Published online Sep 14, 2019. doi: 10.3748/wjg.v25.i34.5152
Peer-review started: May 16, 2019
First decision: July 21, 2019
Revised: July 25, 2019
Accepted: August 19, 2019
Article in press: August 19, 2019
Published online: September 14, 2019
Processing time: 119 Days and 20.3 Hours
The xeroderma pigmentosum group G (XPG) gene at chromosome 13q33 consists of 15 exons, which may be related to the occurrence and development of gastric cancer (GC).
To examine the association of several common single nucleotide polymorphisms (SNPs) of the XPG gene with GC risk and survival.
Five SNPs of XPG (rs2094258, rs751402, rs873601, rs2296147, and rs1047768) were genotyped by PCR restriction fragment length polymorphism in 956 histologically confirmed GC cases and 1012 controls in North China. GC patients were followed for survival status and, if deceased, cause of death. Logistic regression and Cox regression were used for analysing associations of XPG SNPs with risk of GC and prognosis, respectively. For rs2094258, heterozygous model (CT vs CC), homozygous model (TT vs CC), recessive model (TT vs CT + CC), and dominant model (TT + CT vs CC) were analyzed.
None of the examined loci were statistically associated with GC risk, although rs2296147 was marginally associated with GC risk (P = 0.050). GC patients with the rs2094258 CT + CC genotype showed worse survival than those with the TT genotype (log-rank test, P = 0.028), and patients with the CC genotype had a tendency of unfavourable prognosis compared with those with the TT + CT genotype (log-rank test, P = 0.039). The increase in C alleles of rs2094258 [hazard ratio (HR) = 1.19, 95% confidence interval (CI): 1.02-1.45, P = 0.037] were associated with the long-term survival of GC cases. Other risk factors for survival included tumor differentiation (HR = 4.51, 95%CI: 1.99-8.23, P < 0.001), lymphovascular invasion (HR = 1.97, 95%CI: 1.44-3.01, P < 0.001), and use of chemotherapy (HR = 0.81, 95%CI: 0.63-0.98, P = 0.041).
The XPG rs2094258 polymorphism may be associated with overall survival in GC patients.
Core tip: This study investigated the relationships between five functional single nucleotide polymorphisms of the xeroderma pigmentosum group G (XPG) (rs2094258, rs751402, rs2296147, rs1047768, and rs873601) and gastric cancer (GC) risk and survival. The results showed an association between the XPG rs2094258 polymorphism and overall survival in patients with GC. GC patients with the rs2094258 CT + CC genotype showed a worse survival than those with the TT genotype, and patients with the CC genotype had a tendency of unfavourable prognosis compared with those with the TT + CT genotype. The increase in the number of C alleles of rs2094258 was associated with the long-term survival of GC cases.
- Citation: Wang XQ, Terry PD, Li Y, Zhang Y, Kou WJ, Wang MX. Association of XPG rs2094258 polymorphism with gastric cancer prognosis. World J Gastroenterol 2019; 25(34): 5152-5161
- URL: https://www.wjgnet.com/1007-9327/full/v25/i34/5152.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i34.5152
Gastric cancer (GC) is one of the global leading causes of cancer-related death[1]. The World Health Organization (WHO) estimates that 1.03 million people are diagnosed and 783000 die of the disease each year[2]. The highest age standard incidence per 100000 population is observed in Eastern Asia[3], especially in China, at 22.7 per 100000[4]. Multiple factors contribute to the development of this disease, including environmental and genetic factors, such as Helicobacter pylori (H. pylori) infection, drinking, obesity, a high-salt diet, and various genetic factors[5]. Although GC morbidity and mortality have declined in recent years, the social and economic burden of the disease remains high[1,6,7].
DNA repair genes play a key role in maintaining the genomic DNA stability and integrity. Functional genetic variants of DNA repair genes may change the host DNA repair ability and thus affect tumor prognosis[8]. DNA repair genes participate in many crucial pathways including nucleotide excision repair (NER), which is involved in the repair of some types of DNA damage. Recent evidence suggests that NER factors function in processes that facilitate mRNA synthesis or shape three-dimensional chromatin structure[9]. Xeroderma pigmentosum group G (XPG or ERCC5) plays a key role in NER repair, as it can recognize DNA damage and initiate the NER process[10]. XPG has 3’-junction cutting ability on bubble substrates, resulting in 3’-incision in the human double-incision) repair system, and non-catalytically XPG is needed for subsequent 5’-incision by XPF-ERCC1[11].
Some studies have demonstrated that the single nucleotide polymorphisms (SNPs) of XPG may affect the development of cancer, such as lung cancer[12,13], colorectal cancer[14,15], breast cancer[16], neuroblastoma[17], Hodgkin’s lymphoma[18], and oral squamous cell carcinoma[19]. However, only a few studies have explored the associations between XPG gene SNPs and GC, and those studies show inconsistent results. Some studies reported no associations[20-22], while others demonstrated varying degrees of association[23,24]. Furthermore, few studies have explored the potential prognostic importance of XPG SNPs in GC patients[25]. Therefore, we examined GC risk and survival in relation to common functional XPG SNPs in a case-control study in North China.
From September 2010 to June 2013, pathologically confirmed incident GC cases were selected from the First Affiliated Hospital of Xi’an Jiaotong University. During the same time period, controls were recruited from the Physical Examination Centre of the First Affiliated Hospital of Xi’an Jiaotong University. The cases and controls were matched by sex, age (within 5 years), and residential district. Socio-demographic and clinical data were collected during recruitment, such as alcohol consumption and smoking status. TNM staging of GC tumors was done according to the WHO standard. H. pylori infection status was tested by ELISA. The present study protocol was approved by the Institutional Review Board of Health Science Center of Xi’an Jiaotong University. Informed consent was obtained from all study participants.
Cases were followed for survival status and chemotherapy data every 3 mo within the first year and then annually afterwards. Causes and dates of death were recorded, and survival time was calculated from the date of recruitment. Survival time was calculated from time of recruitment to date of death, date of last contact (for those lost to follow-up), or to the last contact with living subjects at the end of the study.
Peripheral blood samples from all cases and controls were collected by the investigators. The TIANamp Blood DNA Kit (Tiangen, Beijing, China) was used for DNA extraction. XPG rs2094258, rs751402, rs873601, rs2296147, and rs1047768 polymorphism genotyping was performed by PCR restriction fragment length polymorphism (PCR-RFLP). The conditions of PCR amplification were: (1) Denaturation at 95 °C for 5 min; (2) 30 cycles of denaturation at 94 °C for 60 s, annealing at 60 °C for 60 s, and extension at 72 °C for 60 s; and (3) Extension at 72 °C for 10 min. PCR products were confirmed by agarose gel electrophoresis. Ten percent of the samples were randomly selected for repeated genotyping and the results were 100 percent consistent.
The socio-demographic data and clinical data between case and control subjects were compared by the chi-square test. A goodness-of-fit chi-squared test was also used to analyze whether the SNP (XPG rs2094258, rs751402, rs2296147, rs1047768 and rs873601) distributions conform to the Hardy-Weinberg equilibrium (HWE) in controls. Odds ratios (ORs) and 95% confidence intervals (CIs) for examined SNPs were analyzed by Logistic regression method and adjusted by age, gender, and H. pylori infection status. The heterozygous model, homozygous model, recessive model, and dominant model were all analyzed for five SNPs. For rs2094258, heterozygous model, homozygous model, recessive model, and dominant model were CT vs CC, TT vs CC, TT vs CT + CC, TT + CT vs CC, respectively. Kaplan-Meier method and log-rank test were used for plotting cases’ survival curves and for comparisons, respectively.
Multivariate Cox regression was used for exploring possible prognostic factors, which included gender, age, drinking, smoking, H. pylori, TNM stage, tumor differentiation, lymphovascular invasion, neural invasion, and chemotherapy. SPSS 24.0 statistical software was used for all statistical analyses (Statistical Package for the Social Sciences, version 24, SSPS Inc, Chicago, IL, United States). All statistical tests were two-sided, with P < 0.05 as the boundary value.
The majority of study participants were male and less than age 60, with no significant differences in these factors between cases and controls (Table 1). All five loci in the control group were consistent with the Hardy-Weinberg equilibrium. The rate of H. pylori infection in GC patients was significantly higher than that in the control group (70.6% vs 53.6%, P < 0.001). None of the associations between studied loci (rs2094258, rs751402, rs2296147, rs1047768, and rs873601) and GC risk was statistically significant (Table 2). However, the rs2296147 CC genotype frequency was higher in the GC case group than that in the control group (5.9% vs 3.9%, P = 0.050). After adjustment for age, sex, and H. pylori status, this genotype was marginally associated with a slightly higher risk of GC (OR = 1.40, 95%CI: 0.97-2.50, P = 0.061) than the TT genotype. The CC genotype was found to be marginally associated with a higher risk of GC (OR = 1.36, 95%CI = 0.99-2.49, P = 0.053) in the recessive model (CC vs CT + TT).
| Case | Control | X2 | P-value | ||
| n = 956 | n = 1012 | ||||
| Age (yr) | |||||
| ≤60 | 569 (59.5) | 637 (62.9) | 2.289 | 0.130 | |
| >60 | 387 (40.5) | 375 (37.1) | |||
| Gender | |||||
| Male | 667 (69.8) | 724 (71.5) | 0.662 | 0.416 | |
| Female | 289 (30.2) | 288 (28.5) | |||
| H. pylori infection | |||||
| Positive | 675 (70.6) | 542 (53.6) | 59.835 | <0.001 | |
| Negative | 281 (29.4) | 470 (46.4) | |||
| Drinking | |||||
| No | 758 (79.3) | 497 (49.1) | 2.787 | 0.095 | |
| Yes | 198 (20.7) | 515 (50.9) | |||
| Smoking | |||||
| No | 560 (58.6) | 517 (51.1) | 1.382 | 0.240 | |
| Yes | 396 (41.4) | 495 (48.9) | |||
| SNP | Genotype | Case | Control | HWE | P-value | OR | 95%CI | P-value | OR1 | 95%CI | P-value |
| n = 956 | n = 1012 | ||||||||||
| Rs2094258 | CC | 390 (40.8) | 437 (43.2) | 0.318 | 0.853 | 1.00 | |||||
| CT | 441 (46.1) | 464 (45.8) | 1.07 | 0.88-1.29 | 0.545 | 1.05 | 0.86-1.31 | 0.626 | |||
| TT | 125 (13.1) | 111 (11.0) | 1.26 | 0.94-1.69 | 0.133 | 1.19 | 0.90-1.20 | 0.249 | |||
| Dominant | 566 (59.2) | 575 (56.8) | 1.10 | 0.92-1.32 | 0.284 | 1.06 | 0.87-1.37 | 0.388 | |||
| Recessive | 831 (86.9) | 901 (89.0) | 1.22 | 0.93-1.60 | 0.150 | 1.19 | 0.91-1.72 | 0.303 | |||
| Rs751402 | CC | 366 (38.3) | 371 (36.7) | 0.183 | 0.913 | 1.00 | |||||
| CT | 467 (48.8) | 476 (47.0) | 0.95 | 0.78-1.16 | 0.618 | 0.98 | 0.77-1.33 | 0.774 | |||
| TT | 123 (12.9) | 165 (16.3) | 0.88 | 0.67-1.15 | 0.341 | 0.90 | 0.61-1.28 | 0.491 | |||
| Dominant | 590 (61.7) | 632 (62.5) | 0.95 | 0.79-1.14 | 0.554 | 0.95 | 0.78-1.22 | 0.711 | |||
| Recessive | 833 (87.1) | 847 (83.7) | 0.90 | 0.71-1.15 | 0.411 | 0.93 | 0.69-1.18 | 0.487 | |||
| Rs2296147 | TT | 599 (62.7) | 640 (63.2) | 0.076 | 0.963 | 1.00 | |||||
| CT | 301 (31.5) | 332 (32.9) | 0.97 | 0.80-1.17 | 0.745 | 0.97 | 0.80-1.19 | 0.674 | |||
| CC | 56 (5.9) | 40 (3.9) | 1.50 | 0.98-2.28 | 0.059 | 1.40 | 0.97-2.50 | 0.061 | |||
| Dominant | 357 (37.3) | 372 (36.8) | 1.03 | 0.85-1.23 | 0.789 | 1.01 | 0.80-1.38 | 0.865 | |||
| Recessive | 900 (94.1) | 973 (96.1) | 1.51 | 1.00-2.29 | 0.050 | 1.36 | 0.99-2.49 | 0.053 | |||
| Rs1047768 | TT | 505 (52.8) | 540 (53.4) | 0.367 | 0.832 | 1.00 | |||||
| CT | 379 (39.6) | 406 (40.1) | 1.00 | 0.83-1.20 | 0.985 | 1.00 | 0.80-1.28 | 0.989 | |||
| CC | 72 (7.6) | 66 (6.5) | 1.17 | 0.82-1.66 | 0.395 | 1.11 | 0.79-1.92 | 0.505 | |||
| Dominant | 451 (47.2) | 472 (46.6) | 1.02 | 0.86-1.22 | 0.812 | 1.01 | 0.81-1.45 | 0.897 | |||
| Recessive | 884 (92.5) | 946 (93.5) | 1.17 | 0.83-1.65 | 0.381 | 1.12 | 0.74-1.84 | 0.561 | |||
| Rs873601 | GG | 271 (28.3) | 288 (28.5) | 0.247 | 0.884 | 1.00 | |||||
| AG | 475 (49.7) | 514 (50.8) | 0.98 | 0.80-1.21 | 0.865 | 0.99 | 0.63-1.88 | 0.931 | |||
| AA | 210 (22.0) | 210 (20.7) | 1.06 | 0.83-1.37 | 0.638 | 1.03 | 0.71-2.01 | 0.807 | |||
| Dominant | 685 (71.7) | 724 (71.5) | 1.12 | 0.92-1.37 | 0.245 | 1.07 | 0.82-1.94 | 0.466 | |||
| Recessive | 746 (78.0) | 802 (79.2) | 1.08 | 0.87-1.33 | 0.511 | 0.98 | 0.88-1.37 | 0.685 |
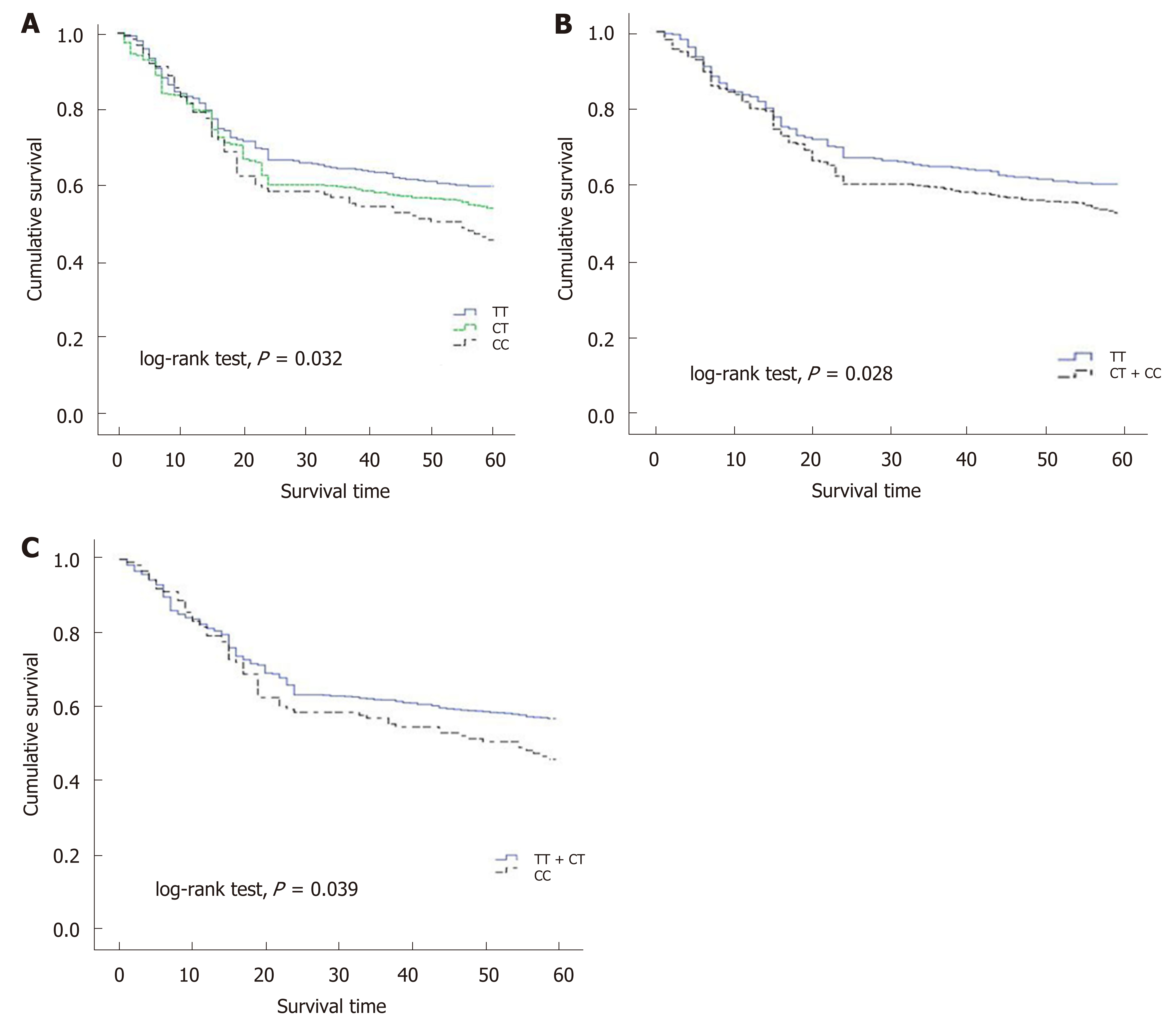
As the number of C alleles of rs2094258 increased, the survival rate of GC cases decreased (log-rank test, P = 0.032) (Table 3). Among all the cases, mortality was 45.4% with rs2094258 TT genotype (no C allele), 53.7% with CT genotype (one C allele), and 59.4% with CC genotype (two C alleles). The other four SNP loci (rs751402, Rs2296147, rs1047768, and rs873601) had no significant correlation with overall survival in patients with GC. For rs2094258, survival curves varied significantly with genotype (log-rank test, P = 0.032) (Figure 1A). GC patients with the rs2094258 CT + CC genotype showed a lower survival than patients with the TT genotype (log-rank test, P = 0.028) (Figure 1B). Cases with the CC genotype had a poorer prognosis than those with the TT + CT genotype (log-rank test, P = 0.039) (Figure 1C).
| SNP | Genotype | Case | Death (%) | 5-yr survival rate (%) | P-value1 |
| Rs2094258 | CC | 388 | 157 (40.5) | 59.4 | 0.032 |
| CT | 440 | 203 (46.1) | 53.7 | ||
| TT | 123 | 68 (55.3) | 45.4 | ||
| Rs751402 | CC | 363 | 162 (44.6) | 55.3 | 0.523 |
| CT | 465 | 216 (46.5) | 53.3 | ||
| TT | 121 | 50 (41.3) | 58.9 | ||
| Rs2296147 | TT | 589 | 281 (47.7) | 52.0 | 0.217 |
| CT | 297 | 124 (41.8) | 57.6 | ||
| CC | 50 | 23 (46.0) | 57.7 | ||
| Rs1047768 | TT | 501 | 243 (48.5) | 51.2 | 0.102 |
| CT | 375 | 152 (40.5) | 59.1 | ||
| CC | 67 | 33 (49.3) | 53.4 | ||
| Rs873601 | GG | 268 | 131 (48.9) | 51.2 | 0.362 |
| AG | 472 | 203 (43.0) | 56.8 | ||
| AA | 207 | 94 (45.4) | 54.7 |
In univariate survival analysis, age (P = 0.011), TNM stage (P < 0.001), no chemotherapy (P < 0.001), poor differentiation (P < 0.001), neural invasion (P < 0.001), and lymphovascular invasion (P < 0.001) were positively associated with the 5-year survival of GC cases (Table 4). Gender and H. pylori status were not significantly associated with the 5-year survival of GC cases. In multivariate analysis, the increase in the number of C alleles of rs2094258 (hazard ratio [HR] = 1.19, 95%CI: 1.02-1.45, P = 0.037), chemotherapy (HR = 0.81, 95%CI: 0.63-0.98; P = 0.041), differentiation (HR = 4.51, 95%CI: 1.99-8.23, P < 0.001), and lymphovascular invasion (HR = 1.97, 95%CI: 1.44-3.01, P < 0.001) were associated with survival in GC cases (Table 5).
| Variable | Classification | n | Death (%) | 5-yr survival rate (%) | P-value1 |
| Gender | Male | 667 | 303 (45.4) | 54.2 | 0.399 |
| Female | 289 | 125 (43.3) | 56.1 | ||
| Age (yr) | ≤60 | 569 | 235 (41.3) | 58.5 | 0.011 |
| >60 | 387 | 193 (49.9) | 49.7 | ||
| Drinking | No | 758 | 335 (44.2) | 55.6 | 0.259 |
| Yes | 198 | 93 (47.0) | 52.9 | ||
| Smoking | No | 560 | 244 (43.6) | 55.9 | 0.417 |
| Yes | 396 | 184 (46.5) | 53.3 | ||
| H. pylori infection | Negative | 281 | 118 (42.0) | 58.1 | 0.284 |
| Positive | 675 | 310 (45.9) | 51.4 | ||
| TNM stage | I | 178 | 18 (10.1) | 87.7 | <0.001 |
| II | 366 | 129 (35.2) | 66.9 | ||
| III | 404 | 281 (69.6) | 29.8 | ||
| Differentiation | Poor | 661 | 329 (49.8) | 50.0 | <0.001 |
| Moderate or high | 280 | 99 (35.4) | 66.7 | ||
| Lymphovascular invasion | Negative | 269 | 57 (21.2) | 79.6 | <0.001 |
| Positive | 675 | 371 (55.0) | 44.8 | ||
| Neural invasion | Negative | 396 | 132 (33.3) | 67.4 | <0.001 |
| Positive | 548 | 296 (54.0) | 45.7 | ||
| Chemotherapy | No | 361 | 281 (77.8) | 22.9 | <0.001 |
| Yes | 592 | 147 (24.8) | 75.1 |
We genotyped five functional SNPs in the XPG gene involved in the NER pathway and assessed their associations with GC risk and survival in China. Although rs2296147 was marginally associated with risk of GC (P = 0.05), we found no statistically significant associations. However, as the number of C alleles of rs2094258 increased, the survival of GC cases showed a decreasing trend. Poor differentiation, lymphovascular invasion, no chemotherapy, and increase in the number of C alleles of rs2094258 were associated with decreased survival among cases.
Few studies have explored the association of XPG SNPs with survival in GC patients. Our results are similar to those of Liu et al[25], who analysed the association between XPG SNPs and survival among 373 GC patients in China. That study found that in univariate model, the survival rate of those with the XPG rs2094258 AG genotype was higher than that of wild-type GG carriers (HR = 0.59, 95%CI: 0.39-0.90, P = 0.014), and that in multivariate analysis, the AA + AG genotype presented a significant survival advantage over the GG genotype (adjusted HR = 0.65, 95%CI: 0.44-0.97)[25]. Liu et al[25] also found that the AA + AG genotype of the XPG rs2094258 polymorphism improved survival in those with the following characteristics: Age more than 60, lymphatic metastasis, TNM stage III-IV, Borrmann III–IV, and diffuse-type gastric tumors[25].
Consistent with our study, a recent meta-analysis did not observe an overall association between the XPG rs2094258 SNPs and GC risk[26]. Some studies have found associations between the rs2094258 polymorphism and GC risk[22,26-28]. For example, Yang et al[23] assessed three XPG SNPs (rs2296147T>C, rs2094258C>T, and rs873601G>A) and found that the rs2094258 C>T polymorphism was associated with an increased GC risk. Meanwhile, H. pylori-infected individuals with the rs2094258 TT genotype had a much higher GC risk (OR = 2.13, 95%CI: 1.22-3.35, P for interaction = 0.030)[23]. However, H. pylori status did not modify associations with this SNP in our data. Varying cofactors among study populations, small sample size, the use of different PCR methods, or the low penetrance of this SNP may account for the inconsistent findings[15].
XPG rs2296147 was not associated with GC in the present study, a finding consistent with those of other studies[24,25]. However, significant associations between rs2296147 polymorphisms and GC risk were observed in Asian populations in numerous studies in one meta-analysis (CT vs TT: OR = 0.93, 95%CI: 0.87-0.99, P = 0.036)[29]. Further, the rs2296147 CC genotype was associated with a reduced risk of GC in a Chinese population (OR = 0.52, 95%CI: 0.27-0.97)[23]. These differences may be due to regional differences, small sample size, or heterogeneity of clinical feature[26].
The NER pathway belongs to the DNA repair pathways, and its functions include removing exogenous or endogenous DNA damage or adduct between chains and recruiting proliferating-cell nuclear antigen to the damage site for the subsequent gap-filling DNA synthesis[30]. DNA repair usually includes two stages of excision and repair synthesis[31]. The XPG gene locating at chromosome 13q33 consists of 15 exons[10], which is considered to cut the DNA at the 3’ terminus, initiate transcription-coupled DNA repair, and participate in RNA polymerase II transcription[32]. In the process of DNA repair, XPG binds to XPB as part of the transcription factor IIH (TFIIH) complex and strongly interacts with the TFIIH complex, a multi-subunit complex located at the intersection of transcription and DNA repair[33], which is involved in the DNA demethylation induced by overexpression of Gadd45a[10]. Meanwhile, XPG-related nucleases are used hierarchically for the excision of double-stranded rDNA break resection[34]. Recent report suggests that XPG incises the R-loop structure and participates in the RAD52-dependent resolution of DNA-RNA hybrids[35]. The rs1047768, rs751402, and rs2296147 are located in the exon 2, proximal promoter, and 5' untranslated region of the gene, respectively[36,37]. Based on the dbSNP database, the rs2094258 located at the XPG gene intron region participates in the initiation of the transcription-coupled DNA repair.
The limitations of this study are as follows. First, we could not explore and determine the exact mechanism by which XPG SNPs influence GC survival. Second, this study only examined five functional SNPs and did not include all the SNPs in the XPG gene that might play a key role in GC development. Third, selection bias and information bias are inherent threats to case-control studies, and cannot be ruled out in our study. Genetic factors have been shown to play key roles in the development, progression, and prognosis of GC. If confirmed by other studies, the results of our study suggest that XPG rs2094258 polymorphisms may serve as genetic biomarkers for GC prognosis, and may provide clues to biological underpinnings of GC progression.
Gastric cancer (GC) is one of the leading causes of cancer-related death worldwide, which causes a high social and economic burden. Xeroderma pigmentosum group G (XPG) or ERCC5 may play a key role in DNA repair, thus affecting cancer prognosis. Studies have shown that XPG SNPs may affect the development of cancer such as lung cancer, colorectal cancer, and breast cancer.
Only a few studies have explored the relationships between XPG gene SNPs and GC, and the results are inconsistent.
To examine GC risk and survival in relation to common functional XPG SNPs through a case-control study, which might improve our understanding of GC and provide new therapeutic targets for this malignancy.
A total of 956 histologically confirmed GC cases and 1012 controls were matched by sex, age (within 5 years), and residential district. Cases were followed and the survival time was recorded. DNA was extracted from peripheral blood samples of all cases and controls. XPG rs2094258, rs751402, rs2296147, rs1047768, and rs873601 polymorphisms were genotyped using PCR-RFLP. Logistic regression and Cox regression were used for analyzing associations of XPG SNPs with risk of GC and prognosis, respectively.
We found that GC patients with the rs2094258 CT + CC genotype showed a worse survival than those with the TT genotype, and patients with the CC genotype had a tendency of unfavorable prognosis compared with those with the TT + CT genotype. The increase in the number of C alleles of rs2094258 was associated with the long-term survival of GC cases. Other risk factors for survival included tumor differentiation, lymphovascular invasion, and use of chemotherapy. However, the exact mechanisms by which XPG SNPs influence GC survival remain to be solved.
The XPG rs2094258 polymorphism may be associated with overall survival in patients with GC.
If confirmed by other studies, the results of our study suggest that XPG rs2094258 polymorphisms may serve as genetic biomarkers for GC prognosis, and may provide clues to biological underpinnings of GC progression.
We thank all the subjects and researchers who participated in this study.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
STROBE Statement: The authors have read the STROBE Statement-checklist of items, and the manuscript was prepared and revised according to the STROBE Statement-checklist of items.
P-Reviewer: Can G, Virgilio E S-Editor: Yan JP L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-2664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1282] [Cited by in RCA: 1467] [Article Influence: 163.0] [Reference Citation Analysis (0)] |
| 2. | World Health Organization. Cancer [cited 12 September 2018]. Available from: http://www.who.int/mediacentre/factsheets/fs297/en/. |
| 3. | Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 274] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 4. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20511] [Article Influence: 2051.1] [Reference Citation Analysis (20)] |
| 5. | Liang J, Xu YY, Zhang C, Xia QR. Association of XPG gene rs751402 polymorphism with gastric cancer risk: A meta-analysis in the Chinese population. Int J Biol Markers. 2018;33:174-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Casamayor M, Morlock R, Maeda H, Ajani J. Targeted literature review of the global burden of gastric cancer. Ecancermedicalscience. 2018;12:883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 7. | Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 502] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 8. | Qian X, Tan H, Zhang J, Liu K, Yang T, Wang M, Debinskie W, Zhao W, Chan MD, Zhou X. Identification of biomarkers for pseudo and true progression of GBM based on radiogenomics study. Oncotarget. 2016;7:55377-55394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Apostolou Z, Chatzinikolaou G, Stratigi K, Garinis GA. Nucleotide Excision Repair and Transcription-Associated Genome Instability. Bioessays. 2019;41:e1800201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Sugasawa K. Xeroderma pigmentosum genes: Functions inside and outside DNA repair. Carcinogenesis. 2008;29:455-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Wakasugi M, Reardon JT, Sancar A. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997;272:16030-16034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 118] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Duan WX, Hua RX, Yi W, Shen LJ, Jin ZX, Zhao YH, Yi DH, Chen WS, Yu SQ. The association between OGG1 Ser326Cys polymorphism and lung cancer susceptibility: A meta-analysis of 27 studies. PLoS One. 2012;7:e35970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Lawania S, Singh N, Behera D, Sharma S. XPG polymorphisms and their association with lung cancer susceptibility, overall survival and response in North Indian patients treated with platinum-based doublet chemotherapy. Future Oncol. 2019;15:151-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Wang F, Zhang SD, Xu HM, Zhu JH, Hua RX, Xue WQ, Li XZ, Wang TM, He J, Jia WH. XPG rs2296147 T>C polymorphism predicted clinical outcome in colorectal cancer. Oncotarget. 2016;7:11724-11732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Su J, Zhu Y, Dai B, Yuan W, Song J. XPG Asp1104His polymorphism increases colorectal cancer risk especially in Asians. Am J Transl Res. 2019;11:1020-1029. [PubMed] |
| 16. | Wang H, Wang T, Guo H, Zhu G, Yang S, Hu Q, Du Y, Bai X, Chen X, Su H. Association analysis of ERCC5 gene polymorphisms with risk of breast cancer in Han women of northwest China. Breast Cancer. 2016;23:479-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | He J, Wang F, Zhu J, Zhang R, Yang T, Zou Y, Xia H. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. 2016;20:1481-1490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 110] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 18. | Al Sayed Ahmed H, Raslan WF, Deifalla AHS, Fathallah MD. Overall survival of classical Hodgkins lymphoma in Saudi patients is affected by XPG repair gene polymorphism. Biomed Rep. 2019;10:10-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 19. | Zavras AI, Yoon AJ, Chen MK, Lin CW, Yang SF. Association between polymorphisms of DNA repair gene ERCC5 and oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114:624-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 20. | Yang LQ, Zhang Y, Sun HF. Investigation on ERCC5 genetic polymorphisms and the development of gastric cancer in a Chinese population. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Lu JJ, Zhang HQ, Mai P, Ma X, Chen X, Yang YX, Zhang LP. Lack of association between ERCC5 gene polymorphisms and gastric cancer risk in a Chinese population. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Feng YB, Fan DQ, Yu J, Bie YK. Association between XPG gene polymorphisms and development of gastric cancer risk in a Chinese population. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Yang WG, Zhang SF, Chen JW, Li L, Wang WP, Zhang XF. SNPs of excision repair cross complementing group 5 and gastric cancer risk in Chinese populations. Asian Pac J Cancer Prev. 2012;13:6269-6272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Chen YZ, Guo F, Sun HW, Kong HR, Dai SJ, Huang SH, Zhu WW, Yang WJ, Zhou MT. Association between XPG polymorphisms and stomach cancer susceptibility in a Chinese population. J Cell Mol Med. 2016;20:903-908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 25. | Liu J, Deng N, Xu Q, Sun L, Tu H, Wang Z, Xing C, Yuan Y. Polymorphisms of multiple genes involved in NER pathway predict prognosis of gastric cancer. Oncotarget. 2016;7:48130-48142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Zhang Z, Yin J, Xu Q, Shi J. Association between the XPG gene rs2094258 polymorphism and risk of gastric cancer. J Clin Lab Anal. 2018;32:e22564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP, Wang YN, Sun MH, Zhou XY, Yang YJ, Wang JC, Jin L, Wei QY, Li J. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235-1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 28. | Hua RX, Zhuo ZJ, Zhu J, Jiang DH, Xue WQ, Zhang SD, Zhang JB, Li XZ, Zhang PF, Jia WH, Shen GP, He J. Association between genetic variants in the XPG gene and gastric cancer risk in a Southern Chinese population. Aging (Albany NY). 2016;8:3311-3320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Han C, Huang X, Hua R, Song S, Lyu L, Ta N, Zhu J, Zhang P. The association between XPG polymorphisms and cancer susceptibility: Evidence from observational studies. Medicine (Baltimore). 2017;96:e7467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Schärer OD. XPG: Its products and biological roles. Adv Exp Med Biol. 2008;637:83-92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Michalska MM, Samulak D, Romanowicz H, Jabłoński F, Smolarz B. Association between single nucleotide polymorphisms (SNPs) of XRCC2 and XRCC3 homologous recombination repair genes and ovarian cancer in Polish women. Exp Mol Pathol. 2016;100:243-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 32. | Clarkson SG. The XPG story. Biochimie. 2003;85:1113-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Kolesnikova O, Radu L, Poterszman A. TFIIH: A multi-subunit complex at the cross-roads of transcription and DNA repair. Adv Protein Chem Struct Biol. 2019;115:21-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 34. | Barnum KJ, Nguyen YT, O'Connell MJ. XPG-related nucleases are hierarchically recruited for double-stranded rDNA break resection. J Biol Chem. 2019;294:7632-7643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 35. | Kato R, Miyagawa K, Yasuhara T. The role of R-loops in transcription-associated DNA double-strand break repair. Mol Cell Oncol. 2019;6:1542244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 36. | Yoon AJ, Kuo WH, Lin CW, Yang SF. Role of ERCC5 polymorphism in risk of hepatocellular carcinoma. Oncol Lett. 2011;2:911-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 37. | Blomquist TM, Crawford EL, Willey JC. Cis-acting genetic variation at an E2F1/YY1 response site and putative p53 site is associated with altered allele-specific expression of ERCC5 (XPG) transcript in normal human bronchial epithelium. Carcinogenesis. 2010;31:1242-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |