Published online Aug 28, 2019. doi: 10.3748/wjg.v25.i32.4715
Peer-review started: March 28, 2019
First decision: April 16, 2019
Revised: July 14, 2019
Accepted: July 19, 2019
Article in press: July 19, 2019
Published online: August 28, 2019
Processing time: 153 Days and 21.4 Hours
Growth arrest-specific gene 2 (GAS2) plays a role in modulating in reversible growth arrest cell cycle, apoptosis, and cell survival. GAS2 protein is universally expressed in most normal tissues, particularly in the liver, but is depleted in some tumor tissues. However, the functional mechanisms of GAS2 in hepatocellular carcinoma (HCC) are not fully defined.
To investigate the function and mechanism of GAS2 in HCC.
GAS2 expression in clinic liver and HCC specimens was analyzed by real-time PCR and western blotting. Cell proliferation was analyzed by counting, MTS, and colony formation assays. Cell cycle analysis was performed by flow cytometry. Cell apoptosis was investigated by Annexin V apoptosis assay and western blotting.
GAS2 protein expression was lower in HCC than in normal tissues. Overexpression of GAS2 inhibited the proliferation of HCC cells with wide-type p53, while knockdown of GAS2 promoted the proliferation of hepatocytes (P < 0.05). Furthermore, GAS2 overexpression impeded the G1-to-S cell cycle transition and arrested more G1 cells, particularly the elevation of sub G1 (P < 0.01). Apoptosis induced by GAS2 was dependent on p53, which was increased by etoposide addition. The expression of p53 and apoptosis markers was further enhanced when GAS2 was upregulated, but became diminished upon downregulation of GAS2. In the clinic specimen, GAS2 was downregulated in more than 60% of HCCs. The average fold changes of GAS2 expression in tumor tissues were significantly lower than those in paired non-tumor tissues (P < 0.05).
GAS2 plays a vital role in HCC cell proliferation and apoptosis, possibly by regulating the cell cycle and p53-dependent apoptosis pathway.
Core tip: This study elucidated the function and mechanism of growth arrest-specific gene 2 (GAS2) in hepatocellular carcinoma (HCC) progression. By overexpression and knockdown approaches, we found that GAS2 inhibited the proliferation of HCC with wild-type p53. The effects of GAS2 on the cell cycle and apoptosis were investigated by flow cytometry, Annexin V apoptosis, and western blot assay, respectively. GAS2 suppressed HCC proliferation by deregulating the cell cycle and p53-dependent apoptosis pathway. Thus, GAS2 is expected to be a promising anti-oncogene and potential therapeutic target in HCC with wild-type p53.
- Citation: Zhu RX, Cheng ASL, Chan HLY, Yang DY, Seto WK. Growth arrest-specific gene 2 suppresses hepatocarcinogenesis by intervention of cell cycle and p53-dependent apoptosis. World J Gastroenterol 2019; 25(32): 4715-4726
- URL: https://www.wjgnet.com/1007-9327/full/v25/i32/4715.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i32.4715
Liver cancer is currently one of the most common causes of cancer-related deaths globally. Liver cancer has become a major health problem in developing countries, where the highest incidence and mortality rates are in Eastern and Southeastern Asia and parts of Africa[1,2]. Particularly, approximately 50% of liver cancer cases and deaths worldwide are in China. Among these, hepatocellular carcinoma (HCC) accounts for about 70% to 90% of liver cancers worldwide[3,4]. The prognosis of HCC is poor with high mortality because of limited effective treatment options[5].
Growth arrest-specific (GAS) genes encode proteins implicated directly in reversible growth arrest. This family contains seven genes (GAS1–GAS7) that encode proteins exhibiting distinct biochemical properties[6-8]. Among these genes, the protein encoded by GAS2 is a cell death substrate of caspase-3 that exerts some effects on modulating microfilament and cellular morphological variation during apoptosis[9,10]. Through microfilament alterations regulated by GAS2, the actin cytoskeleton and cell shape are quickly reset to respond to growth arrest induced by environmental stimuli such as apoptosis, different proliferative stimuli with various growth factors. Therefore, GAS2 as a component of the microfilament system functions in mitogenesis, cell cycle, cell growth, apoptosis, and cell survival.
GAS2 protein is broadly expressed in many normal tissues, and is particularly highly expressed in the liver; however, it is not expressed in some tumor tissues such as prostate and breast[11,12]. Although it has a potentially important role in cell survival, the role of GAS2 in HCC is largely unexplored. We hypothesized that GAS2 possesses anti-oncogenic properties in HCC cells.
Clinical liver specimens were derived from 54 HCC patients with surgical treatment at The University of Hong Kong–Shenzhen Hospital (Guangdong Sheng, China). HCC tissue samples were obtained from curative hepatic tumor resection except for necrotic and hemorrhagic areas. The paired adjacent non-tumor tissues were more than 5 cm away from the tumor edge, where was estimated to have no tumor invasion. All tissue samples were snap-frozen immediately after resection and stored in a -80°C nitrogen canister until use. This study was executed according to the ethical guidelines of the 1975 Declaration of Helsinki and was authorized by the Institutional Review Boards at The University of Hong Kong–Shenzhen Hospital [(2014)84]. All HCC patients provided signed informed consent giving approval for the use of clinical specimens for research purposes.
Human normal liver cell lines (LO2 and MIHA) and HCC cell lines (SK-Hep1, PLC5, Huh 7, and Hep3B) were obtained from the American Type Culture Collection (Manassas, VA, United States). All cells were routinely maintained in high-glucose Dulbecco’s Modified Eagle’s medium (Gibco, Gaithersburg, MD, United States) with 100 mL/L fetal bovine serum (Thermo Scientific HyClone, Buffalo, NY, United States) and 10 mL/L MEM Non-Essential Amino Acids (Gibco) at 37°C in a humidified incubator containing 50 mL/L CO2.
A total of 1 μg RNA sample extracted from samples by TriZol reagent (Invitrogen, Carlsbad, CA, United States) was mixed with DNase I (Invitrogen) and then synthesized to cDNA by SuperScript II reverse transcriptase (Invitrogen). Quantitative real-time PCR (qPCR) was performed on the 7500 Real-Time PCR System apparatus (Applied Biosystems, Framingham, MA, United States) to detect gene transcripts in the cDNA template mixed with SYBR Green (Applied Biosystems). The relative mRNA level of GAS2 (F5’-TG CAAATGCCCAAACAAGTTC-3’; GAS2-R5’-TTCTCCCACTCGGTATCTTCC TT-3’) was evaluated by relative quantification of an internal control gene expression based on a similar amplification efficiency. Statistical analyses were carried out by the two-tailed t-test.
pDEST40-GAS2 and pDEST40-CTRL were given by Prof. Yutaka Kondo (Nagoya, Japan)[10]. A density of 1-3 × 105 cells per well were seeded in a 6-well plate overnight to achieve 60%-80% confluency. Plasmids were transfected into cells using FuGene 6 reagent (Roche, Basel, Switzerland) at a ratio of 1:3 as per the manufacturer’s instructions. The transfected cells were incubated daily for 3 d. The optimal transfection efficiency was monitored in 2 d.
According to the manufacturer’s (HiPerfect; QIAGEN, Hilden, Germany) protocols, 50 nM small interfering RNAs (siRNAs) against GAS2, 100 nM siRNAs against p53 (ON-TARGET plus SMART pool; Thermo Fisher Scientific Ltd., Waltham, MA, United States), and a control sequence (siCtrl: 5’-UUCUCCGAACGUGUCACGU-3’) were transfected into the cell samples. These transfectants were added into a mixture of 100 μL serum-free culture medium with 12 μL transfection reagent (Hiper-Fect) and cultured daily for 3 d. The optimum interference efficiency was observed in 2 d.
Total protein lysates from tissue and cell samples were extracted by T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific) and lysis buffer containing a protease inhibitor cocktail (Roche), respectively. The protein concentration was measured by the Bradford method (Bio-Rad Laboratories, Hercules, CA, United States). Protein lysates (50 μg) were separated on an SDS-PAGE gel, and then transferred onto polyvinylidene difluoride membranes for western blot analysis. The various antibodies used were mouse anti-GAS2, mouse anti-P53, mouse anti-β-actin (Santa Cruz Biotechnology, CA, United States), rabbit anti-poly (ADP-ribose) polymerase (PARP), and rabbit anti-caspase3 (Cell Signaling Technology, Danvers, MA, United States). Signals were quantified by scanning densitometry.
Cell proliferation was assessed by the cell counting and MTS assays (Promega Biotech Co., Ltd., Madison, WI, United States). In cell counting, the number of cells was measured by Trypan blue dye exclusion daily for 5 consecutive days. In the MTS experiment, the cells subject to different transfections were incubated in a 96-well plate sextuplicate for 5 consecutive days. In daily counting, the MTS mixture with 100 μL fresh culture medium and 20 μL MTS solution was added into the cultured cells. The absorbance of the colorimetric products formed was determined by the Quant Microplate Spectrophotometer (BioTek Instruments, Inc., Winooski, VT, United States) at a 490 nm wavelength. All data were determined from three independent experiments.
The cells transfected with the labeled plasmids by FuGene 6 reagent was incubated in 6-well plates for 2 wk in G418-selective medium (Invitrogen Life Technologies, Carlsbad, CA, United States). The formative drug-resistant colonies stained by 2 mL/L crystal violet were counted under the microscope. All experiments were performed in three independent experiments.
Collected cell pellets fixed in ice-cold 700 mL/L ethanol-phosphate-buffered saline were stained with propidium iodide solution (50 μg/mL, Sigma-Aldrich, St. Louis, MO, United Stated). Cellular DNA content was measured by a flow cytometer with fluorescence-activated cell sorting (FACS) caliber (BD Biosciences, San Jose, CA, United States). The cell cycle profiles were analyzed using WinMDI2.9 software (WinMDI Version 2.9-Windows 3.95/DOS 5.0) in three independent experiments.
After treatment, cells collected were mixed with 5 μL Annexin V-APC and 5 μL 7-AAD according to the protocol of the APC Annexin V Apoptosis Detection Kit I (BD Pharmingen). Finally, the abovementioned mixture was added into 400 μL of 1× binding buffer and measured by flow cytometry in 1 h. These data were analyzed by WinMDI 2.9 software.
The data of cellular proliferation, cell cycle distribution, colony formation, apoptosis, and gene expression were determined by the independent Student’s t-test. The clinical relevance of GAS2 expression in HCCs and the matched non-tumorous liver tissues was analyzed by the non-parametric Wilcoxon’s matched pairs test. Scatterplot and related statistical analyses were performed using GraphPad Software (version 5.0). P values less than 0.05 were considered statistically significant.
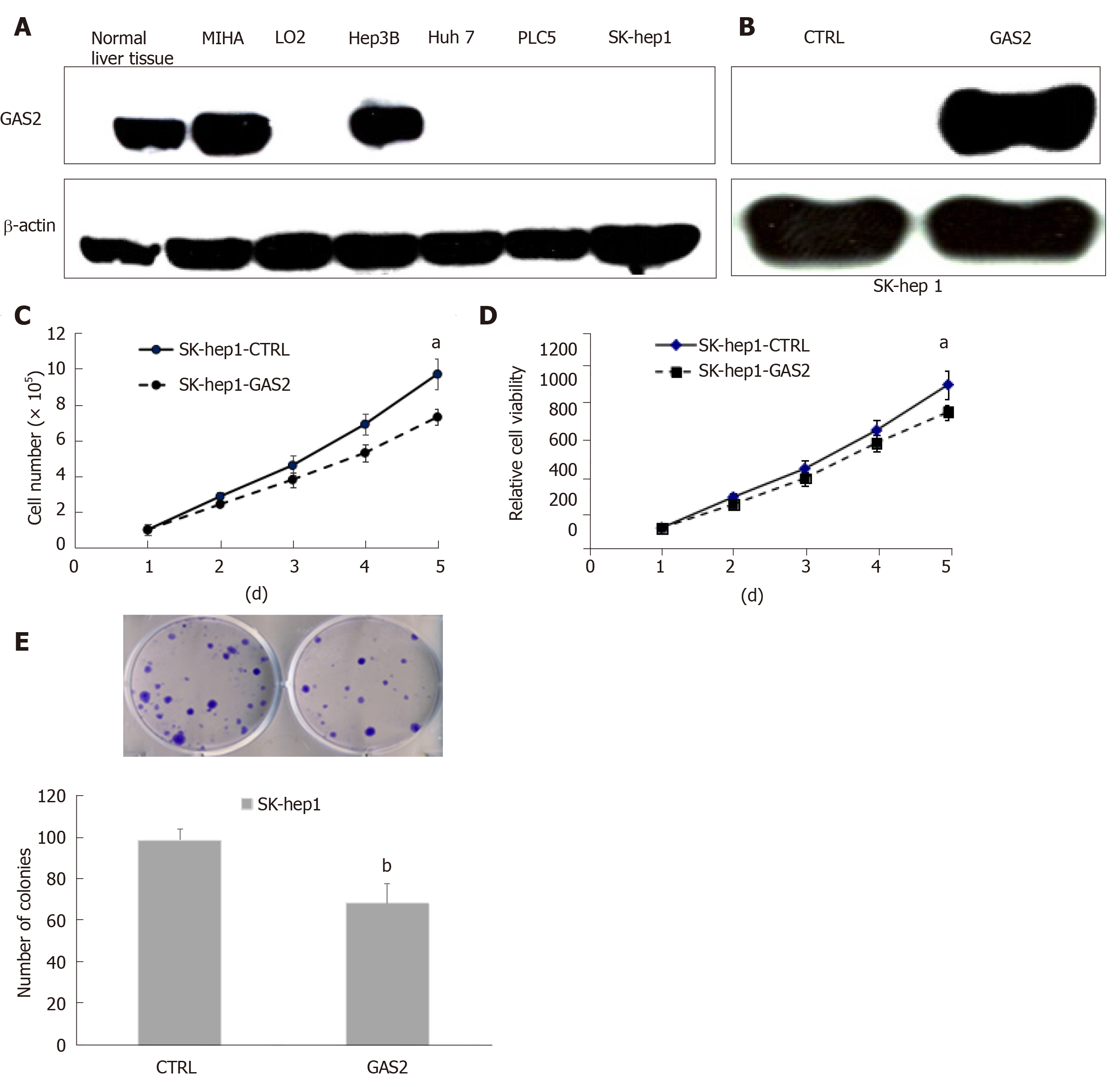
To investigate the roles of GAS2 in HCC, we first examined GAS2 expression in the liver normal and tumor tissues and its related cell lines. We found that GAS2 was highly expressed in most normal liver tissues and MIHA hepatocytes, while GAS2 was depleted in most tumor tissues and some HCC cell lines such as Huh7, PLC5, and SK-hep1 cells, with the exception of Hep3B (Figure 1A).
To further explore the functions of GAS2 in HCC development, we investigated the effect of ectopic expression of GAS2 on cell proliferation. We overexpressed GAS2 in HCC cells without endogenous GAS2, for example SK-hep1/Huh7/PLC5 (Figure 1B and Supplementary Figures 1A and Supplementary Figures 2A) and then analyzed cell viability by counting, MTS and colony formation assays. After a 48-h transfection, introduction of GAS2 suppressed cell growth rate in a time-dependent fashion compared with the empty vector control (P < 0.05; Figure 1C and D). Furthermore, GAS2 overexpression also notably reduced colony formation ability compared with the control (P < 0.01; Figure 1E). However, there were no significant differences in the proliferation of Huh7 and PLC5 cells (P > 0.05; Supplementary Figures 1B-D and Supplementary Figures 2B-D).
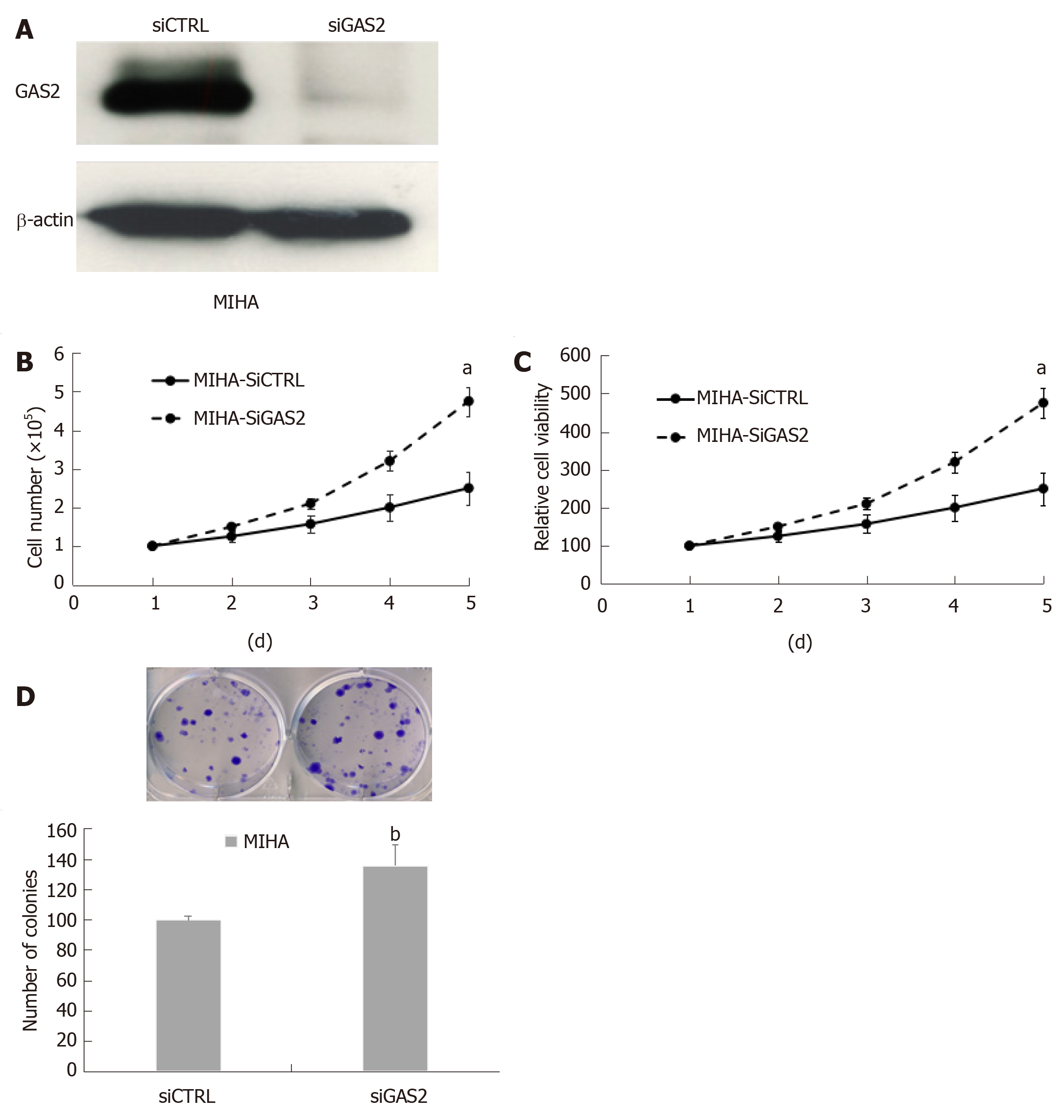
On the other hand, we used specific siRNA to knock down endogenous GAS2 expression in the MIHA and Hep3B cells, and then assessed the cell proliferation (Figure 2A and Supplementary Figures 3A). The results showed that the downregulation of GAS2 in MIHA cells caused a higher increase in cell viability (P < 0.05; Figure 2B and C) and colony formation ability in a time-dependent fashion compared with the control cells (P < 0.01, Figure 2D). But Hep3B without GAS2 did not grow significantly faster than the control cells (P > 0.05; Supplementary Figure 3B-D).
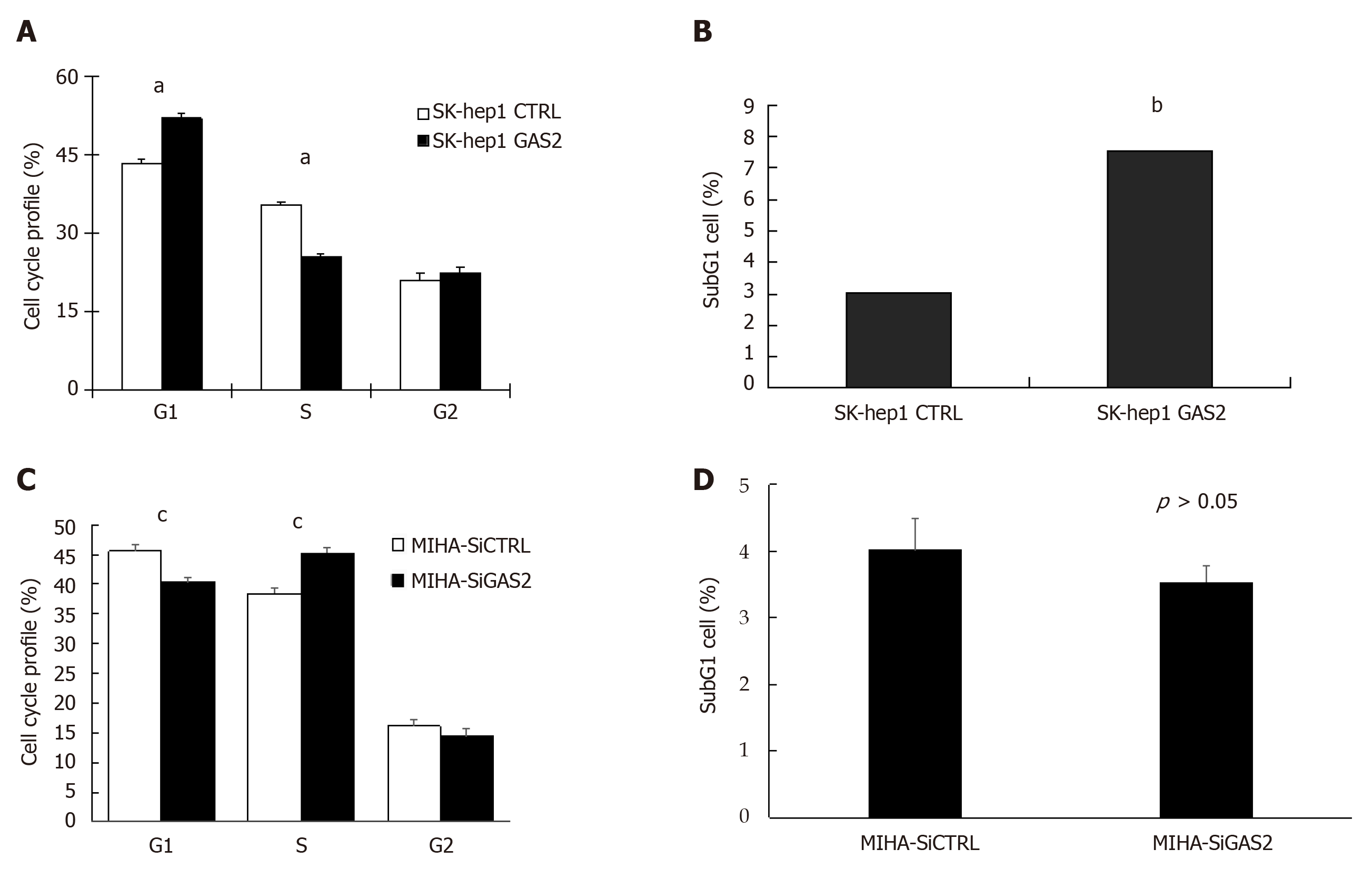
The growth-suppressive effect of GAS2 may depend on its activity to cell cycle progression. FACS analysis of GAS2-transfected SK-hep1cells revealed a significant increase in the population of G0/G1 phase cells compared to the control cells. Moreover, there was also a significant decrease in the population of S phase cells although the extent appears minimal (P < 0.01; Figure 3A). Therefore, this also implies GAS2 overexpression can impede G1-to-S cell cycle transition and arrest more G1 cells. More importantly, the significant elevation of subG1 cell population upon overexpression of GAS2-transfected SK-hep1 cells had a significantly more than 2-fold difference compared with the control SK-hep1 cells (P < 0.01; Figure 3B), suggesting that the growth inhibition caused by GAS2 was primarily related to apoptosis.
However, on the other hand, FACS analysis of GAS2 downregulation in MIHA cells showed that there was a decrease in the population of G0/G1 phase cells and an increase in the population of S phase cells compared to the control cells (P < 0.05; Figure 3C). There wasn’t the significant difference between G0/G1 phase and S phase, let alone subG1 phase (P > 0.05; Figure 3D).
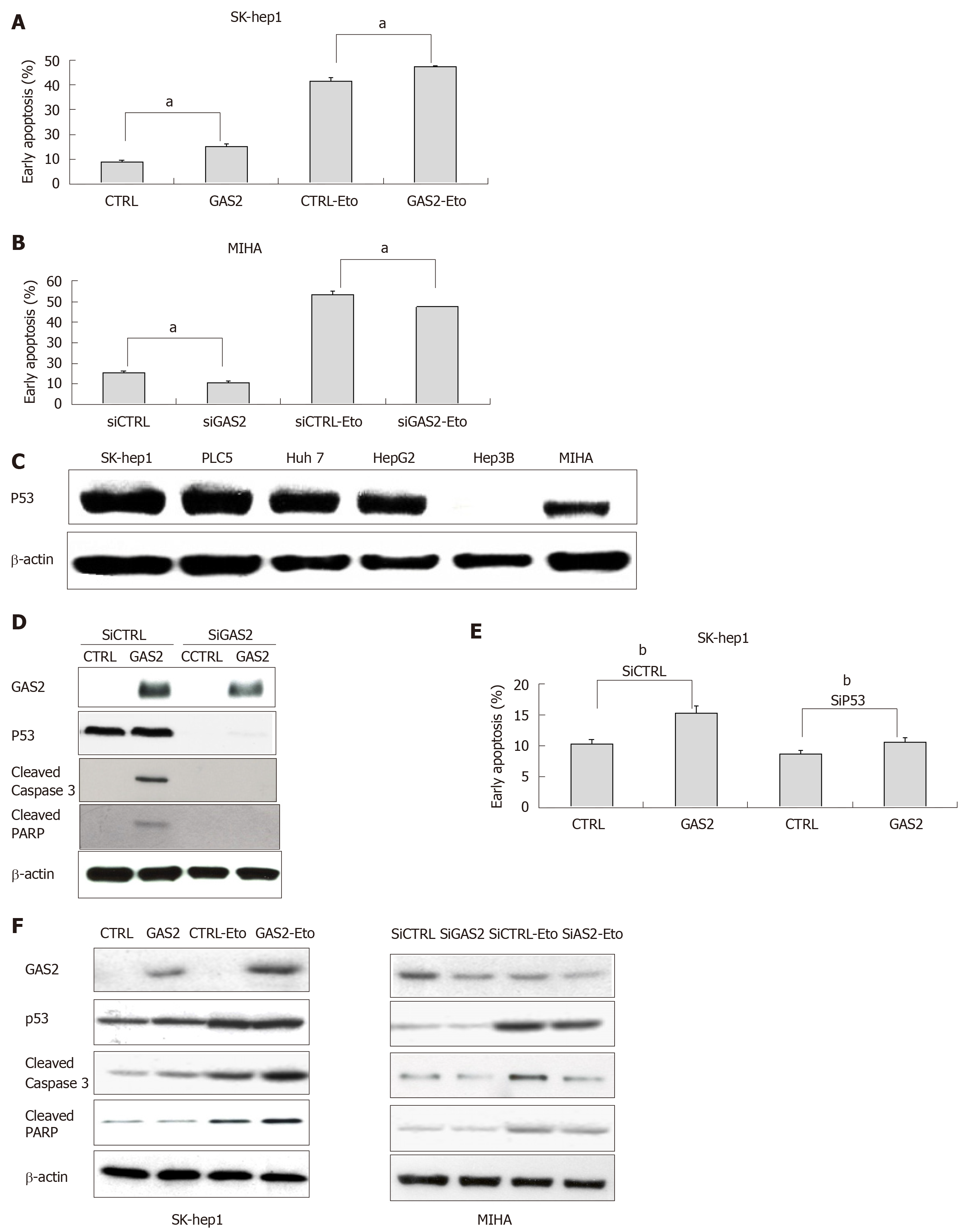
To determine if GAS2-dependent growth inhibition was related to apoptosis, additional examinations were performed on the functionally characterized SK-hep1 and MIHA cells, without or with endogenous GAS2 expression. Annexin V apoptosis assay showed that ectopic overexpression of GAS2 significantly increased the population of early apoptotic cells, and such promotion was further enlarged by etoposide, an activator promoter of p53-mediated apoptosis[13] (P < 0.05; Figure 4A). By contrast, downregulation of GAS2 in MIHA cells with specific siRNA to abolish endogenous GAS2 expression decreased early apoptosis (P < 0.05; Figure 4B). These findings showed that the anti-proliferative function of GAS2 was exerted through induction of apoptosis.
Because p53 plays an important role in apoptotic response, we next explored the possible function of p53 in GAS2-mediated apoptosis. Western blotting showed that p53 protein was expressed in normal hepatocyte cells (MIHA) and most HCC cell lines except Hep3B (Figure 4C). Treatment of siRNA against p53 (siP53) was significantly identified in SK-hep1-control and SK-hep1-GAS2 cells by western blotting. The levels of p53 and apoptosis markers, that is, cleaved caspase-3 and cleaved PARP were diminished after knockdown of p53 (Figure 4D). Annexin V apoptosis assay demonstrated that knockdown of p53 significantly inhibited early apoptosis in GAS2-SK-hep1 cells compared with control cells with abundant p53 expression (P < 0.05; Figure 4E). At the same time, in addition to p53, we also checked the apoptosis markers in the GAS2-overexpressed SK-Hep1 and GAS2-ablated MIHA cells with or without etoposide treatment. Overexpression of GAS2 and the treatment of etoposide can increase the levels of p53, cleaved caspase-3, and cleaved PARP. In addition, upregulation of GAS2 further enlarged the expression of p53 and apoptosis markers induced by etoposide treatment. Conversely, downregulation of GAS2 diminished the expression of p53 and apoptosis markers and the effect of etoposide treatment, relative to the corresponding controls (Figure 4F). Measurements of the resulting cells showed that the GAS2-dependent stimulation of apoptosis. These data indicate the existence of a p53-GAS2-caspase cascade and its function in cell growth retardation.
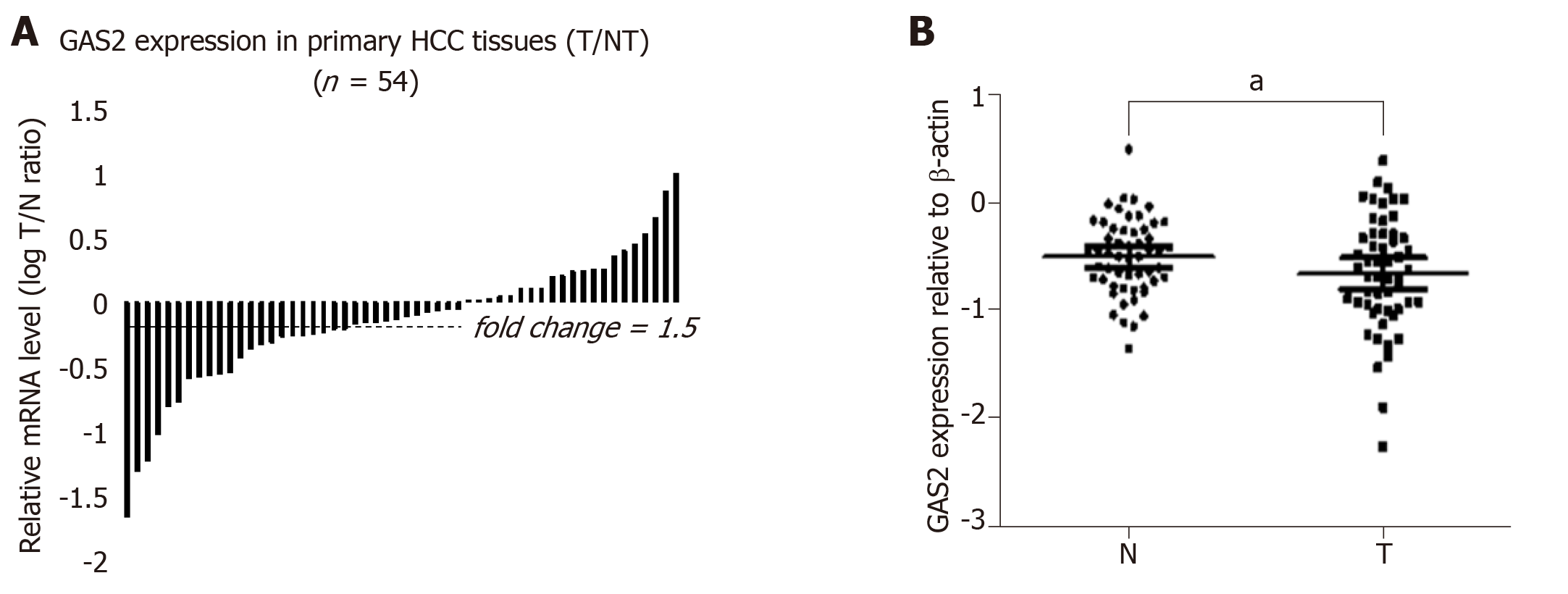
To further investigate the clinical relevance of GAS2 in human HCC tissues and their corresponding non-tumor liver tissues, we first examined the expression of GAS2 in 54 paired HCC and non-tumor liver samples by qPCR and western blot analysis. GAS2 expression was downregulated in 61.1% of HCC tissues (33 of 54) compared to the high basal levels in normal liver tissues. In contrast to the matched non-tumor tissues, marked downregulation (defined as greater than 1.5-fold change) of GAS2 was detected in 27of 54 samples (50%) (P < 0.05; Figure 5A). Regarding the relative GAS2 protein expression levels, the average fold changes of GAS2 expression in tumor tissues were significantly lower than those in the matched non-tumor tissues (P < 0.05; Figure 5B).
GAS2 is a member of the GAS gene family, which plays a role in modulating reversible growth arrest, mitogenesis, cell cycle, apoptosis, and cell survival. In 1988, Schneider et al[14] and Brancolini et al[15] found that GAS2 protein was highly expressed in serum-starved 3T3 fibroblasts and was phosphorylated by cysteine proteases during growth arrest. Its hyperphosphorylation is specifically relocalized to the appearance of membrane ruffles formed at the edges of the cells during G0-G1 transition[16]. In the course of apoptosis, the carboxyl-terminal domain of GAS2 polypeptide is phosphorylated by caspase enzymes. This proteolytical process is triggered by caspase-3 but not by caspase-2[9,17]. These aspartic-specific cysteine proteases are fundamental effectors of the apoptotic program, resulting in apoptosis by cleaving specific cellular targets or death substrates[18,19]. Removal of the carboxyl terminal domain of GAS2 dramatically performs the potential cell shape changes of the affected cells through reorganizing the microfilaments system in order to response to the growth arrest induced by environmental stimuli such as apoptosis[20]. As a consequence, some study thought that the processing of GAS2 throughout apoptosis by caspase enzymes could exert some critical effects on cell death.
The GAS2 gene maps to the human chromosome 11p15.2-p14.3, coding for a protein of 314 amino acids with a molecular weight of approximately 36 kDa[21]. It is expressed in many normal tissues, mostly in the liver, while it is absent in several cancer cell lines such as prostate cancer cells and breast cancer cells (MCF7, HCC1954)[11,12]. Although recent findings have revealed the functional links of GAS2 in cell survival, the mechanisms of GAS2 in HCC remain incompletely defined.
In our study, we found that GAS2 protein was expressed in most normal liver tissues and liver cell lines, while it was depleted in some HCC cell lines, for example, Huh7, PLC5 and Sk-hep1. In order to better define the effect of GAS2 in HCC development, we examined the functional consequences of GAS2 overexpression in a null-GAS2 expressing HCC cell line, SK-hep1, which carries wild-type functional p53. Although the lack of GAS2 and the presence of p53 were also found in the PLC5 and Huh7 cells, p53 mutation was reported in these two cell lines. PLC5 displayed Arg249Ser mutations in p53[22-24], while Tyr220Cys mutation of p53 was observed in Huh7[25-28]. Our subsequent functional analyses, by way of overexpression and knockdown approaches, illustrated that GAS2 inhibited the proliferation of HCC cells with wild-type functional p53. Furthermore, FACS analysis revealed overexpression of GAS2 induced more cell arrest in the G0/G1 phase and less cell population in the S phase, particularly the elevation of subG1, which may be primarily related to apoptosis.
The imbalance between cellular proliferation and apoptosis may contribute to carcinogenesis[29-31]. The interruption of apoptosis is likely one of the key mechanisms for promoting the transition from benign cells to cancer cells. The induction of apoptosis in SK-hep1 cells by GAS2 was also accompanied by the inhibition of cellular proliferation. It had been reported that GAS2 was a death substrate cleaved by caspase-3 and can efficiently increase cell susceptibility to apoptosis following UV irradiation, etoposide, and MMS treatments, which are dependent on increased p53 stability and transcription activity[32]. Our study showed that the combined GAS2 overexpression and etoposide treatment had an additive effect in promoting apoptosis in SK-hep1 cells with wild-type functional p53 compared with cells treated with etoposide alone. The presence of etoposide might accelerate the proteolysis of GAS2. The molecular basis of p53-dependent apoptotic pathway in HCC cells was also analyzed by the expression of apoptosis markers, that is, cleaved caspase-3 and cleaved PARP functioned in the execution of the intrinsic mitochondrial apoptotic pathway. The proteolytic cleavage of PARP facilitated cellular disassembly and undergone apoptosis[33,34]. According to our data, with the presence of p53, overexpression of GAS2 could increase the level of cleaved caspase-3 and cleaved PARP induced by etoposide. Without p53, overexpression of GAS2 was invalid in the level of cleaved caspase-3 and cleaved PARP induced by etoposide. Likewise, without GAS2 overexpression, these apoptosis markers were attenuated. This implies that p53-dependent apoptotic pathway induced by GAS2 triggers a p53-GAS2-caspase cascade effect that initiates the cell death program.
Furthermore, further validation of GAS2 in a clinic 54 pairs of HCC and their matched adjacent non-tumor liver tissues subset is warranted. We found that GAS2 was downregulated in more than 60% of HCCs. Their average fold changes of GAS2 expression in tumor tissues were significantly lower than those in the matched non-tumor tissues. Of note, HCC is an extremely heterogenous disease, displaying extensive histologic, transcriptomic and genetic diversity. On the genetic level, the most common mutant genes are TP53 (encoding p53 protein) and CTNNB1 (encoding-catenin protein), both mutated in 20%-40% of HCCs. Particularly, the frequency of TP53 mutation in HCC ranges from 22% to 33%[35-38]. The p53 protein manipulates various molecular functions in cell such as DNA synthesis and repair, cell cycle arrest, senescence, and apoptosis[39]. Since the role of GAS2 in cell proliferation, cell cycle, and apoptosis was dependent on wild-type p53, together with our clinical data, we speculated that the signal pathway of p53-GAS2 molecular axis might be a primary tumorigenic mechanism in HCCs with wild-type p53. Of course, there other silencing GAS2-targeted signal pathways in HCC development need to be considered.
Together, our multiple functional experiments reveal that the anti-proliferative nature of GAS2 may be one of the major hepatocarcinogenesis mechanisms. We further characterized that GAS2 inhibited HCC cell proliferation possibly via enhancing susceptibility to p53-dependent apoptosis. Accordingly, our findings not only enhance our understanding of the mechanisms of liver carcinogenesis, but also provide potential therapeutic targets for this aggressive malignancy.
Hepatocellular carcinoma (HCC) is the most common primary liver cancer, and is a leading cause of cancer-related mortality in China. The prognosis of HCC is poor with high mortality because of limited options of effective treatment. Thus, new therapeutic targets that may confer survival benefit are urgently needed in HCC.
Growth arrest-specific gene 2 (GAS2) is a member of the GAS gene family, which is universally expressed in most normal tissues, particularly in the liver, but is depleted in some tumor tissues. However, the functional mechanisms of GAS2 in HCC are not fully defined.
The aim of this study was to investigate the role of GAS2 in the liver and HCC and its underlying mechanism.
GAS2 expression was examined by real-time PCR and western blotting in tissues and cells. The proliferation of GAS2 expression was analyzed by counting, MTS, and colony formation assays. Cell cycle analysis was performed by flow cytometry. Cell apoptosis was investigated by the Annexin V apoptosis assay.
GAS2 protein expression was more downregulated in HCC than in normal tissues. Overexpression of GAS2 inhibited the proliferation of HCC cells with wild-type p53 and knockdown of GAS2 showed the opposite effects. The more arrested G1 cells in the cell cycle and p53-GAS2 caspase cascade might be involved in the oncogenic function of GAS2 in HCC.
The study showed that GAS2 suppressed the proliferation and apoptosis of HCC cells, and the possible mechanism was by regulating the cell cycle and p53-dependent apoptosis pathway. Thus, GAS2 is expected to be an important anti-oncogene and potential therapeutic target in HCC.
The function and mechanism of GAS2 in HCC development has been confirmed, and the significance of GAS2 as a promising therapeutic target for HCC with wild-type p53 is highlighted.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ozaki I, Ratnasari N S-Editor: Yan JP L-Editor: Filipodia E-Editor: Qi LL
| 1. | Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev. 2016;25:16-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2004] [Cited by in RCA: 2488] [Article Influence: 248.8] [Reference Citation Analysis (0)] |
| 2. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21366] [Article Influence: 2136.6] [Reference Citation Analysis (3)] |
| 3. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20511] [Article Influence: 2051.1] [Reference Citation Analysis (20)] |
| 4. | Zhu RX, Seto WK, Lai CL, Yuen MF. Epidemiology of Hepatocellular Carcinoma in the Asia-Pacific Region. Gut Liver. 2016;10:332-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 266] [Cited by in RCA: 362] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 5. | Testino G, Leone S, Borro P. Alcohol and hepatocellular carcinoma: A review and a point of view. World J Gastroenterol. 2014;20:15943-15954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 79] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (2)] |
| 6. | Sarkar S, Poon CC, Mirzaei R, Rawji KS, Hader W, Bose P, Kelly J, Dunn JF, Yong VW. Microglia induces Gas1 expression in human brain tumor-initiating cells to reduce tumorigenecity. Sci Rep. 2018;8:15286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Shan S, Liu Z, Guo T, Wang M, Tian S, Zhang Y, Wang K, Zheng H, Zhao X, Zuo P, Wang Y, Li D, Liu C. Growth arrest-specific gene 6 transfer promotes mesenchymal stem cell survival and cardiac repair under hypoxia and ischemia via enhanced autocrine signaling and paracrine action. Arch Biochem Biophys. 2018;660:108-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Xu Q, Liu X, Wang X, Hua Y, Wang X, Chen J, Li J, Wang Y, Stoeger T, Chen S, Huang N. Growth arrest-specific protein 7 regulates the murine M1 alveolar macrophage polarization. Immunol Res. 2017;65:1065-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Cui J, Chen B, Wang H, Han Y, Chen X, Zhang W. Glucosidase II β-subunit, a novel substrate for caspase-3-like activity in rice, plays as a molecular switch between autophagy and programmed cell death. Sci Rep. 2016;6:31764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Huang W, Bei L, Hjort EE, Eklund EA. Decreased calpain activity in chronic myeloid leukemia impairs apoptosis by increasing survivin in myeloid progenitors and xiap1 in differentiating granulocytes. Oncotarget. 2017;8:50629-50641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 476] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 12. | Allegrucci C, Rushton MD, Dixon JE, Sottile V, Shah M, Kumari R, Watson S, Alberio R, Johnson AD. Epigenetic reprogramming of breast cancer cells with oocyte extracts. Mol Cancer. 2011;10:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 13. | Sun B, Ross SM, Rowley S, Adeleye Y, Clewell RA. Contribution of ATM and ATR kinase pathways to p53-mediated response in etoposide and methyl methanesulfonate induced DNA damage. Environ Mol Mutagen. 2017;58:72-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 724] [Cited by in RCA: 796] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 15. | Brancolini C, Bottega S, Schneider C. Gas2, a growth arrest-specific protein, is a component of the microfilament network system. J Cell Biol. 1992;117:1251-1261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Brancolini C, Schneider C. Phosphorylation of the growth arrest-specific protein Gas2 is coupled to actin rearrangements during Go-G1 transition in NIH 3T3 cells. J Cell Biol. 1994;124:743-756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Brancolini C, Lazarevic D, Rodriguez J, Schneider C. Dismantling cell-cell contacts during apoptosis is coupled to a caspase-dependent proteolytic cleavage of beta-catenin. J Cell Biol. 1997;139:759-771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 176] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 18. | Cieplak P. Letter to the Editor: Caspase cleavage sites in the human proteome: CaspDB, a database of predicted substrates. Apoptosis. 2015;20:421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 19. | Kumar S, van Raam BJ, Salvesen GS, Cieplak P. Caspase cleavage sites in the human proteome: CaspDB, a database of predicted substrates. PLoS One. 2014;9:e110539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 20. | Brancolini C, Benedetti M, Schneider C. Microfilament reorganization during apoptosis: The role of Gas2, a possible substrate for ICE-like proteases. EMBO J. 1995;14:5179-5190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 184] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 21. | Collavin L, Buzzai M, Saccone S, Bernard L, Federico C, DellaValle G, Brancolini C, Schneider C. cDNA characterization and chromosome mapping of the human GAS2 gene. Genomics. 1998;48:265-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 22. | Perdrix A, Najem A, Saussez S, Awada A, Journe F, Ghanem G, Krayem M. PRIMA-1 and PRIMA-1 Met (APR-246): From Mutant/Wild Type p53 Reactivation to Unexpected Mechanisms Underlying Their Potent Anti-Tumor Effect in Combinatorial Therapies. Cancers (Basel). 2017;9:pii: E172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 23. | Zhang B, Zhang Y, Zou X, Chan AW, Zhang R, Lee TK, Liu H, Lau EY, Ho NP, Lai PB, Cheung YS, To KF, Wong HK, Choy KW, Keng VW, Chow LM, Chan KK, Cheng AS, Ko BC. The CCCTC-binding factor (CTCF)-forkhead box protein M1 axis regulates tumour growth and metastasis in hepatocellular carcinoma. J Pathol. 2017;243:418-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 24. | Fang SC, Hsu CL, Lin HT, Yen GC. Anticancer effects of flavonoid derivatives isolated from Millettia reticulata Benth in SK-Hep-1 human hepatocellular carcinoma cells. J Agric Food Chem. 2010;58:814-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Dixit U, Pandey AK, Liu Z, Kumar S, Neiditch MB, Klein KM, Pandey VN. Correction for Dixit et al., "FUSE Binding Protein 1 Facilitates Persistent Hepatitis C Virus Replication in Hepatoma Cells by Regulating Tumor Suppressor p53". J Virol. 2017;91:pii: e01609-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Guo J, Ozaki I, Xia J, Kuwashiro T, Kojima M, Takahashi H, Ashida K, Anzai K, Matsuhashi S. PDCD4 Knockdown Induces Senescence in Hepatoma Cells by Up-Regulating the p21 Expression. Front Oncol. 2019;8:661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Gomes AR, Abrantes AM, Brito AF, Laranjo M, Casalta-Lopes JE, Gonçalves AC, Sarmento-Ribeiro AB, Botelho MF, Tralhão JG. Influence of P53 on the radiotherapy response of hepatocellular carcinoma. Clin Mol Hepatol. 2015;21:257-267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 28. | Nazim UM, Park SY. Attenuation of autophagy flux by 6-shogaol sensitizes human liver cancer cells to TRAIL-induced apoptosis via p53 and ROS. Int J Mol Med. 2019;43:701-708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Matsuura K, Canfield K, Feng W, Kurokawa M. Metabolic Regulation of Apoptosis in Cancer. Int Rev Cell Mol Biol. 2016;327:43-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 128] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 30. | Belotti EM, Stassi AF, Velázquez MML, Díaz PU, Marelli BE, Rey F, Notaro US, Ortega HH, Salvetti NR. Changes in the Proliferation/Apoptosis Balance in the Bovine Ovary: A Key Early Event in Follicular Persistence. Cells Tissues Organs. 2017;204:314-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Conover CA. The IGF-p53 connection in cancer. Growth Horm IGF Res. 2018;39:25-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Benetti R, Del Sal G, Monte M, Paroni G, Brancolini C, Schneider C. The death substrate Gas2 binds m-calpain and increases susceptibility to p53-dependent apoptosis. EMBO J. 2001;20:2702-2714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 85] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 33. | Matsuno Y, Hyodo M, Fujimori H, Shimizu A, Yoshioka KI. Sensitization of Cancer Cells to Radiation and Topoisomerase I Inhibitor Camptothecin Using Inhibitors of PARP and Other Signaling Molecules. Cancers (Basel). 2018;10:pii: E364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 34. | Zheng CC, Hu HF, Hong P, Zhang QH, Xu WW, He QY, Li B. Significance of integrin-linked kinase (ILK) in tumorigenesis and its potential implication as a biomarker and therapeutic target for human cancer. Am J Cancer Res. 2019;9:186-197. [PubMed] |
| 35. | Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 713] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 36. | Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 536] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 37. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1340] [Article Influence: 134.0] [Reference Citation Analysis (0)] |
| 38. | Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu.; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1729] [Article Influence: 216.1] [Reference Citation Analysis (1)] |
| 39. | Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5100] [Cited by in RCA: 5114] [Article Influence: 204.6] [Reference Citation Analysis (0)] |