC02407 played a role in GC through the LINC02407-miR-6845-5p/miR-4455-adhesion G protein-coupled receptor D1 (ADGRD1) pathways, and thus, it may be an important oncogene and has potential value in GC diagnosis and treatment.
The authors demonstrated that LINC02407 is overexpressed in GC tissues and cell lines, which could provide more evidence for the clinical use of lncRNAs as biomarkers in GC. We also confirmed that LINC02407 plays a role in GC through the LINC02407-miR-6845-5p/miR-4455-ADGRD1 pathways. Overexpression of LINC02407 may reflect a promising treatment strategy for GC, which calls for more validated data in the future.
Through this study, we will have a deeper understanding of the role and mechanism of lncRNAs in GC in the future. And members of the LINC02407-miR-6845-5p/miR-4455-ADGRD1 axis can be useful targets for future prevention and treatment innovations in GC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Dinç T, Ierardi E, Virgilio E S-Editor: Tang JZ L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Weng W, Li H, Goel A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer. 2019;1871:160-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 2. | Rynkeviciene R, Simiene J, Strainiene E, Stankevicius V, Usinskiene J, Miseikyte Kaubriene E, Meskinyte I, Cicenas J, Suziedelis K. Non-Coding RNAs in Glioma. Cancers (Basel). 2018;11:pii: E17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 3. | Deng SJ, Chen HY, Zeng Z, Deng S, Zhu S, Ye Z, He C, Liu ML, Huang K, Zhong JX, Xu FY, Li Q, Liu Y, Wang C, Zhao G. Nutrient Stress-Dysregulated Antisense lncRNA GLS-AS Impairs GLS-Mediated Metabolism and Represses Pancreatic Cancer Progression. Cancer Res. 2019;79:1398-1412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 4. | Wen X, Liu X, Mao YP, Yang XJ, Wang YQ, Zhang PP, Lei Y, Hong XH, He QM, Ma J, Liu N, Li YQ. Long non-coding RNA DANCR stabilizes HIF-1α and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. 2018;8:5676-5689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 5. | Gong N, Teng X, Li J, Liang XJ. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl Mater Interfaces. 2019;11:37-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 6. | Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y, Zhang C, Zhang Y, Lin Z, Li J, Yao W, Deng W, Zhang Y, Meng M, Pan X, Li C, Yin D, Bi X, Li G, Lin DC. Super-Enhancer-Associated Long Noncoding RNA HCCL5 Is Activated by ZEB1 and Promotes the Malignancy of Hepatocellular Carcinoma. Cancer Res. 2019;79:572-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 7. | Liu SJ, Lim DA. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 8. | Gil N, Ulitsky I. Production of Spliced Long Noncoding RNAs Specifies Regions with Increased Enhancer Activity. Cell Syst. 2018;7:537-547.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 9. | Zhan Y, Chen Z, Li Y, He A, He S, Gong Y, Li X, Zhou L. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37:273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 10. | Schor IE, Bussotti G, Maleš M, Forneris M, Viales RR, Enright AJ, Furlong EEM. Non-coding RNA Expression, Function, and Variation during Drosophila Embryogenesis. Curr Biol. 2018;28:3547-3561.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 11. | Hinger SA, Cha DJ, Franklin JL, Higginbotham JN, Dou Y, Ping J, Shu L, Prasad N, Levy S, Zhang B, Liu Q, Weaver AM, Coffey RJ, Patton JG. Diverse Long RNAs Are Differentially Sorted into Extracellular Vesicles Secreted by Colorectal Cancer Cells. Cell Rep. 2018;25:715-725.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 12. | Shi X, Liu Z, Liu Z, Feng X, Hua F, Hu X, Wang B, Lu K, Nie F. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177-187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 13. | Ding Z, Lan H, Xu R, Zhou X, Pan Y. LncRNA TP73-AS1 accelerates tumor progression in gastric cancer through regulating miR-194-5p/SDAD1 axis. Pathol Res Pract. 2018;214:1993-1999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | He W, Zhang D, Li X, Wu J, Yang X, Wang Q, Lu W, Jiang J, Wu C. TCGA datasetbased construction and integrated analysis of aberrantly expressed long noncoding RNA mediated competing endogenous RNA network in gastric cancer. Oncol Rep. 2018;40:3511-3522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 15. | Zhou Z, Lin Z, He Y, Pang X, Wang Y, Ponnusamy M, Ao X, Shan P, Tariq MA, Li P, Wang J. The Long Noncoding RNA D63785 Regulates Chemotherapy Sensitivity in Human Gastric Cancer by Targeting miR-422a. Mol Ther Nucleic Acids. 2018;12:405-419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 16. | Luo Y, Tan W, Jia W, Liu Z, Ye P, Fu Z, Lu F, Xiang W, Tang L, Yao L, Huang Q, Xiao J. The long non-coding RNA LINC01606 contributes to the metastasis and invasion of human gastric cancer and is associated with Wnt/β-catenin signaling. Int J Biochem Cell Biol. 2018;103:125-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 317] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 18. | Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31:893-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 147] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 19. | Li CY, Liang GY, Yao WZ, Sui J, Shen X, Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, Zhang WH, Yin LH, Pu YP. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016;48:1965-1976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 20. | Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680-11687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 147] [Cited by in RCA: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 21. | Lü MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang SM, He FT, Yang SM. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524-3534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 152] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 22. | Zhang J, Fan D, Jian Z, Chen GG, Lai PB. Cancer Specific Long Noncoding RNAs Show Differential Expression Patterns and Competing Endogenous RNA Potential in Hepatocellular Carcinoma. PLoS One. 2015;10:e0141042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 23. | Hu Y, Tian H, Xu J, Fang JY. Roles of competing endogenous RNAs in gastric cancer. Brief Funct Genomics. 2016;15:266-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P, Xiao B, Guo J. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 130] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 25. | Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H, Teng L. LncRNAs: emerging biomarkers in gastric cancer. Future Oncol. 2015;11:2427-2441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 26. | Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 236] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 27. | Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F, Yan D. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323-1332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 28. | Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol. 2015;36:479-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 29. | Zheng L, Zhang Y, Fu Y, Gong H, Guo J, Wu K, Jia Q, Ding X. Long non-coding RNA MALAT1 regulates BLCAP mRNA expression through binding to miR-339-5p and promotes poor prognosis in breast cancer. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 30. | Zhang L, Xue Z, Yan J, Wang J, Liu Q, Jiang H. LncRNA Riken-201 and Riken-203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 2019;52:e12573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 31. | Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML, Tian YF, Peng CH, Shen BY, Chen YL, Wang YD. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019;446:49-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |