Published online Sep 21, 2019. doi: 10.3748/wjg.v25.i35.5300
Peer-review started: May 16, 2019
First decision: June 16, 2019
Revised: July 11, 2019
Accepted: July 19, 2019
Article in press: July 19, 2019
Published online: September 21, 2019
Processing time: 129 Days and 0.3 Hours
Circular RNAs (circRNAs) are considered to be highly stable due to the closed structure, which are predominately correlated with the development and progression of a wide variety of cancers. Colon cancer is one of the most common malignancies worldwide. A recent study demonstrated the upregulated expression of circPIP5K1A in non-small cell lung cancer. However, few studies have investigated the relationship between circ_0014130 level and colon cancer. Therefore, elucidating the underlying mechanisms of circPIP5K1A’s role may help with the identification of novel diagnostic and therapeutic targets for colon cancer.
To investigate the status of circPIP5K1A in colon cancers and its effects on the modulation of cancer development.
The expression level of circPIP5K1A in tissue and serum samples from colon cancer patients, as well as human colonic cancer cell lines was detected by real-time quantitative reverse transcription-polymerase chain reaction. Following the transfection of specifically synthesized small interfering RNA (siRNA) into colon cell lines, we used Hoechst staining assay to measure the ratio of cell death in the absence of circPIP5K1A. Moreover, we also used the Transwell assay to assess the migratory function of colon cells overexpressing circPIP5K1A. Additionally, we employed a series of bioinformatics prediction programs to predict the potential of circPIP5K1A-targeted miRNAs and mRNAs. The miR-1273a vector was constructed, and then transfected with or without circPIP5K1A vector into colon cancer cells. Afterwards, the expression of activator protein 1 (AP-1), interferon regulating factor 4 (IRF-4), caudal type homeobox 2 (CDX-2), and zinc finger of the cerebellum 1 (Zic-1) was detected by western blotting.
CircPIP5K1A was significantly upregulated in colon cancer tissue relative to their adjacent normal tissues. Knockdown of circPIP5K1A in colon cancer cells impaired cell viability and suppressed cell invasion and migration, while enforced expression of circPIP5K1A exhibited the opposite effects on cell migration. Bioinformatics prediction program predicted that the association of circPIP5K1A with miR-1273a, as well as AP-1, IRF-4, CDX-2, and Zic-1. Subsequent studies showed that overexpression of circPIP5K1A augmented the expression of AP-1 but attenuated the expression of IRF-4, CDX-2, and Zic-1. Reciprocally, overexpression of miR-1273a abrogated the oncogenic function of circPIP5K1A in colon cancers.
Overall, our data demonstrate the oncogenic role of circPIP5K1A-miR-1273a axis in regulation of colon cancer development, which provides a novel insights into colon cancer pathogenesis.
Core tip: We found that circular RNA PIP5K1A (circPIP5K1A) was selectively upregulated in colon cancer. Through sponging miR-1273a, circPIP5K1A promoted cell survival and enhanced the invasive and migratory functions of colon cancer cells, eventually exacerbating malignant transformation.
- Citation: Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol 2019; 25(35): 5300-5309
- URL: https://www.wjgnet.com/1007-9327/full/v25/i35/5300.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i35.5300
As the third most common malignant disease, colon cancer is the leading cause of mortality worldwide[1]. To date, conventional and effective therapeutic methods for colon cancer include surgery, adjuvant radiation therapy or chemotherapy, and molecular targeted therapy[2]. Despite the extensive improvements in diagnosis and treatments, poor survival and unsatisfactory prognosis remain an issue due to delayed diagnosis and adverse drug effects[2]. Therefore, investigation of novel diagnostic and therapeutic methods, as well as the underlying molecular mechanisms of colon cancer is required.
Accumulating studies have shown that the roles of noncoding RNAs such as circular RNAs (circRNAs) and microRNAs (miRNAs) are predominately correlated with the development and progression of a wide variety of cancers[3]. CircRNAs, one class of noncoding RNA with covalent closed loops, are widely spread in eukaryotes and are mainly formed by back-splicing without 3’- and 5’-ends[4]. CircRNAs are considered to be highly stable due to the closed structure, which has garnered interest from scholars in determining whether they can be used as novel biomarkers in many diseases[5]. Several studies have identified many possible functions of circRNAs[6] including miRNA sponges[7], regulating gene transcription and splicing[8], and forming RNA-protein complexes[9]. It is widely acknowledged that some circRNAs may participate in disease progression by interfering with miRNA, thereby influencing target gene expression[10]. Remarkably, the prevalence of high-throughput sequencing technologies make diverse bioinformatics analyses available, and more studies have revealed dysregulated expression of circRNAs in many cancers including laryngeal cancer[11], hepatocellular carcinoma[12], gastric cancer[13], gliomas[14], and esophageal cancer[15]. A previous study demonstrated that circ_001988 is downregulated in colon cancer, and may be a novel diagnostic biomarker in colon cancer[16]. Another study showed that circ_000984 induces cell growth and metastasis in colon cancer[17]. A recent study demonstrated the upregulated expression of circ_0014130 in non-small cell lung cancer[18]. However, few studies have investigated the relationship between circ_0014130 level and colon cancer. Based on the circBase database (http://www.circbase.org/), hsa_circ_0014130 is located at chr1:151206672-151212515, and the corresponding gene symbol is PIP5K1A. Thus, we denoted hsa_circ_0014130 as circPIP5K1A.
Here we found that circPIP5K1A was selectively upregulated in cancerous tissues from patients with colon cancer, which in turn suppressed cell death and augmented cancer cell invasion and migration. Through sponging miR-1273a, circPIP5K1A augmented the expression of factor activator protein 1 (AP-1), but attenuated the expression of interferon regulating factor 4 (IRF-4), caudal type homeobox 2 (CDX-2), and zinc finger of the cerebellum 1 (Zic-1), which are crucial for tumorigenesis.
This study obtained ethical approval from the ethics committee of Hubei Cancer Hospital (Hubei Sheng, China). Twenty paired tumor samples and adjacent normal samples were collected from colon cancer patients in our hospital. The specimens were confirmed by hematoxylin and eosin staining and stored in RNAlater. Written informed consent was obtained from all participants.
Human keratinocyte line HaCaT, normal human colon epithelial cell line HCoEpiC, human colon cancer cell lines HCT-116, SW620, SW480, and COLO320DM, as well as human gastric cancer cell line HGC-27, human esophageal cancer cell line TE-10, human lung cancer cell line A549, and human hepatoma cell line HepG2 were obtained from Obio Technology Co., Ltd (Shanghai, China). The cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Carlsbad, CA, United States) containing 10% fetal bovine serum (FBS; Gibco) with standard incubation conditions (5% CO2 and 37 °C).
The synthesis of three small interfering RNAs (siRNAs) and relative si-NC for circPIP5K1A, as well as the construction of overexpressing circPIP5K1A vector and overexpressing miR-1273a vector were performed by Biosyntech Co., Ltd (Suzhou, China). The siRNA or overexpressing vector was transfected into COLO320DM cells by Lipofectamine 2000 (Invitrogen, Gaithersburg, MD, United States). Silence or overexpression effect was detected by real-time quantitative polymerase chain reaction (RT-qPCR) 48 h after transfection. The most effective siRNA was used for subsequent experiments.
Total RNA was obtained by Trizol (Invitrogen) according to the manufacturer’s instructions. RNA concentration and purity were measured using a UV spectrophotometer (BD, Franklin Lakes, NJ, United States), and RNA with A260/A280 ratios of 1.8-2.0 was considered high-quality RNA without protein and DNA contamination. Then high-quality RNA was reverse transcribed into complementary DNA with a Reverse Transcription Kit (TaKaRa, Dalian, China). PCR amplification was performed by a SYBR Premix Ex Taq TM II (Takara). The primers sequences are listed in Table 1. The PCR program was: 95 °C for 3 min, 40 cycles of 95 °C for 10 s and 59 °C for 20 s, using the ABI Stepone plus Real-time PCR system (Applied Biosystems, Foster City, CA, United States). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 served as the internal control for measuring circPIP5K1A and PIP5K1A levels. Data were analyzed with 2-ΔΔCt method. To identify the cyclic structure of circPIP5K1A, RNA was treated with or without RNase R at 37 °C for 15 min, purified by phenol–chloroform extraction, and then subjected to RT-qPCR.
| RNA | Primers sequences |
| circPIP5K1A | F: 5‘-AGATTCCCTAACCTCAACCAGA-3' |
| R: 5‘-CGAATGTTCTTGCCACCTGC-3' | |
| PIP5K1A | F: 5‘-CCTCATGCAAGATTTCTACGTGG-3' |
| R: 5‘-GGCCGGATACCAAATAGCTCC-3' | |
| GAPDH | F: 5‘-AGAAGGCTGGGGCTCATTTG-3' |
| R: 5‘-AGGGGCCATCCACAGTCTTC-3' | |
| U6 | F: 5‘-AGGGGCCATCCACAGTCTTC-3' |
| R: 5‘-AACGCTTCACGAATTTGCGT-3' | |
| circ-PIP5K1A siRNA-1 | 5'-UUCUUCUAAGGGAUUGGAGUU-3' |
| circ-PIP5K1A siRNA-2 | 5'-UUCUAAGGGAUUGGAGUUGGU-3' |
| circ-PIP5K1A siRNA-3 | 5'-UAAGGGAUUGGAGUUGGUCUU-3' |
| Relative si-NC | 5'-AAUUCUCCGAACGUGUCACGU-3' |
The potential miRNAs and mRNAs bound to the promoter regions of circPIP5K1A were predicted, and then the potential miRNAs and mRNAs bound to circPIP5K1A sequence were further predicted using RegRNA2.0 (http://www.regrna.mbc.nctu. edu.tw/).
Hoechst staining kit (Beyotime Institute of Biotechnology, Shanghai, China) was used to evaluate cell apoptosis. Cells treated with circPIP5K1A siRNA were collected and fixed in 4% paraformaldehyde, followed by staining with Hoechst 33258 as manufacturer's instructions. The apoptotic cells were calculated by fluorescence microscope.
Transwell inserts (Corning, New York, NY, United States) were used to detect cell migration ability. First, the cells transfected with circPIP5K1A siRNA or overexpressing circPIP5K1A vector were plated onto the upper chamber containing serum-free medium. Meanwhile, DMEM with 10% FBS was added to the bottom chamber for 48 h. Next, the cells in the bottom chamber insert were treated with 4,6-diamidino-2-phenylindole for 5 min. The number of migrated cells was calculated with an inverted microscope (Olympus, Tokyo, Japan).
COLO320DM cells were transfected with overexpressing circPIP5K1A vector, overexpressing miR-1273a vector, or the combination of overexpressing circPIP5K1A vector and overexpressing miR-1273a vector. The cells with various treatments were collected, and then lysed by RIPA lysis buffer (Gibco) supplemented with phenylmethanesulfonyl fluoride (1 mM; Sigma, St. Louis, MO, United States). Protein was extracted by centrifugation and detected by the BCA kit (Shanghai Sangon Biotech Co., Ltd, China). The protein sample was separated on an SDS-PAGE gel, and electrotransferred to polyvinylidene fluoride membranes, followed by blocking in 5% nonfat milk for 1 h. Next, the membrane was probed with primary antibodies for transcription factor AP-1, IRF-4, CDX-2, Zic1, or GAPDH (1:1000; Abcam, Cambridge, MA, United States), followed by incubation with secondary antibody (1:1000; Beyotime) for 2 h at room temperature, respectively. Finally, enhanced chemiluminescence (Millipore, Burlington, MA, United States) was used to detect the protein levels.
Statistical analysis was conducted using SPSS Statistics software 22.0 (Chicago, IL, United States). Data are presented as mean ± standard deviation (SD) and analyzed by one-way analysis of variance followed by Dunnett’s post-hoc t-test. aP < 0.05 was considered statistically significant.
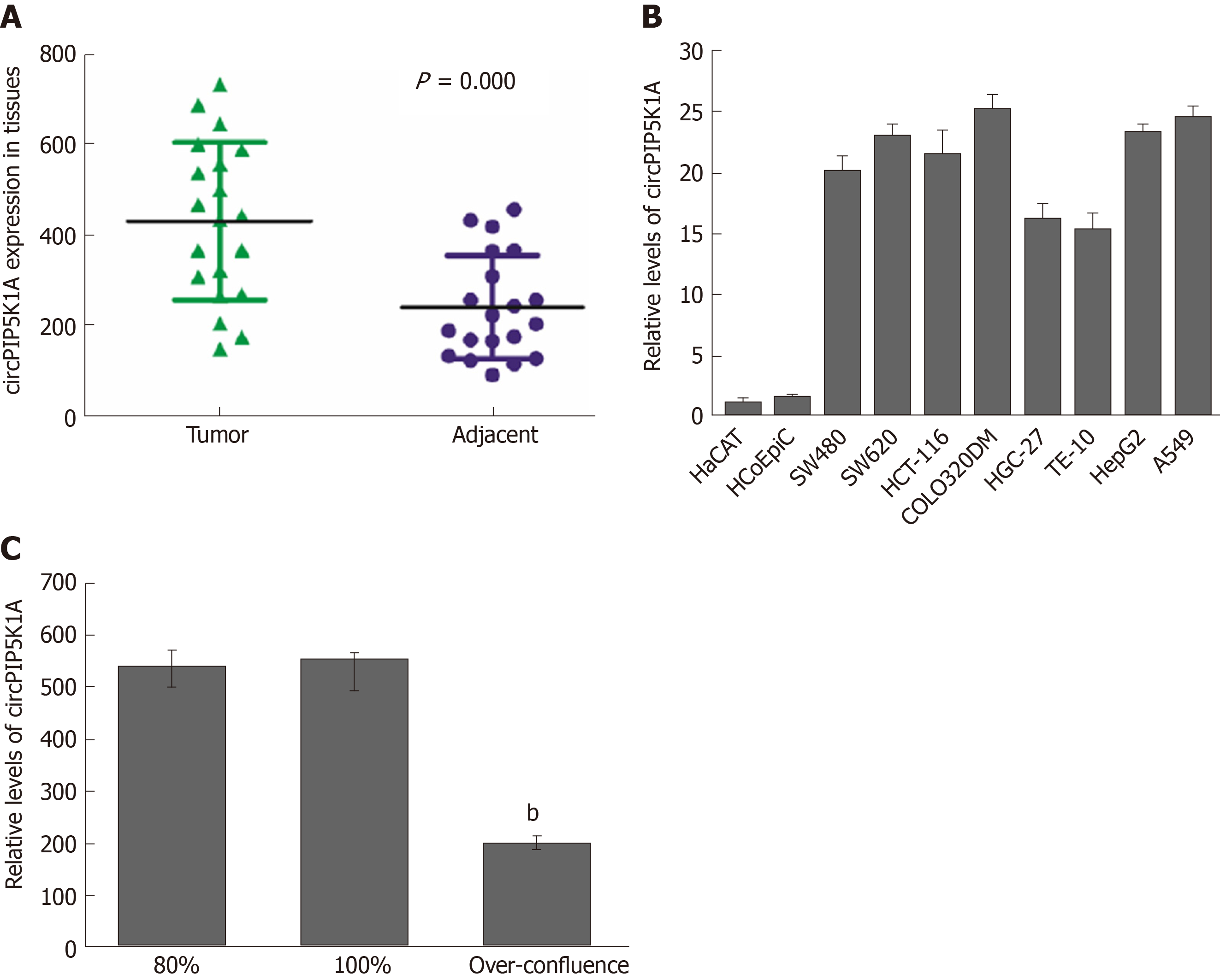
To assess the status of circPIP5K1A in colon cancer development, we employed the RT-qPCR assay to detect the mRNA level of CircPIP5K1A in clinical samples. As shown in Figure 1A, circPIP5K1A was markedly upregulated in colon cancer tissues relative to their adjacent normal tissues. Similar results were detected in in several human cancer cell lines including SW480, SW620, HCT-116, COLO320DM, HGC-27, TE-10, HepG2, and A549M, while the mRNA level of circPIP5K1A in normal cell line HCoEpiC and HaCaT was undetectable (Figure 1B). Moreover, we also found that cell over-confluence led to conspicuously inhibited circPIP5K1A expression (Figure 1C), which suggested that circPIP5K1A was involved in colon cancer development.
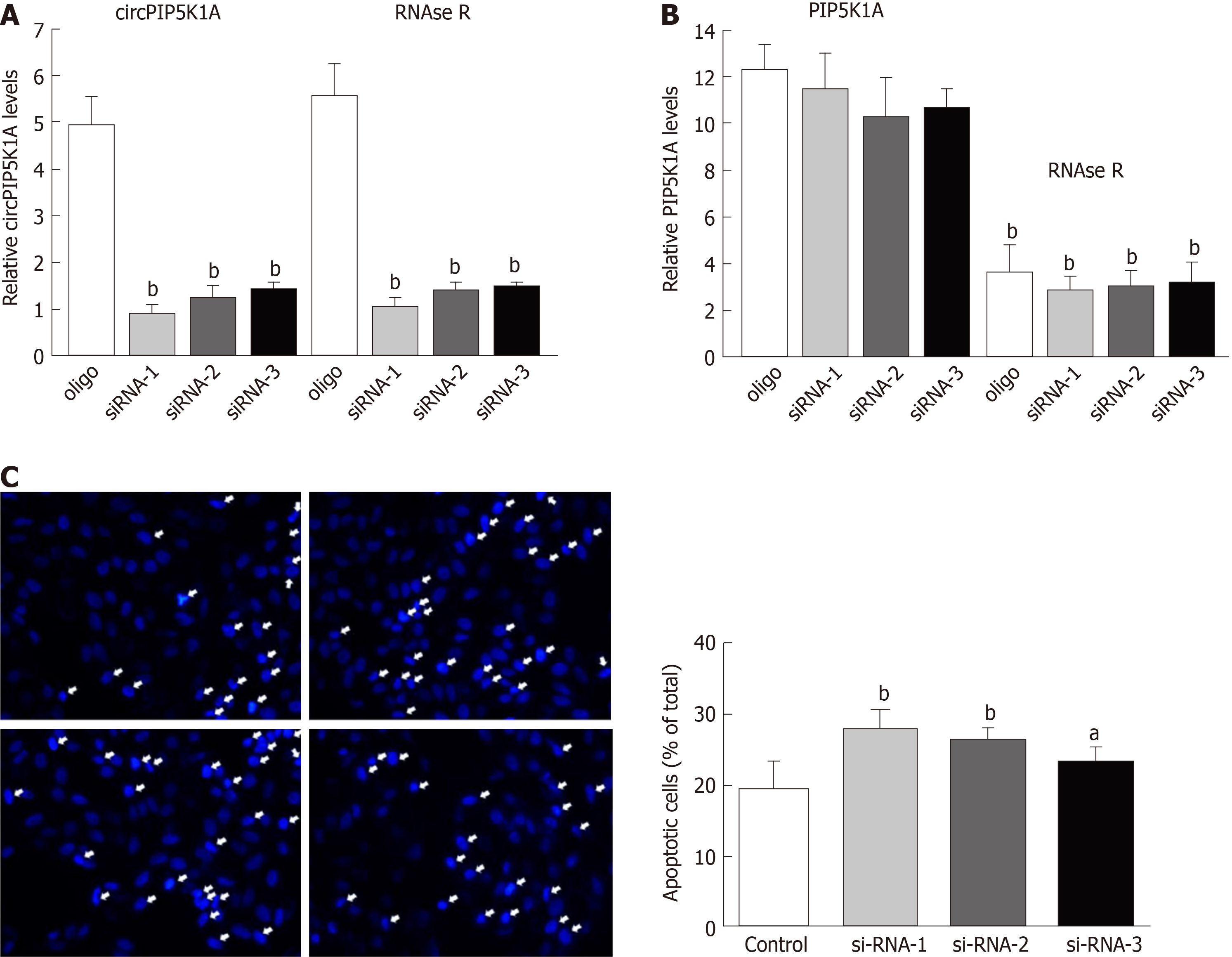
To investigate the role of circPIP5K1A in colon cancer progression, we transfected circPIP5K1A-targeted siRNA1, 2, and 3 into COLO320DM cells, which expressed the highest level of circPIP5K1A among the cancer cell lines. Our data revealed that the expression of circPIP5K1A was dramatically suppressed in the cells transfected with circPIP5K1A-targeted siRNA compared with cells transfected with scrambled siRNA. Moreover, the downregulation of circPIP5K1A by siRNA was resistant to RNase R digestion (bP < 0.01, Figure 2A). Conversely, circPIP5K1A-targeted siRNA hardly affected the endogenous expression of PIP5K1A (Figure 2B), which confirmed the specificity of circPIP5K1A-targeted siRNA. Notably, we found that the absence of circPIP5K1A resulted in obviously increased cell death compared with cells transfected with scrambled siRNA (Figure 2C).
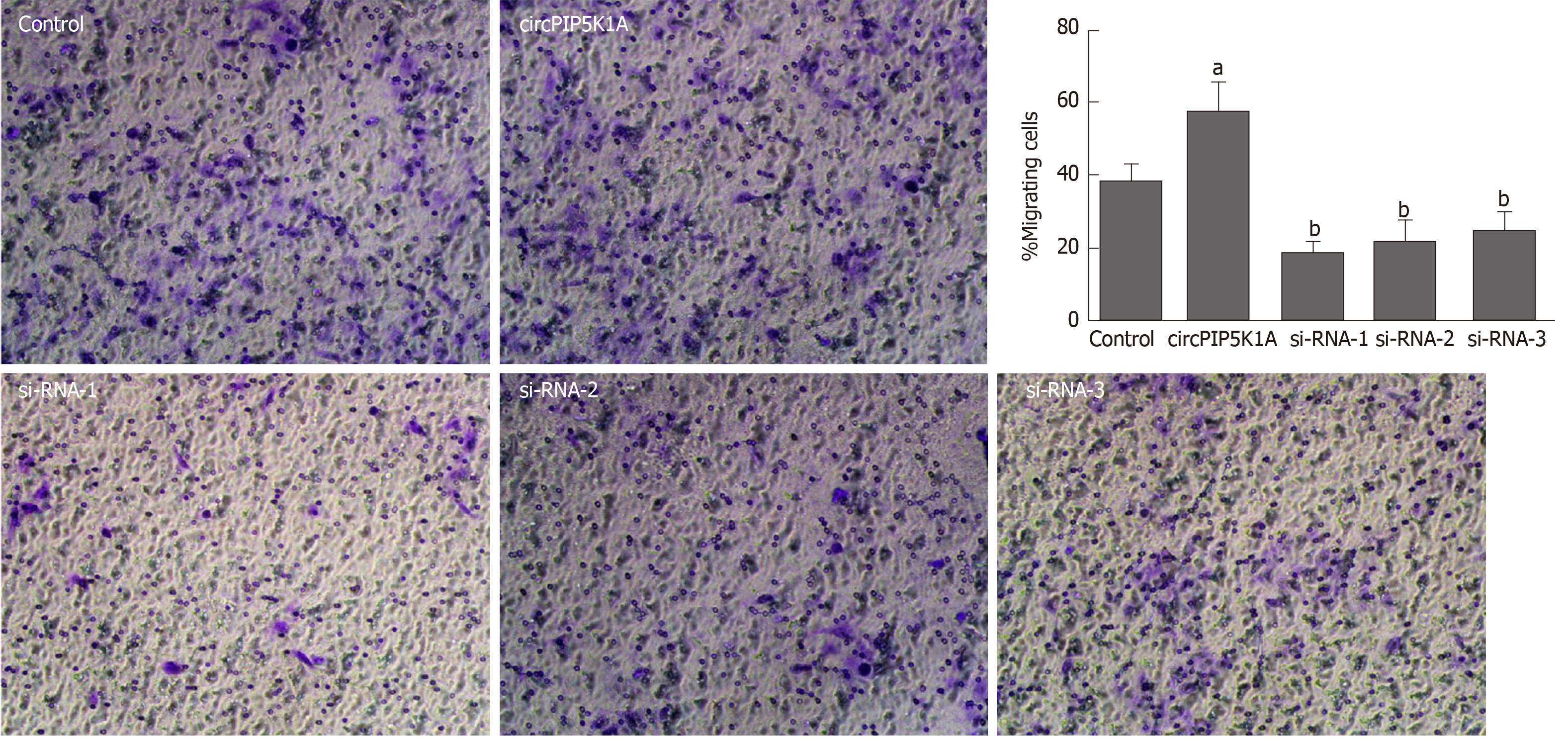
To determine whether circPIP5K1A can regulate cancer migration, we utilized the Transwell assay, and found that enforced expression of circPIP5K1A significantly augmented cell invasion and migration relative to mock-transfected cells. Reciprocally, silencing of endogenous circPIP5K1A exhibited opposite effects on cell migration (Figure 3). Taken together, our data demonstrate that upregulation of circPIP5K1A promotes cell viability and migration.
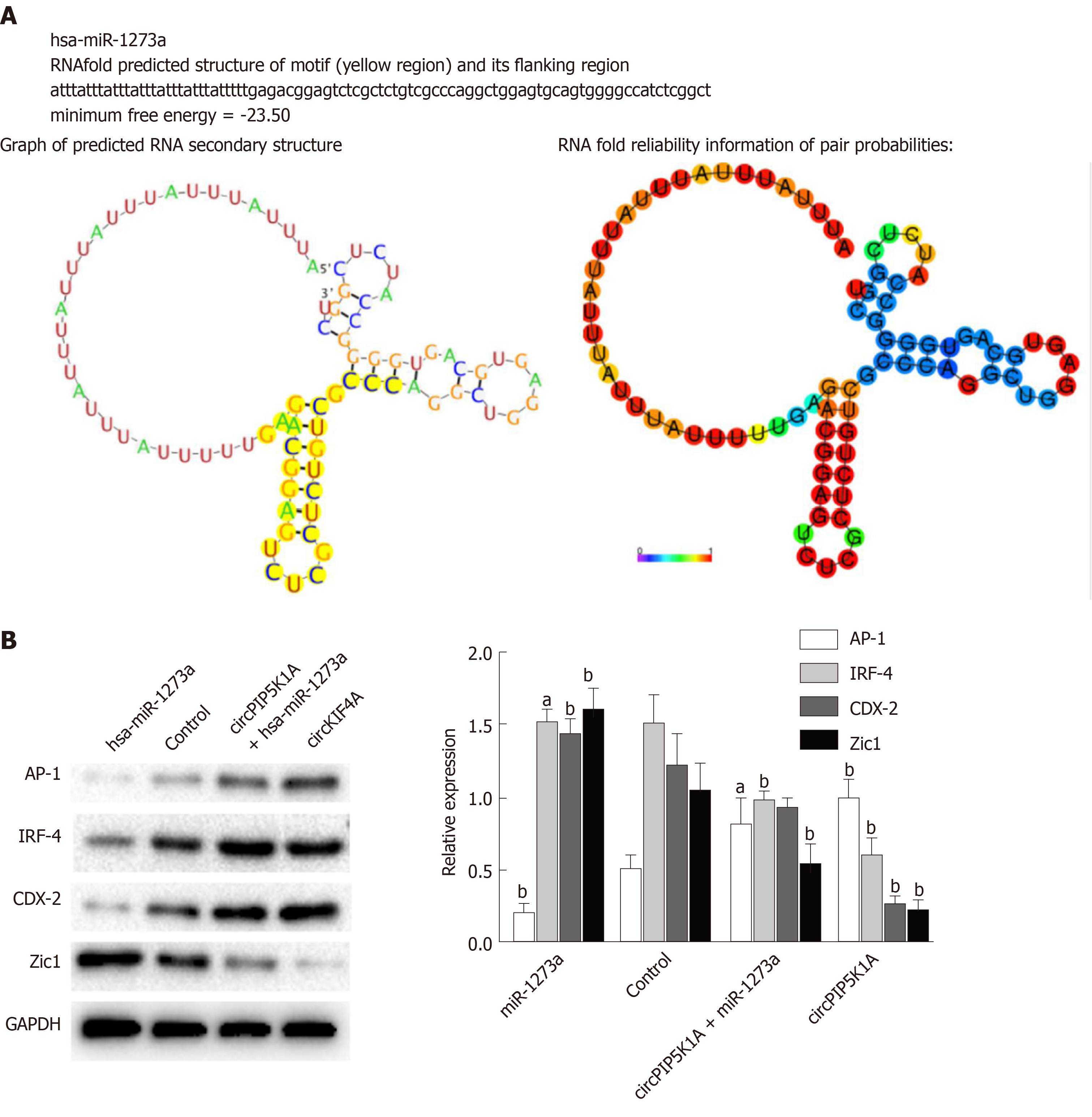
To investigate the molecular function of circPIP5K1A in colon cancer development, we used RegRNA to predict circPIP5K1A-associated RNAs. The highest score of RNAs were miR-1273a as well as traditional mRNAs such as AP-1, IRF-4, CDX-2, and Zic-1 (Figure 4A). Further study revealed that overexpression of circPIP5K1A significantly upregulated both the mRNA and protein level of AP-1, while attenuating the expression of IRF-4, CDX-2, and Zic-1 (Figure 4B and C). Conversely, overexpression of miR-1273a obviously alleviated circPIP5K1A-mediated suppression of IRF-4, CDX-2 and Zic-1 expression (Figure 4B and C).
Our present study showed that circPIP5K1A was significantly upregulated in colon cancer tissue and cell lines. Silencing of circPIP5K1A in COLO320DM cells induced cell death and suppressed cell migration. Moreover, bioinformatics predicted that circPIP5K1A could bind to miR-1273a as well as AP-1, IRF-4, CDX-2, and Zic-1. Overexpression of circPIP5K1A augmented AP-1 expression but attenuated the expression of IRF-4, CDX-2, and Zic-1, while overexpression of miR-1273a inhibited the oncogenic role of circPIP5K1A in the progression of colon cancer development.
Accumulating studies have identified circRNAs as novel biomarkers in cancer diagnosis due to the differential expression between normal and tumor tissues. For example, hsa_circ_0000520 level is increased in gastric cancer tissues and cells, which is negatively related to TNM stage, indicating that circ_0000520 may be a novel biomarker in gastric cancer[19]. Circ_0001649 is reportedly negatively correlated with colorectal cancer pathological differentiation, and is considered a new biomarker for the diagnosis of colorectal cancer[20]. A recent study reported the upregulation of hsa_circ_0014130 (circPIP5K1A) in non-small cell lung cancer, and it is considered a potential biomarker in non-small cell lung cancer[18]. In this study, we found that circPIP5K1A was selectively upregulated in colon cancer tissues and cells. It is well known that circRNAs are involved in various cellular biological processes and exert important regulatory roles in the progression of multiple cancers. A previous study demonstrated that hsa_circ_0001649 exerted tumor-suppressive effects by inducing cell apoptosis, as well as inhibiting cell proliferation and migration in cholangiocarcinoma cells[21]. In addition, it has been reported that the downregulation of circARHGAP26 inhibits cell proliferation and promotes cells apoptosis, indicating that circARHGAP26 may have tumor-promoting effects[22]. Here, we found that circPIP5K1A elicited stimulatory effects on cell proliferation and migration. These findings suggest that circPIP5K1A might contribute to the development of colon cancer.
Similarly to long non-coding RNA, the main role of circRNAs is to decrease miRNA abundance by sponging miRNA in the cytoplasm, and then affecting the expression of related genes. For example, hsa_circ_001564 knockdown suppresses osteosarcoma progression by acting as an miRNA sponge[23]. In this study, miR-1273a, AP-1, IRF-4, CDX-2, and Zic-1 were predicted to bind to circPIP5K1A. AP-1, encoding transcription factor AP-1, plays a pivotal role in tumorigenesis[24]. In cancers, AP-1 activity is increased, and inhibiting AP-1 activity can block cell proliferation and invasion[24]. A previous study reported that inhibition of AP-1 signaling may be a therapeutic target for colon cancer[25]. IRF-4, as a member of the interferon regulating factor family of transcription factors, is considered to be an oncogene in lymphoid malignancy and multiple myeloma[26]. Notably, recent studies have demonstrated that IRF-4 is a tumor suppressor in breast cancer[27] and lung cancer[28]. The homeodomain transcription factor CDX-2 has been investigated in colon cancer by many studies, which have suggested that CDX-2 loss is associated with the poor prognosis of colon cancer[29-32]. Zic-1 is a member of zinc finger of the cerebellum family, which encodes zinc-finger transcription factors[33]. In colorectal cancer, Zic-1 is downregulated and considered a tumor inhibitor[34]. Consistent with these studies, circPIP5K1A overexpression clearly upregulated the expression of AP-1, as well as downregulated the expression of IRF-4, CDX-2, and Zic-1. Notably, miR-1273a overexpression rescued the effects of circPIP5K1A on the expression of AP-1, IRF-4, CDX-2, and Zic-1 in colon cancer cells. Thus, we speculate that miR-1273a may inhibit the regulatory role of circPIP5K1A in colon cancer cells. However, the relationship between circPIP5K1A and miR-1273a should be investigated in further studies. In conclusion, our data demonstrate that circPIP5K1A is selectively increased in colon cancer and exerts oncogenic effects on tumorigenesis, which may be regulated by miR-1273a.
Circular RNAs (circRNAs) are considered to be highly stable due to their closed structure, which are predominately correlated with the development and progression of a wide variety of cancers. A recent study demonstrated the upregulated expression of circPIP5K1A in non-small cell lung cancer. However, few studies have investigated the relationship between circ_0014130 level and colon cancer. Therefore, elucidating the underlying mechanisms of circPIP5K1A’s role may help identify novel diagnostic and therapeutic targets for colon cancer.
It is necessary to explore whether circPIP5K1A regulates miR-1273a to affect cell death, cell invasion, and migration in colon cancer. Recent studies have demonstrated that circPIP5K1A exerts an oncogenic role in non-small cell lung cancer, which made it a good lead for further studies regarding the mechanism of circPIP5K1A regulation during colon cancer development.
In this study, we evaluated the expression level of circPIP5K1A in clinical tumor samples and colon cancer cells, and then investigated the effects of circPIP5K1A on colon cell apoptosis and migration in vitro by gain- and loss-of–function approaches. Moreover, we explored whether circPIP5K1A promotes colon cancer development through sponging miR-1273a. Our study provides significant insights into the mechanism of circPIP5K1A during colon cancer development that may contribute to the future design of more effective therapies.
First, circPIP5K1A level was detected in colon cancer tissue and cell lines by RT-qPCR assay. Then gene transfection or silencing experiments were conducted to construct stably expressed or depleted circPIP5K1A cell lines to complete subsequent functional studies. A series of in vitro experiments, such as cell apoptosis assay and Transwell assays, were performed to explore the effects of circPIP5K1A on cell apoptosis, invasion and migration. The potential miRNAs and mRNAs bound to the promoter regions of circPIP5K1A were predicted, and then the potential miRNAs and mRNAs bound to circPIP5K1A sequence were further predicted using RegRNA2.0. MiR-1273a vector was constructed, and then transfected with or without circPIP5K1A vector into colon cancer cells. Then the expression of AP-1, IRF-4, CDX-2, and Zic-1 was detected by western blotting.
CircPIP5K1A was significantly upregulated in colon cancer tissue relative to their adjacent normal tissues. The results of in vitro experiments showed a positive role of circPIP5K1A in the proliferation, invasion, and migration of colon cancer cells. Bioinformatics prediction program predicted that the association of circPIP5K1A with miR-1273a, as well as AP-1, IRF-4, CDX-2, and Zic-1. Further study revealed that the overexpression of circPIP5K1A significantly upregulated both the mRNA and protein level of AP-1, while attenuating the expression of IRF-4, CDX-2, and Zic-1. Conversely, overexpression of miR-1273a clearly alleviated the circPIP5K1A-mediated suppression of IRF-4, CDX-2 and Zic-1 expression.
In conclusion, circPIP5K1A is selectively increased in colon cancer, and circPIP5K1A plays a positive role in colon cancer cell proliferation, invasion, and migration. In addition, overexpression of circPIP5K1A augmented AP-1 expression but attenuated the expression of IRF-4, CDX-2, and Zic-1, while overexpression of miR-1273a inhibited the oncogenic role of circPIP5K1A in progression of colon cancer development.
Our study illuminates the role and molecular mechanism of circPIP5K1A in the regulation of colon cancer development, and demonstrates the oncogenic role of the circPIP5K1A-miR-1273a axis in regulating colon cancer development. The findings of this study provide novel insights into colon cancer pathogenesis.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bordonaro M, Christodoulou DK S-Editor: Yan JP L-Editor: Filipodia E-Editor: Ma YJ
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15468] [Article Influence: 2578.0] [Reference Citation Analysis (2)] |
| 2. | Muro K. Systemic chemotherapy for metastatic colorectal cancer -Japanese Society for Cancer of the Colon and Rectum (JSCCR) Guidelines 2016 for treatment of colorectal cancer. Nihon Shokakibyo Gakkai Zasshi. 2017;114:1217-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 3. | Zhang Z, Xie Q, He D, Ling Y, Li Y, Li J, Zhang H. Circular RNA: New star, new hope in cancer. BMC Cancer. 2018;18:834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 4. | Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1021] [Cited by in RCA: 1288] [Article Influence: 117.1] [Reference Citation Analysis (0)] |
| 5. | Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 833] [Cited by in RCA: 1157] [Article Influence: 144.6] [Reference Citation Analysis (0)] |
| 6. | Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 1295] [Article Influence: 143.9] [Reference Citation Analysis (0)] |
| 7. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6020] [Article Influence: 501.7] [Reference Citation Analysis (0)] |
| 8. | Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 1750] [Article Influence: 145.8] [Reference Citation Analysis (0)] |
| 9. | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3481] [Cited by in RCA: 3414] [Article Influence: 284.5] [Reference Citation Analysis (0)] |
| 10. | Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, Zhang W, Li X, Li X, Li Y, Li G, Zeng Z, Xiong W. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 308] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 11. | Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T, Wang X, Li Q, Tian L, Liu M, Sun Y. Circular RNA: A novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Hu J, Li P, Song Y, Ge YX, Meng XM, Huang C, Li J, Xu T. Progress and prospects of circular RNAs in Hepatocellular carcinoma: Novel insights into their function. J Cell Physiol. 2018;233:4408-4422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 13. | Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 216] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 14. | Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 255] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 15. | Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, Jin X, Xie C. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 162] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 16. | Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, Zheng W. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020-16025. [PubMed] |
| 17. | Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ, Liu XQ, Wu WD. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674-91683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 18. | Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, Yuan H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8:2878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 19. | Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, Liu Z, Cao H, Cao X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 20. | Ji W, Qiu C, Wang M, Mao N, Wu S, Dai Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497:122-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 21. | Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L, Cui Y, Jiang X. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 22. | Wangxia LV, Fang Y, Liu Y, Zhao Y, Shi Z, Zhong H. Circular RNA ARHGAP26 is over-expressed and its downregulation inhibits cell proliferation and promotes cell apoptosis in gastric cancer cells. Saudi J Gastroenterol. 2019;25:119-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495:2369-2375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 24. | Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Curr Cancer Drug Targets. 2007;7:317-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 108] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Vaiopoulos AG, Papachroni KK, Papavassiliou AG. Colon carcinogenesis: Learning from NF-kappaB and AP-1. Int J Biochem Cell Biol. 2010;42:1061-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Gualco G, Weiss LM, Bacchi CE. MUM1/IRF4: A Review. Appl Immunohistochem Mol Morphol. 2010;18:301-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 27. | Heimes AS, Madjar K, Edlund K, Battista MJ, Almstedt K, Gebhard S, Foersch S, Rahnenführer J, Brenner W, Hasenburg A, Hengstler JG, Schmidt M. Prognostic significance of interferon regulating factor 4 (IRF4) in node-negative breast cancer. J Cancer Res Clin Oncol. 2017;143:1123-1131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 28. | Wu YY, Hwang YT, Perng WC, Chian CF, Ho CL, Lee SC, Chang H, Terng HJ, Chao TY. CPEB4 and IRF4 expression in peripheral mononuclear cells are potential prognostic factors for advanced lung cancer. J Formos Med Assoc. 2017;116:114-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Mallo GV, Soubeyran P, Lissitzky JC, André F, Farnarier C, Marvaldi J, Dagorn JC, Iovanna JL. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J Biol Chem. 1998;273:14030-14036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 131] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 30. | Hinoi T, Loda M, Fearon ER. Silencing of CDX2 expression in colon cancer via a dominant repression pathway. J Biol Chem. 2003;278:44608-44616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 31. | Hinkel I, Duluc I, Martin E, Guenot D, Freund JN, Gross I. Cdx2 controls expression of the protocadherin Mucdhl, an inhibitor of growth and β-catenin activity in colon cancer cells. Gastroenterology. 2012;142:875-885.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 32. | Olsen J, Eiholm S, Kirkeby LT, Espersen ML, Jess P, Gögenür I, Olsen J, Troelsen JT. CDX2 downregulation is associated with poor differentiation and MMR deficiency in colon cancer. Exp Mol Pathol. 2016;100:59-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 33. | Ali RG, Bellchambers HM, Arkell RM. Zinc fingers of the cerebellum (Zic): Transcription factors and co-factors. Int J Biochem Cell Biol. 2012;44:2065-2068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 34. | Gan L, Chen S, Zhong J, Wang X, Lam EK, Liu X, Zhang J, Zhou T, Yu J, Si J, Wang L, Jin H. ZIC1 is downregulated through promoter hypermethylation, and functions as a tumor suppressor gene in colorectal cancer. PLoS One. 2011;6:e16916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |