Published online Aug 21, 2019. doi: 10.3748/wjg.v25.i31.4300
Peer-review started: April 15, 2019
First decision: May 16, 2019
Revised: May 31, 2019
Accepted: July 19, 2019
Article in press: July 19, 2019
Published online: August 21, 2019
Processing time: 130 Days and 7.6 Hours
Methionine adenosyltransferases (MATs) are essential enzymes for life as they produce S-adenosylmethionine (SAMe), the biological methyl donor required for a plethora of reactions within the cell. Mammalian systems express two genes, MAT1A and MAT2A, which encode for MATα1 and MATα2, the catalytic subunits of the MAT isoenzymes, respectively. A third gene MAT2B, encodes a regulatory subunit known as MATβ which controls the activity of MATα2. MAT1A, which is mainly expressed in hepatocytes, maintains the differentiated state of these cells, whilst MAT2A and MAT2B are expressed in extrahepatic tissues as well as non-parenchymal cells of the liver (e.g., hepatic stellate and Kupffer cells). The biosynthesis of SAMe is impaired in patients with chronic liver disease and liver cancer due to decreased expression and inactivation of MATα1. A switch from MAT1A to MAT2A/MAT2B occurs in multiple liver diseases and during liver growth and dedifferentiation, but this change in the expression pattern of MATs results in reduced hepatic SAMe level. Decades of study have utilized the Mat1a-knockout (KO) mouse that spontaneously develops non-alcoholic steatohepatitis (NASH) and hepatocellular carcinoma (HCC) to elucidate a variety of mechanisms by which MAT proteins dysregulation contributes to liver carcinogenesis. An increasing volume of work indicates that MATs have SAMe-independent functions, distinct interactomes and multiple subcellular localizations. Here we aim to provide an overview of MAT biology including genes, isoenzymes and their regulation to provide the context for understanding consequences of their dysregulation. We will highlight recent breakthroughs in the field and underscore the importance of MAT’s in liver tumorigenesis as well as their potential as targets for cancer therapy.
Core tip: In this review we provide the most comprehensive guide to methionine adenosyltransferases discussing their structures, functions and consequences of dysregulation in liver cancers emphasizing their potential as prognostic biomarkers for liver cancers and as targets for liver cancer therapy.
- Citation: Murray B, Barbier-Torres L, Fan W, Mato JM, Lu SC. Methionine adenosyltransferases in liver cancer. World J Gastroenterol 2019; 25(31): 4300-4319
- URL: https://www.wjgnet.com/1007-9327/full/v25/i31/4300.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i31.4300
This review examines the roles of methionine adenosyltransferases (MATs) in liver cancers with a focus on how dysregulation of these proteins contributes to their pathogenesis. Hepatocellular carcinoma (HCC) is the most common primary liver cancer and currently the second leading cause of cancer-related death worldwide[1]. In the majority of the cases, HCC develops in patients with underlying chronic liver disease and cirrhosis, which are mainly derived from viral hepatitis infection, alcohol abuse and non-alcoholic fatty liver disease (NAFLD)[2]. The threat of HCC is expected to continue rising due to increasing cases of NAFLD given the obesity epidemic occurring worldwide[3]. Even more alarming are reports of HCC from NAFLD that occurred in the absence of cirrhosis[4]. Cholangiocarcinoma (CCA) is the second most common primary liver cancer, which typically occur in the setting of chronic biliary inflammation. MATs exert very similar roles in both types of liver cancers.
MATs (EC 2.5.1.6) belong to a family of enzymes that are essential to life as they catalyze the biosynthesis of SAMe, the main methyl donor of the cell. Apart from methylation, SAMe is also important as a precursor in glutathione (GSH) and polyamine synthesis[5]. As SAMe is used for the methylation of biomolecules including DNA, RNA, proteins, biological amines and phospholipids, any change in its biosynthesis and catabolism within the cell has a profound effect on cellular processes such as growth, differentiation and response to injury. Over the past decade SAMe-independent roles of MAT enzymes have emerged, most notably, their ability to function as bona fide transcription factors as well as their ability to form parts of scaffold complexes which have been implicated in cancer development[6-9].
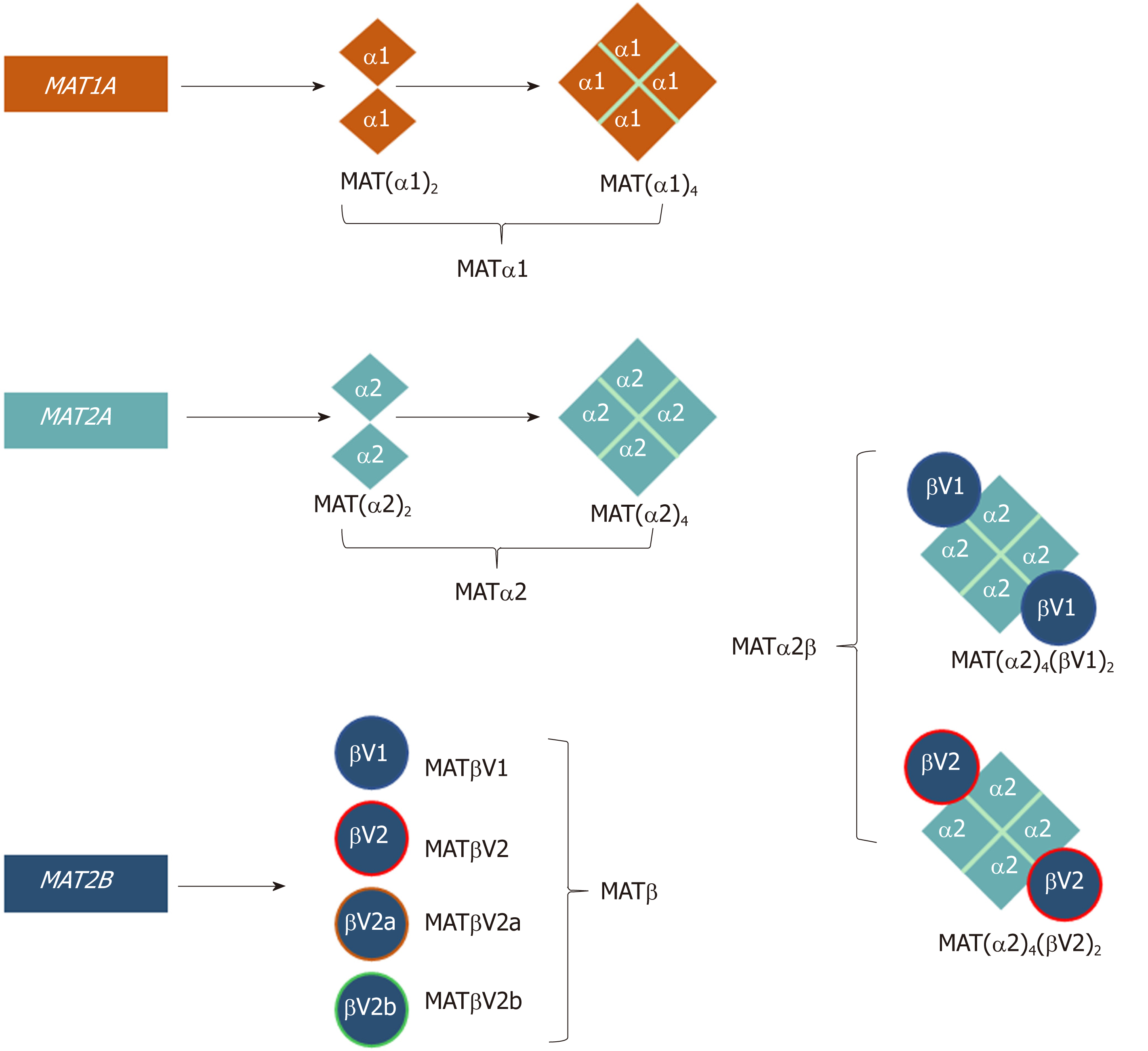
In mammals, three distinct genes, MAT1A, MAT2A, and MAT2B give rise to the protein products MATα1, MATα2, and MATβ, respectively (Table 1, Figure 1)[10]. MAT1A encodes for a 395-amino acid (396 in mouse, and 397 in rat) catalytic subunit in humans that is mainly expressed in hepatocytes (parenchymal cells of the liver) as well as bile duct epithelial cells and pancreatic acinar cells[8,11]. MATα1 can oligomerize to form a homotetramer (MATIII) or homodimer (MATI). The MATα2 subunit, the extrahepatic catalytic protein (395 amino acids), that has an 84% sequence identity to MATα1, can also form dimers and tetramers but with a bias towards the dimer conformation[10,12,13]. MATα2 is expressed in non-parenchymal cells of the liver as well as in all extrahepatic tissues. The regulatory subunit MATβ has four known isoforms, MATβV1, MATβV2, MATβV2a, and MATβV2b with the former two being the major splice variants[14]. MATβV2a and MATβV2b are expressed at very low levels compared to MATβV2 and have not been studied in detail[14]. MATβV1 is expressed in fetal liver, prostate, lung, brain, thyroid and the adrenal gland, whilst MATβV2 is expressed in skeletal muscle and heart. Both isoforms are expressed in the kidney and thymus[14]. MATβV1 and MATβV2 are 331 and 323 amino acids long, respectively, differ only by 20 amino acids at their N-terminus and share little sequence identity (7%) to the catalytic MAT proteins. MATβ has a binding pocket for the cofactor NADP+, which can interact with both major isoforms[12,15].
| Gene | Protein | Amino acids | Alternative names | Oligomeric state | Regulatory subunit | Co-factors | Km for methionine | Km for ATP | Ki for SAMe | X-ray structure |
| MAT1A | MATα1 | 395 | MATIII MAT(α1)2 | Dimer | No | 210 μmol/L-7 mmol | 1-3 mmol | None | 20BV | |
| MATI MAT(α1)4 | Tetramer | No | 23 μmol/L-1 mmol | 0.2-0.5 mmol | 400 μmol/L | |||||
| MAT2A | MATα2 | 395 | MAT(α2)2 | Dimer | 4-10 μmol/L | 70 μmol/L | 60 μmol/L | 5A19 | ||
| MAT(α2)4 | Tetramer | 5A1I | ||||||||
| MAT2B | MATβV1 | 334 | Monomeric | NADP+ | ||||||
| MATβV2 | 323 | Monomeric | NADP+ | 2YDX |
MAT enzymes were first thought to function as SAMe producing factories in the cytosol, and that SAMe would be delivered to the specific compartments such as the nucleus or mitochondria for methylation reactions[16]. However, a decade ago MATα1 was reported to be present within the nucleus[17]. This was followed by publications describing MATα2, MATβV1 and MATβV2 to also be within this organelle[6,18]. These publications showed that a nuclear location of MAT proteins was associated with enhanced histone H3K27 methylation, an epigenetic modification that leads to gene silencing[17]. Most recently MATα1 was reported to be present in the mitochondrial matrix of hepatocytes, enhancing mitochondrial function and negatively regulating cytochrome P450 2E1 (CYP2E1) through methylation[19]. These accumulating publications reinforce the concept that MATs are recruited to subcellular compartments to provide a local source of SAMe.
To date there are numerous crystal structures of MAT proteins from different species in different active site conformations, complexed with substrates, products, or their analogues[12,20-23]. The catalytic proteins have a three-domain organization that is conserved amongst other MAT family subunits[21,22,24]. Monomeric MAT enzymes are incapable of producing SAMe as they do not contain a complete active site. Upon dimerization, both monomers contribute residues to form two active sites. The large hydrophobic surface of monomeric MAT constitutes the site of the monomer-monomer interface. A common feature of MAT enzymes is that they contain a “gating loop” (in human MATα1 and α2 residues 113-131) that flanks the active site, which has been hypothesized to move dynamically to allow access to the active site[21]. When the active site is occupied, the loop closes to form a gate over the active site, but when the active site is empty, the loop becomes disordered or open. MATα1 dimers can also form tetramers through the central domain of the subunits[24]. Recent work has shown that mutation to residues of the gating loop can reduce enzyme activity and SAMe formation[25].
The regulatory β‐subunit is structurally very different from the α‐subunits, unable to produce SAMe by itself. MATβ proteins contain signature motifs of the SDR (short-chain dehydrogenase/reductase) superfamily including a Rossman fold that can bind FAD+ or NADP+ although MATβ favors the latter[22]. MATα2 and MATβ interact to give rise to the MATα2β complexes (also referred to as MATII)[12,26] (Figure 1, Table 1). To date only the structure of a MATα2βV2 complex has been solved and it consists of a MATα2 tetramer flanked by two MATβV2 subunits (MATα2(4)βV2(2)). This showed that MATα2 can exist and function as a tetramer in the presence of MATβ[12]. The oligomeric state of this crystal structure, confirmed with small angle x-ray scattering[12], is different from the suggested tetrameric form [MAT(α2)2(β)2][27] or the proposed computational model in which MATαβ was assumed to be a trimer [MAT(α2)2(β)1][15]. Mutational analysis showed by gel filtration that the minimum motif required for the formation of the MAT(α2)4(βV2)2 complex comprises three residues at the end of the C-terminus of MATβV2 (V321F322H323)[12]. Several publications, using recombinant purified proteins, have shown that MATα1 can also interact with MATβV1, although this interaction is several orders of magnitude weaker than that of MATα2 and MATβ[12,25]. The MATα1βV1 complex is not likely to occur within the cell as MATα1 and MATβ are generally not expressed at the same time.
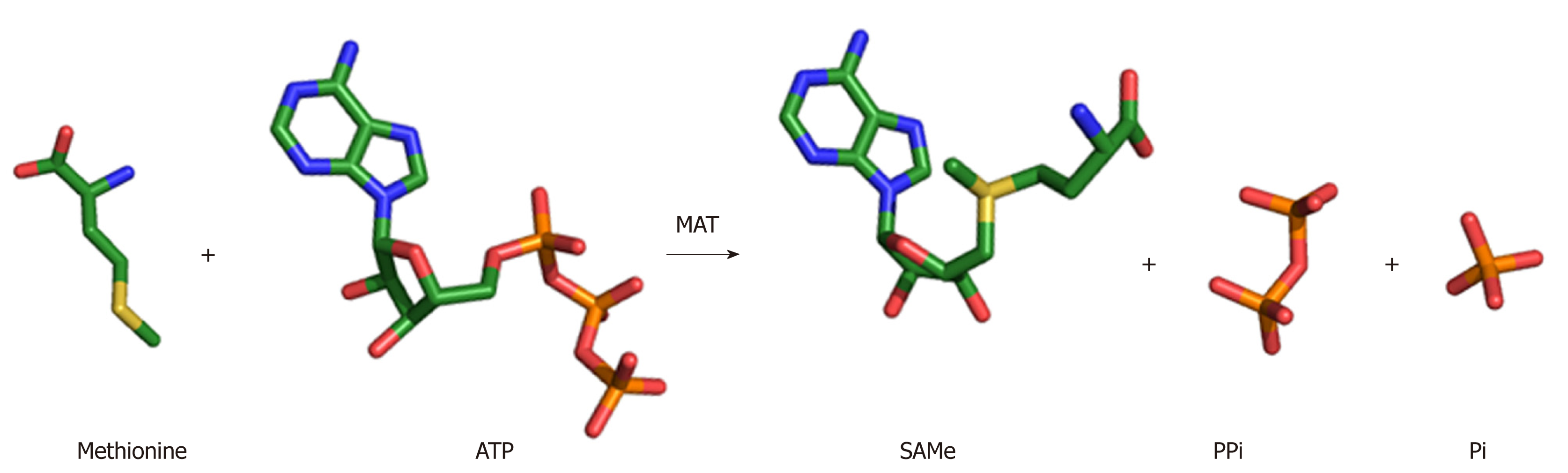
The wealth of structures available for MAT enzymes, as well as the range of biochemical evidence, has provided terrific detail and insight into the enzymatic mechanism. SAMe is produced by the addition of the amino acid methionine to the energy molecule ATP (Figure 2). Upon entry of the substrates ATP and methionine into the active site, the flexible gating loop becomes well-ordered closing the active site. The synthesis of SAMe follows an SN2 catalytic mechanism[28,29], whereby the sulphur atom of methionine attacks the C5’ atom of ATP displacing the tripolyphosphate (PPPi) moiety to form SAMe. The PPPi is then hydrolyzed giving rise to pyrophosphate (PPi) and orthophosphate (Pi), providing the energy to facilitate product release by dislodging the gating loop[30]. For a detailed mechanism, see Komoto et al[21] 2004 and Murray et al[23] 2016.
Despite high sequence identity MAT enzymes exhibit different kinetic and regulatory properties for methionine, ATP, and SAMe. The Km for the methionine is lowest in MATα2 followed by MAT(α1)2 and is the highest for MAT(α1)4[5]. The Km for ATP is also highest for MAT(α1)4 (1–2 mmol), intermediate for MAT(α1)2 (0.2–0.5 mmol), and lowest for MATα2 (70 μM)[31,32]. SAMe, the product produced by MAT, can act as a feedback inhibitor to some of these enzymes[33]. MATα2 is the most sensitive to SAMe with a 50% inhibitory concentration (IC50) of 60 μM which equates to the normal physiological liver levels. MAT(α1)4 is minimally inhibited by SAMe (IC50 = 400 μmol/L) whilst MAT(α1)2 can be stimulated eight-fold by high SAMe levels (500 μmol/L)[33]. These differences between MATα1 and MATα2 are important to allow MATα1 to maintain a high SAMe production in the liver (produces 6-8 g/day) compared with MATα2, which does not contribute significantly to this SAMe pool[34]. Indeed, by expressing MATα1 the liver is able to catabolize 50% of methionine intake via conversion to SAMe and allow an up to 10-fold rise in SAMe level following a methionine rich meal[35]. Consistent with this, cells that express MATα1 have much higher steady-state SAMe levels than cells that express MATα2[36]. When either major isoform of MATβ interacts with MATα2 they increase the kcat (turnover rate of an enzyme-substrate complex) of the MATαβ complexes[12,37,38] and increase the susceptibility to feedback inhibition by SAMe[37].
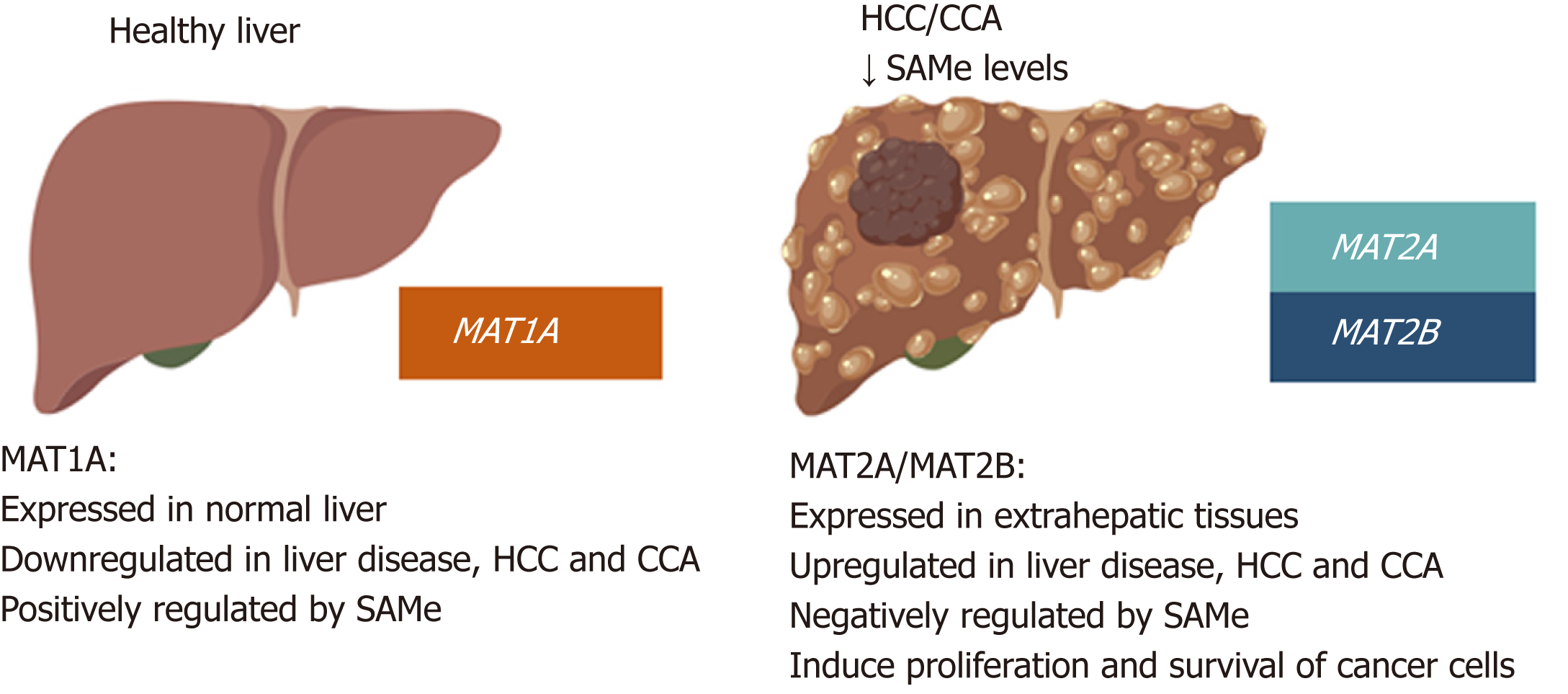
Many studies have demonstrated that MAT proteins play important roles in chronic liver disease and hepatocarcinogenesis and a switch in their expression pattern is a frequent event in liver cancers. MAT1A, which is mainly expressed in the liver and maintains the differentiated state of hepatocytes, is downregulated in most cirrhotic patients[39], in patients with alcoholic hepatitis[40], during de-differentiation and in HCC[41,42]. Conversely, MAT2A and MAT2B, which are normally expressed only by non-parenchymal cells of the liver and extrahepatic tissues, are induced in HCC[14,42,43]. This MAT1A to MAT2A/MAT2B switch contributes to reduced SAMe levels and is an important determinant of liver injury, fibrosis and liver cancer development in both rodents and humans[5].
While MAT1A is a marker for normal differentiated liver, MAT2A and MAT2B are markers for rapid liver growth and de-differentiation. MAT2A and MAT2B are transcriptionally induced in human HCC, during rapid liver growth, de-differentiation, and in response to ethanol feeding in rodents[44-46]. Reduced hepatic MAT activity has also been observed in cirrhotic patients, which explains why many cirrhotic patients have hypermethioninemia[39]. In human HCC, the MAT1A:MAT2A expression ratio has been inversely correlated with cell growth and genomic instability and directly correlated with HCC apoptosis and overall DNA methylation; a reduced ratio is a prognostic marker of more malignant and lower survival HCCs[47]. MATs are regulated at transcriptional, post-transcriptional and post-translational levels by a complex network of mechanisms (Table 2). Many of these are dysregulated in HCC and participate to alter MATs expression.
| MAT1A | MAT2A | MAT2B | |
| Transcriptional regulation | Glucocorticoids (+) | CpG hypomethylation (+) | AP-1 (+) |
| C/EBP (+) | Histone H4 acetylation (+) | NFκB (+) | |
| CpG hypermethylation (-) | c-MYB (+) | SIRT1 (+) | |
| Histone H4 deacetylation (-) | SP-1 (+) | ||
| c-MYC (-) | AP-1 (+) | ||
| MAFG (-) | NFκB (+) | ||
| c-MAF (-) | HIF1α (+) | ||
| PPARγ (-) | |||
| PPARβ (+) | |||
| HBx (+) | |||
| CREB (+) | |||
| Post-transcriptional regulation | AUF1 (-) | HuR (+) | HuR (+) |
| miR-485-3p (-) | Methylated-HuR (-) | miR-21-3p (+) | |
| miR-495 (-) | miR-21-3p (+) | ||
| miR-664 (-) | miR-34a (+) | ||
| miR-34b (+) | |||
| Post-translational regulation | Phosphorylation (-) | Phosphorylation (+) | Phosphorylation (+) |
| Nitrosylation (-) | Sumoylation (+) | GIT1 interaction (+) | |
| Oxidation (-) | Acetylation (-) | MATα2 interaction (+) | |
| MATβ interaction (+) |
The MAT1A promoter contains binding sites for multiple transcription factors, including hepatocyte nuclear factor (HNF), activator protein 1 (AP-1), CCAAT enhancer binding protein (C/EBP), c-MYC and glucocorticoids[48]. Some of these factors are determinants of liver-specific gene expression of MAT1A, such as HNF and C/EBP, with the latter also control MAT1A expression by promoter regulation[48,49]. Prohibitin 1 (PHB1), which is highly expressed in normal hepatocytes and downregulated in most HCCs, positively regulates MAT1A mRNA levels[50]. Finally, c-MYC, MAFG and c-MAF, transcription factors that are overexpressed in human HCC and CCA, have been shown to bind to a repressive E-box element in the human MAT1A promoter to downregulate MAT1A transcription[8,9].
MAT1A expression is also regulated by DNA epigenetic modifications. Lower MAT1A expression in HCC has been associated with promoter hypermethylation and histone H4 deacetylation of its promoter[47,51]. Further investigation revealed a 750-base pair (bp) region upstream of the MAT1A transcriptional start site for these epigenetic modifications[51]. In HepG2 cells and cirrhotic human livers, hypermethylation of sites +10 and +88, relative to the transcriptional start site have been reported to also downregulate MAT1A transcription[52]. Low MAT1A mRNA levels and hyp-ermethylation of both the MAT1A promoter and coding regions were also reported in patients with advanced NAFLD[53].
Binding of AU-rich RNA binding factor (AUF1) to the 3’-untranslated region (UTR) of MAT1A mRNA negatively regulates its stability. There is an inverse correlation between AUF1 and Mat1a expression; de-differentiation of rat hepatocytes in culture increases the expression of AUF1, contributing to the fall in Mat1a mRNA levels, whereas during liver development AUF expression falls, which coincide with increased Mat1a expression; AUF1 is highly expressed in human HCC and its knockdown increased MAT1A mRNA levels[54].
MAT1A mRNA is also regulated by microRNAs (miRNAs) in HCC[55,56]. Injection of 2-acetylaminofluorene in rats resulted in preneoplastic liver lesions, induction of both miR-22 and miR-29b and inhibition of Mat1a mRNA expression[55]. MiR-485-3p, miR-495, and miR-664, which are increased in human HCC and responsible for the induction of LIN28B, an oncoprotein that is overexpressed in HCC and represses the tumor suppressor Let-7, have been shown to negatively regulate MAT1A. These specific miRNAs, through the downregulation of MAT1A, lowered nuclear SAMe levels, leading to hypomethylation of the LIN28B promoter region and increased LIN28B expression[56]. Inhibition of these miRNAs reduced tumor growth in vitro and in vivo by recovering MAT1A expression and inducing apoptosis[56].
MAT2A transcription is upregulated during liver growth and de-differentiation[57-59]. Like MAT1A, MAT2A is also regulated by multiple transcription factors including c-MYB, E2F and specificity protein 1 (SP-1), all of which increase its promoter activity[57,59]. The MAT2A promoter can also be induced by tumor necrosis factor-α (TNF-α) via nuclear factor κβ (NF-κβ) and AP-1 elements in the promoter region[60]. Transforming growth factor β1 (TGF-β1) also increases the activity of the MAT2A promoter via NF-κβ in hepatic stellate cells (HSC)[61]. Multiple PPAR response elements (PPRE) that bind nuclear receptors including peroxisome-proliferator activated receptors (PPAR) are present in the rat Mat2a promoter[62]. In normal liver PPARγ is a marker of HSC quiescence, whilst PPARβ is induced in activated HSCs during liver fibrogenesis[63,64]. Both PPARγ and PPARβ occupy the same site on the Mat2a promoter to cause opposite effects[62]. PPARγ negatively regulate Mat2a transcription but during HSC activation PPARγ expression and activity fall, allowing PPARβ to bind instead and induce Mat2a expression[62]. The MAT2A promoter is also regulated by methylation and acetylation and in human HCC, MAT2A promoter is hypomethylation and associate with higher histone H4 acetylation[58]. Expression of MAT2A is also induced in a hypoxic tumor environment because hypoxia-inducible factor-1α (HIF-1α) binds to a consensus sequence within the MAT2A promoter activating its expression in human hepatoma cells[65]. Finally, hepatitis B X protein (HBx) was shown to activate MAT2A gene transcription by facilitating NF-κB and CREB binding to the MAT2A promoter, explaining MAT2A induction in HBV-associated HCC[66].
Mechanism of MAT2B transcriptional regulation remains poorly characterized. In the human liver cancer cell line HepG2, TNF-α can upregulate MAT2B-V1 mRNA but not MAT2B-V2 through an AP-1 and NF-κβ dependent mechanisms[14]. Sirtuin 1 (SIRT1), a NAD+-dependent deacetylase, can also induce the expression of MAT2B[67]. MAT2B-V1 mRNA expression has also been found to be regulated by leptin in HepG2 cells by mechanisms that involve extracellular signal regulated kinase (ERK) and AKT[68].
The stability of MAT2A mRNA can be influenced by the human RNA-binding (HuR) protein and its methylated form, methyl-HuR[54]. The function of HuR depends on the methylated state of the protein with methyl-HuR destabilizing target mRNAs and HuR stabilizing them[54]. During hepatocyte de-differentiation as well as HCC, HuR is induced but there is a decline in methyl-HuR, which results in a higher HuR/methyl-HuR ratio. For MAT2A this causes increased binding of HuR to MAT2A mRNA stabilizing it in HCC and de-differentiated hepatocytes[54]. HuR has also been shown to stabilize MAT2B mRNA in a similar manner as MAT2A mRNA[67].
Drug-induced miRNAs including miR-21-3p have been shown to control MAT2A and MAT2B stability in HepG2 cells. Either treatment with the anticancer drug berberine, which induces miR-21-3p, or overexpression of miR-21-3p itself causes apoptosis and inhibition of growth by downregulating both MAT2A and MAT2B[69]. Most recently miR-34a and miR-34b, tumor suppressor miRNAs that are down-regulated in multiple cancers including HCC, were shown to directly target MAT2A mRNA and lower its expression[70].
The N6-methyladenosin (m6A) methyltransferase METTL16, has been shown to methylate the 3’UTR of MAT2A hairpins in HEK293T and HeLa cells[71,72]. In high SAMe conditions methylation of hairpin 1 (hp1) of the MAT2A 3’UTR promotes intron retention and nuclear degradation whilst low SAMe levels promote the enhanced binding of METTL16 to the hp1 of MAT2A 3’UTR leading to increased splicing and MAT2A translation[71,72]. Regulation of MAT2A by METTL16 has not been reported in HCC.
The activity and stability of MAT proteins can be altered by post-translational modifications including nitrosylation, phosphorylation and sumoylation. MATα1 has cysteine at position 121 (C120 in human), which lies within the flexible gating loop, can be both nitrosylated or oxidized leading to enzyme inactivation[73]. GSH, an antioxidant, and other thiol-reducing agents can prevent and reverse this inactivation[74]. MATα2 cannot be inhibited in this manner as glycine is at this position. Pho-sphorylation of MATα1 at threonine 342 (T341 in human) by protein kinase C was reported 25 years ago, and this modification does not alter the kinetic properties of this enzyme[75]. However, treatment with alkaline phosphatase to dephosphorylate T342 lowered the activity of both MAT(α1)2 and MAT(α1)4in vitro[75]. Whether this is true in vivo has not been examined.
MATα2 and MATβ have also been reported to be phosphorylated. During liver fibrosis and HSC activation, MATα2 and MATβ are phosphorylated via mitogen activated protein kinase/ERK kinase (MEK) and ERK, respectively, which leads to their stabilization[76]. Analysis revealed Y371/Y374 in MATα2 and T257/Y259 in MATβ to be the sites of phosphorylation and importantly, mutation of these residues inhibited HSC activation[76]. The use of in vitro kinase assays, gene silencing, and chemical inhibitors have shown that MEK could be responsible for the phosphorylation of MATα2, whilst MATβ may be modified by ERK[76].
Sumoylation is a post-translational modification that involves the conjugation of proteins with a small ubiquitin modifier (SUMO) that can lead to changes in the target protein’s stability, localization, tertiary interaction and activity[77]. Attachment of SUMO-1 to a protein is achieved through the sole E2-conjugating enzyme, ubiquitin-conjugating enzyme 9 (UBC9)[78]. The addition of SUMO-1 to a protein is generally associated with stability, and it has been shown that MATα2 has three SUMO-1 modifications at the lysine residues K340, K369 and K394, that increase the stability of MATα2 and also enhance its interaction with oncoproteins such as B-Cell CLL/lymphoma 2 (BCL-2)[77,79]. It has been shown that treatment with SAMe in liver cancer cells reduces the expression and activity of UBC9[80].
While in normal liver MATα2 acetylation at K81 by the E1A binding protein (P300) causes its ubiquitination and degradation, in liver disease a lack of acetylation stabilizes the protein, and this has been associated with HCC development that is attributed to the deacetylase HDAC3[81].
While the expression of MATs can influence the steady-state levels of SAMe[36], SAMe level can, in return, influence MAT expression. During de-differentiation of primary hepatocytes in culture MAT1A expression falls while MAT2A expression is induced[82]. This effect is due to a fall in SAMe level since it can be blocked by the addition of SAMe. Consistently, a fall in SAMe level (by restricting L-methionine in medium) leads to rapid induction of MAT2A expression that is blocked upon the addition of SAMe[58,83]. Treatment with SAMe results in higher methyl-HuR level leading to enhanced mRNA destabilization of MAT2A which may explain its negative effect on MAT2A expression[54]. SAMe also inhibits MAT2B expression at baseline and prevents leptin induced induction in hepatoma cells[68].
MAT proteins exhibit distinct interactomes in normal and diseased liver. All three MATs interact with a variety of proteins regulating their expression and contributing to liver injury and carcinogenesis.
MATα1 can act as a transcription co-factor by interacting with other E-box binding regulatory proteins. MATα1 can heterodimerize with MAX in HCC to repress E-box-driven promoter activity, which results in the negative regulation of the transcription factors c-MYC, MAFG and c-MAF, and their oncogenic activity[9,50]. MATα1 has also been found to interact with CYP2E1 to negatively regulate its protein expression by inducing its proteasomal degradation[19]. Interestingly, MATα1 also interacts with p53 and DNA damage-regulated gene 1 (PDRG1) in hepatoma cells and in a mouse model of acute liver injury, which exhibited reduced total MATα1 expression but accumulation of nuclear MATα1[84]. PDRG1 is an oncogene that is upregulated in bladder, breast and colon cancer[85]. Interaction of PDGR1 with MATα1 in the nucleus resulted in a decrease in MAT activity and DNA hypomethylation[84]. The role of PDRG1 in HCC remains unknown.
MATα2 not only binds to and stabilizes BCL-2 protein, it also enhances BCL-2 transcription in liver and colon cancer cell lines by binding to its promoter[79]. MATβ is known to interact with HuR; when either of the MAT2B variants is overexpressed, cytosolic HuR content increases leading to higher mRNA levels of HuR targets such as cyclin D1 and cyclin A and proliferation[18]. MATβ also interacts with SIRT1, and resveratrol increases this interaction by stabilizing them[67]. MATβ also interacts with G-protein-coupled receptor kinase-interacting protein 1 (GIT1) to form a scaffold complex that interacts and activates all components of the RAS/RAF/MEK/ERK signaling pathway in liver cancer cells, promoting growth in vitro and in vivo[7]. Interaction between MATβ and GIT1 appear to also stabilize MATβ[86]. Finally, both MATα2 and MATβ are often overexpressed in parallel in multiple cancers and part of the reason is that their interaction stabilizes both proteins[70].
MAT genes deregulation has been widely associated with alterations that contribute to liver disease and the development of HCC. MAT1A downregulation increases oxidative stress, progenitor cells expansion, genomic instability and other mechanisms implicated in tumorigenesis, whilst MAT2A and MAT2B, which are induced in HCC, confer growth and survival advantages to cancer cells (Reviewed in Lu 2012)[5]. In humans, the fall in MAT activity observed in cirrhotic patients is thought to contribute to the pathogenesis and progression of the disease as well as predisposition to HCC[39].
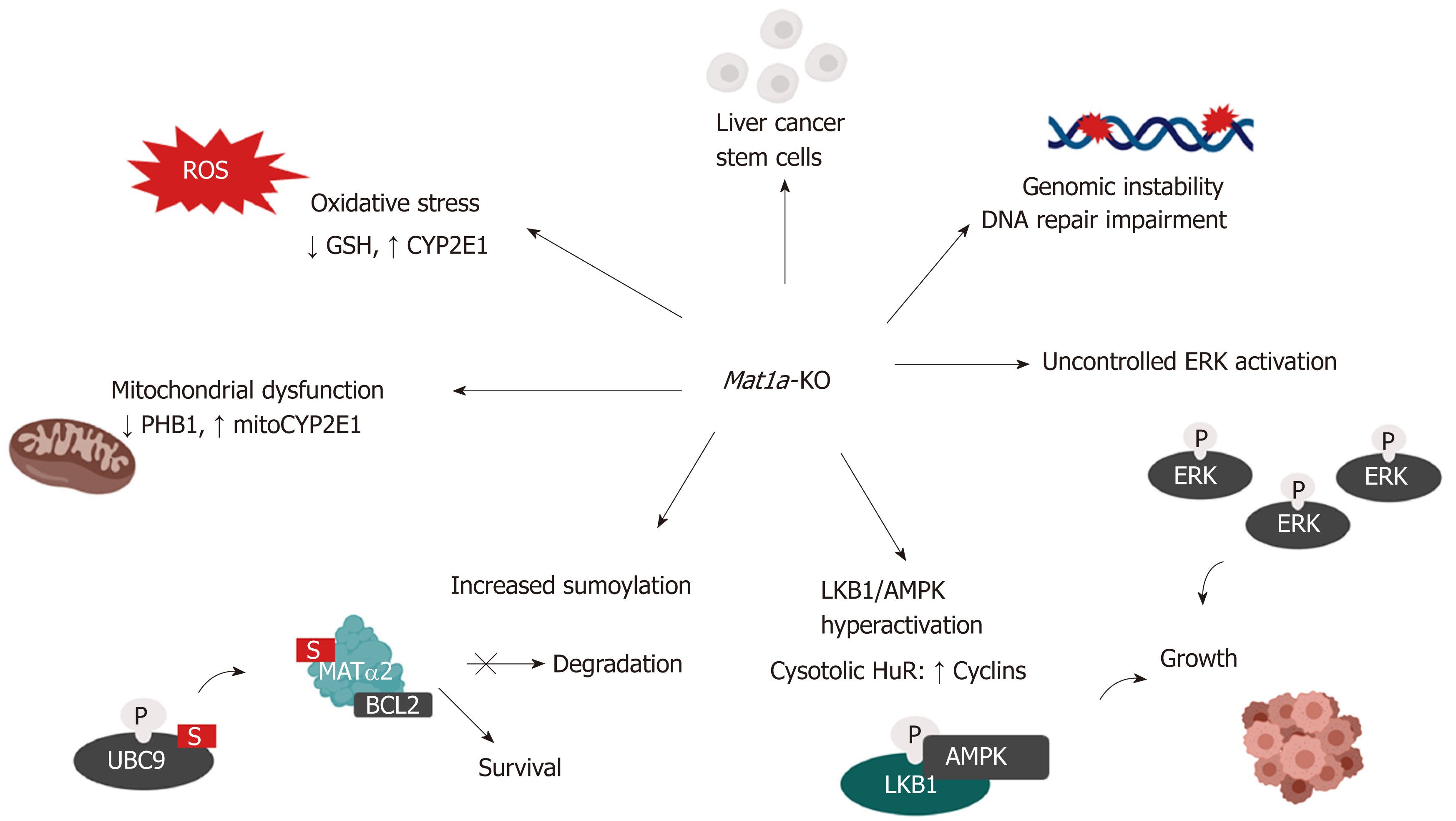
The Mat1a-KO mouse model has provided important insights into the mechanisms of how MAT1A might influence HCC development. This model is relevant to human liver disease since MAT1A expression is markedly reduced in the majority of cirrhotic patients[87]. Mice lacking Mat1a have markedly increased serum methionine levels and chronically reduced hepatic SAMe (70% lower) and GSH (40% lower) levels. By three months Mat1a-KO mice develop hepatic hyperplasia and are more susceptible to liver steatosis in response to a choline-deficient diet. By eight months, Mat1a-KO mice spontaneously develop NASH on a normal diet and by 18 to 20 months they develop HCC[88]. The livers of Mat1a-deficient mice also exhibit oxidative stress caused by low GSH levels, impaired mitochondrial function and increased expression of CYP2E1[34]. CYP2E1 is the principal P-450 enzyme responsible for the metabolism of hepatotoxins such as alcohol, acetaminophen and CCl4 in the liver and has a critical role in the generation of reactive oxygen species (ROS). Because of this, Mat1a-KO mice are more susceptible to CCl4–induced liver injury[34]. Mitochondrial dysfunction is another important mechanism that sensitizes Mat1a-KO mice to liver injury, which was mainly attributed to low levels of both the mitochondrial chaperone PHB1 and oxidative stress[34,89]. Our recent work adds loss of mitochondrial MATα1, an important regulator of mitochondrial function, as another mechanism[19].
Genomic instability, which arises from the large number of genomic mutations, chromosomal aberrations, duplications, deletions and replication errors that cancer cells carry, is a characteristic of most cancers including HCC and is considered an early step in carcinogenesis[90,91]. Genomic instability may contribute to initiation of the malignant transformation, progression of the cancer and even resistance to therapy, influencing the overall prognosis of the cancer[92].
MAT1A regulates DNA methylation via SAMe and so MAT1A deficiency leads to DNA hypomethylation, which may contribute to genomic instability. Interestingly, alterations in the activity of MAT proteins and global DNA hypomethylation are prognostic markers for human HCC possibly through genomic instability[93]. These results suggest that early dysregulation of MAT proteins could influence the progression from preneoplastic lesions to cancer. In addition, the Apurinic /Apyrimidinic Endonuclease 1 (APEX1) protein, which participates in the base excision repair of premutagenic apurinic/apyrimidinic (AP) sites, the most frequent DNA lesion in cells, is markedly downregulated in Mat1a-KO livers. APEX1 protein level is reduced in de-differentiated hepatocytes with reduced Mat1a expression and low SAMe levels and is recovered by SAMe treatment[94]. Taken together, these findings demonstrate that hepatic SAMe depletion promotes genomic instability by different mechanisms contributing to malignant transformation.
Along with oxidative stress, mitochondrial dysfunction represents an important trigger to hepatocarcinogenesis[95,96]. Although mitochondrial metabolism in malignant cells is controversial, deregulated cellular energetics associated with mitochondrial dysfunction are considered a common event in cancer[95]. As described above Mat1a-KO livers exhibit mitochondrial dysfunction from multiple mechanisms, including lower PHB1 expression, loss of mitochondrial MATα1 and increased expression of CYP2E1.
A proteomics study identified several mitochondrial proteins to be downregulated in Mat1a-KO mice from birth until the development of NASH[89]. PHB1 is a well-known mitochondrial chaperone that stabilizes newly synthesized mitochondrial proteins and maintains the organization and stability of mitochondrial nucleoids. PHB1 is essential for mitochondrial function and was found significantly downregulated in the livers of Mat1a-KO mice as compared to wild-type animals. In vitro experiment suggested low SAMe level promoted increased PHB1 degradation, as SAMe addition prevented the fall in PHB1 protein during culture[89]. However, a recent study showed PHB1 and MAT1A exert reciprocal positive regulation on each other at the mRNA level[50], adding another mechanism to low PHB1 expression in the Mat1a-KO liver.
The generation of a liver specific Phb1-KO mouse model confirmed that PHB1 hepatic deficiency predisposes to liver injury and malignant transformation. Liver-specific Phb1-KO mice have liver injury at a very young age, abnormal mitochondria and increased oxidative stress. Mice lacking Phb1 develop progressive fibrosis and multi-focal HCC by 8-10 mo of age[97]. Although HCC could have developed as a consequence of chronic inflammation, accumulating evidence support a tumor suppressor function of PHB1 in the liver. For instance, PHB1 silencing in murine non-transformed AML12 cells increased cyclin D1, H19, and IGF2 expression and enhanced E2F binding to the cyclin D1 promoter and proliferation[97]. PHB1 can cooperate with CCCTC-binding transcription factor (CTCF) to negatively regulate H19 and IGF2 expression, both of which are induced in HCC[98]. Reduced PHB1 expression has also been shown to induce IL-8 transcription by activating NF-κB and AP-1, resulting in enhanced IL-8 expression and release to promote tumorigenesis[99]. Finally, PHB1 also acts as a negative regulator of WNT signaling, and its downregulation causes the induction of multiple WNT ligands and downstream activation of canonical WNT ‐β‐-catenin signaling in murine liver and human HCC cells, in part through E2F[100].
It should be noted that the tumor suppressor role of PHB1 is highly controversial since PHB1 expression is increased in other cancers. PHB1 is also found in the nucleus, where it has been shown to interact with Rb and p53 among other proteins to bring about a change in transcriptional activities of E2F and p53[101]. Different subcellular localizations and post-translation modifications may explain the contradictory tumor regulatory activities of PHB1 in different cell types.
Recently MATα1 was shown to be present in the mitochondrial matrix in hepatocytes[19]. Mat1a-KO hepatocytes had reduced mitochondrial membrane potential and higher mitochondrial ROS, both of which were normalized when MAT1A was overexpressed. Oxygen consumption rate, ATP production and maximal and spare respiratory capacities were all reduced in Mat1a-deficient mitochondria, supporting the negative effect that MAT1A deficiency has on mitochondrial function. Another important finding that may contribute to mitochondrial dysfunction and liver injury in Mat1a-KO mice is the interaction of MATα1 and CYP2E1 in the mitochondria. Mat1a deficiency leads to higher CYP2E1 mitochondrial levels. Mitochondrial CYP2E1 also regulates the production of ROS and is known to contribute to liver injury and mitochondrial dysfunction[102,103]. MATα1 negatively regulates CYP2E1 expression at mRNA and protein levels, with the latter being the dominant mechanism that involves methylation of CYP2E1 R379, promoting its proteasomal degradation[19].
MATα1 was found to also interact with important mitochondrial proteins including several subunits of the electron transport chain complexes, which raises the possibility that MATα1 could be regulating mitochondrial function in multiple ways. These findings highlight a critical role of MATα1 in regulating mitochondrial function and could provide a novel target for the treatment of different liver diseases where mitochondrial dysfunction plays a key role. Taken together, reduced mitochondrial MATα1 could play a key role in the mitochondrial dysfunction that is often observed in HCC[104].
Cancer stem cells, also known as tumor‐initiating cells, are known to play a central role in tumor development, metastasis and recurrence and are considered key therapeutic target for cancer treatment. Hepatic oval cells are the cancer stem cells of the liver and important contributors of hepatocarcinogenesis[105]. They are quiescent in normal adult liver and low in number and expand during severe and prolonged injury as seen in various models of experimental carcinogenesis[106].
Methyl-deficient diets have been used to induce oval cell proliferation and HCC formation in susceptible models such as p53−/− mice[107]. Mat1a-KO livers contain increased populations of liver cancer stem cells (or CD133+/CD49f+ oval cells), as they age[108]. These cells have increased expression of several oncogenes and are tumorigenic in vivo. Interestingly, Mat1a-KO’s cancer stem cells show increased MAPK signaling with enhanced ERK activity, like Mat1a-KO’s hepatocytes, and increased oncogenic signaling (K-Ras and Survivin)[108]. Constitutive ERK activation makes these cells resistant to the apoptotic effect of TGF-β, a well-known growth inhibitor in hepatocytes[109,110]. How SAMe deficiency allows expansion of oval cells remains unknown.
Increasing evidence indicates that the deregulation of various signaling pathways progressively increase with HCC progression and has a prognostic value. The MAT1A to MAT2A/MAT2B switch is also associated with activation of multiple signaling pathways including RAS/ERK[76,86,111], IκB kinase (IKK)/NF-kB[14,60], Phosphoinositide 3-kinase (PI3K)/AKT[68,112,113] and Liver kinase B1 (LKB1)/AMPK-activated protein kinase (AMPK)[114]. Increased sumoylation[80] and c-MYC expression[8], which are well-known contributors of hepatocarcinogenesis, have also been associated with reduced hepatic SAMe levels.
ERK signaling is tightly regulated in normal cells but uncontrollably active in cancer cells, being one of the several growth signals associated with highly malignant HCCs[115,116]. ERK activity is regulated by the dual-specificity MAPK phosphatase (DUSP1). DUSP1 is a member of a family of dual-specificity MAPK phosphatases that dephosphorylates both serine/threonine and tyrosine residues[117]. Interestingly, there is a reciprocal regulation between DUSP1 and ERK[111]. Transient activation of ERK leads to catalytic activation DUSP1, which in turn inhibits ERK activity by dephosphorylation[118]. DUSP1 feedback inhibits ERK and this activity of DUSP1 is crucial for the regulation of ERK activity in liver cells. However, prolonged ERK activation induces the phosphorylation of DUSP1 at the Ser296 residue rendering the DUSP1 protein susceptible to proteasomal degradation[119]. In human HCC, DUSP1 expression is negatively correlated with proliferation and microvessel density and positively with survival[118].
Hepatic DUSP1 expression is decreased in Mat1a-KO mice both at the mRNA and protein levels, being more pronounced at the protein level and was normalized after SAMe treatment[111]. SAMe increased DUSP1 mRNA level by enhancing p53 binding to its consensus element in the DUSP1 promoter, which is known to activate DUSP1 transcriptionally[111]. Increased binding of p53 to the DUSP1 promoter in SAMe fed mice was due to the fact that SAMe stabilizes APEX1, which is a known trans-activator of p53[94]. DUSP1 lower protein level was attributed to its faster degradation due to increased proteasomal activity in Mat1a-KO mice. SAMe treatment normalized proteasomal activity, increasing DUSP1 protein level and normalized ERK activity in Mat1a-KO mice[111]. These findings suggest that SAMe deficiency leads to uncontrolled ERK activation due at least in part to decreased DUSP1 expression during HCC development. ERK signaling was also found to be regulated by MAT2B since its knockdown inhibited the activation of MAPK/ERK pathway induced by leptin in liver cancer cell lines[68]. Finally, as mentioned earlier, the MATβ-GIT1 complex efficiently binds to MEK and ERK leading to their activation. Consistent with this, overexpression of MAT2B or GIT1 in the HCC cell line Huh7 enhanced tumor growth and metastasis in a mouse orthotopic HCC model[86].
Even though LKB1 is considered a tumor suppressor in a variety of cancers[120], its role in liver carcinogenesis remains controversial given the fact that both reduced and increased LKB1 levels have been reported in human HCC correlating with prognosis[121-124]. In recent years, several publications have supported the oncogenic role of LKB1 in liver cancer[122-125].
LKB1 phosphorylates and activates AMPK, a central metabolic sensor, to control cell growth in response to environmental nutrient changes. Indeed, the LKB1/AMPK signaling pathway has tumor suppressor activity since it serves as a metabolic checkpoint arresting cell growth in conditions of nutrient deprivation or low intracellular ATP levels[126]. In the liver the activation of LKB1 and AMPK leads to the production of nitric oxide (NO) and is required for hepatocyte proliferation, as seen in regenerating livers after partial hepatectomy and hepatocyte growth factor (HGF) treated rat hepatocytes[114]. Interestingly, early during liver regeneration hepatic SAMe level falls and exogenous SAMe inhibits regeneration[127]. The thought is that SAMe level needs to fall in order to release the inhibitory tone it exerts on mitogenic pathways. Although Mat1a–KO mice have higher basal proliferation, they exhibit impaired liver regeneration because hepatic SAMe level remained unchanged[127]. Consistent with this, in the presence of SAMe, protein phosphatase 2A (PP2A) interacts with AMPK, leading to its dephosphorylation and inactivation. One explanation for the fall in hepatic SAMe level early in liver regeneration is due to increased NO formation, which can inactivate MATα1[128,129].
Activation of the LKB1/AMPK pathway may contribute to hepatocarcinogenesis through other mechanisms as well. Increased LKB1 and AMPK activity results in nuclear to cytoplasmic HuR translocation and the subsequent stabilization of several cyclin mRNAs enhancing cell proliferation[130]. LKB1/AMPK activation is also required for survival of HCC cells derived from Mat1a-KO livers, named SAMe-D[114]. LKB1 can regulate AKT-mediated cell survival independent of PI3K, AMPK, and mTOR2 (mammalian target of rapamycin complex). LKB1 is hyperactivated in SAMe-D cells and can control survival through the phosphorylation and cytoplasmic retention of p53. In normal cells, p53 expression is maintained at a low level and is inactivated by Mdm2-mediated nuclear to cytosol transportation and proteasomal degradation. In response to oncogenic insults, p53 translocates to the nucleus to exert its tumor suppressor activity and activate pathways associated with DNA repair, cell cycle arrest and apoptosis. Notably, HuR nucleocytoplasmic shuttling also stabilizes p53 mRNA. Supporting these findings, increased cytoplasmic staining of p53 and phospho-LKB1 were found in the Mat1a-KO livers and in livers from human HCC derived from both NASH and alcoholic steatohepatitis[131]. Figure 3 summarizes the mechanisms of HCC development in the Mat1a-KO mouse model.
CCA is, after HCC, the second most common primary hepatic malignancy and like HCC, its development involves MAT genes deregulation. MAT1A is highly expressed also in normal bile duct epithelial cells and is repressed during chronic cholestasis and in murine and human CCA[8]. There are common mechanisms of MAT genes deregulation between HCC and CCA. For example, hypermethylation of the MAT1A promoter has also been observed in CCA. The transcription factors c-MYC, MAFG and c-MAF, which are all induced both in HCC and CCA, also negatively regulate MAT1A transcription in CCA by binding to its repressor E-box promoter region[8]. PHB1, which is also downregulated in most human CCAs, positively regulates MAT1A while suppressing c-MYC, MAFG, and c-MAF expression in mice[9,50]. Consistently, reduced PHB1 expression predisposes to the development of cholestasis-induced CCA[8,50]. Figure 4 summarizes changes in MATs and SAMe in HCC and CCA as compared to normal liver.
The Mat1a-KO mouse model demonstrates that the switch from Mat1a to Mat2a leads to hepatic SAMe deficiency and hypermethioninemia[88]. Patients with human liver cirrhosis[39], alcoholic hepatitis[40], advanced NAFLD[53], as well as HCC [42] all have lower MAT1A mRNA level. The restoration of hepatic SAMe level appears to be an obvious therapy to overcome the fall in SAMe level due to the MAT1A/MAT2A switch. There have been very few human trials that examined SAMe in chronic liver disease. One study showed that SAMe treatment (1,200 mg orally per day in three divided doses) for two years improved the survival or delayed liver transplantation in patients who had alcoholic liver cirrhosis with less advanced liver disease (Child’s class A and B)[132]. However, this was a post-hoc analysis, so it remains to be confirmed. Another study in patients with hepatitis B-related advanced-stage (stages B-C) HCC who received 1000 mg of SAMe two hours before surgery intravenously and continued for five consecutive postoperative days showed a reduction of alanine aminotransferase and aspartate transaminase levels, delayed recurrence and a greater 24-mo survival rate as well as a lower risk of complications[133]. However, analysis by the Cochrane Hepato-Biliary group using a total of 330 alcoholic liver disease patients treated with SAMe in 8 clinical trials did not confirm the beneficial effects of SAMe in human liver cirrhosis although they were small with variable quality, which resulted in the meta-analysis failing to show a significant benefit of SAMe treatment[134]. At the present time the verdict on the effectiveness of SAMe in chronic liver disease is still out and its utility in chemoprevention of HCC remains to be studied.
In CCl4-induced liver injury SAMe treatment prevented the activation of HSCs by inhibiting the collagen promoter as well as downregulating TGF-β-induced extracellular matrix protein, α-smooth muscle actin (α-SMA)[135]. It has also been demonstrated that SAMe is selectively pro-apoptotic in liver cancer cells but anti-apoptotic in normal hepatocytes[136,137]. This property makes SAMe particularly attractive as a chemopreventive agent. Indeed, SAMe can chemoprevent against HCC in several preclinical models[5]. However, SAMe was ineffective at treating established tumors in an orthotopic HCC model[138]. Investigation into this revealed that buildup of SAMe in the liver was prevented by the compensatory induction of glycine-N methyl transferase (GNMT), an important enzyme abundantly expressed in the liver that is responsible for catabolizing SAMe and keeping SAMe level within a tight range[5,138]. Although acute pharmacologic SAMe treatment can transiently increase liver SAMe level by 10-fold, prolong SAMe treatment induced GNMT expression so that SAMe level increased only 30%, which is insufficient to exert a pro-apoptotic effect[138]. However, in human cirrhosis and HCC GNMT express is often downregulated[139], so the efficacy of SAMe for HCC treatment in human warrants further investigation. At present, there are two SAMe ongoing clinical trials in HCC treatment. One (NCT03178929) is designed to investigate SAMe treatment (2000 mg, P.O.) in HCC patients (Barcelona Clinic Liver Cancer 0/A) after radical treatment, where tumor recurrence is the endpoint. The other (NCT02586285) aims to study the SAMe treatment (500-1000 mg, iv, per day) in HCC patients after curative treatment. Although targeting SAMe has been the central focus for treatment, therapies that target the MAT proteins themselves could also offer potential benefit to which some of these are described below.
Overexpressing MAT1A in the liver cancer cell line Huh7 resulted in a stable increase in SAMe levels, an induction in tumor suppressor genes, downregulation of angiogenesis genes, reduced cell growth and increased apoptosis in vitro and in vivo[140]. This is a proof of principle that raising MAT1A expression in liver cancer could be an effective treatment strategy. While delivering MAT1A gene therapy to HCC cells is not a suitable approach, targeting miRNAs that downregulate MAT1A expression might be feasible. MiR-485-3p, miR-495, and miR-664 are three miRNAs that are increased in human HCC that directly target MAT1A at the 3’UTR[56]. Treatment with siRNA against any of the miRNAs resulted in higher MAT1A expression, reduced HCC growth in an orthotopic HCC model[56]. Higher SAMe level as a result can also downregulate MAT2A/MAT2B to slow HCC growth. Taken together, raising endogenous MAT1A expression and hence SAMe level in the HCC cells is an attractive treatment approach that deserves further study.
Therapeutic agents to target MATα2 have been proposed for many years. In the 1970s methionine analogs were suggested to act as substrate-competitive inhibitors and proposed as chemotherapeutic agents[141]. Stilbene derivatives (FIDAS agents) have been proposed as inhibitors of MATα2, but these compounds are unlikely to specifically inhibit MATα2 because of their ability to bind multiple proteins targets[142]. Using molecules that directly target the active site of MATα2 may be problematic as this protein is expressed in most extrahepatic cells which require MATα2 as the SAMe generator. In addition, the high level of sequence and structural identity between MATα1 and MATα2 makes it difficult to design active site molecules that would only target one these proteins. One strategy is to target the interaction of the MATα2β complexes as these complexes provide a proliferative advantage to HCC cells by lowering steady state SAMe level, stabilizing each other as well as activating several mitogenic pathways[7,43,68,86]. A novel allosteric inhibitor has been shown to bind to the interacting site of these proteins, which demonstrated promise in lung cancer cells[13]. It remains to be examined in liver cancer models.
Targeting a posttranslational modification could also be a good strategy to only target MAT proteins that are contributing to the pathogenesis of HCC. Designing a molecule for a specific modification on MATα1, MATα2, or MATβ may be possible to stabilizes MATα1 whilst destabilizing MATα2 and MATβ. For instance, sumoylation on MATα2 facilitated BCL-2 induction in HCC, and induced chemoresistance in HCC cells[79]. Using small molecules to block the addition of SUMO to MATα2 would lower its expression as well as that of BCL-2. Similarly, MATα2 and MATβ are both stabilized by phosphorylation at specific residues which facilitated HSC activation[76]. HSC activation is an important mediator of liver fibrosis, and therefore blocking the site-specific phosphorylation of these MAT proteins could be a novel treatment for liver fibrosis. As mentioned before, MATβ has been implicated to promote cell survival in HCC due to its interaction with the MEK/ERK/MAPK pathway[68,86]. The discovery that MATβ interacted with GIT1 and caused the amplification of RAS-mediated MAPK activation provides another novel target where a molecule could block this protein-protein interaction to inhibit MAPK signaling and cell proliferation[7].
Epigenetic alterations including DNA methylation are recognized as a major characteristic in HCC and may serve as diagnostic and prognostic biomarkers[143]. MATα1 to MATα2 switch and low SAMe level are associated with HCC development and strongly predict patients’ survival[47,144]. High MAT1A expression in human HCC is negatively correlated to tumor size (> 5 cm), whilst up-regulation of MAT2A is positively correlated to serum alpha-fetoprotein level and one-year recurrence after hepatectomy[145]. Moreover, the MATα1 to MATα2 switch may promote HCC invasion and metastasis and lower patient recurrence-free survival[146]. All these suggest MATs may serve as diagnostic and prognostic molecular markers in HCC and likely CCA as well.
Taken together, accumulating evidence demonstrates the importance of MAT genes in liver tumorigenesis. Part of the mechanism is related to a lowering in the steady state SAMe level. However, all three MATs are found in the nucleus where they regulate gene expression via epigenetics as well as bona fide transcription factor and cofactors. They also have distinct interactomes and affect a myriad of signaling pathways. SAMe is an attractive chemopreventive and possibly therapeutic agent in liver cancer and its efficacy in both warrant investigation. Small molecules that inhibit interactions between MATβ-GIT1 or MATα2-β, both of which provide proliferative advantages to liver cancer cells, and targeting posttranslational modifications that can increase MAT1A and/or lower MAT2A/2B are all exciting future directions for translating the knowledge gained from understanding mechanisms of MAT dysregulations to therapy in liver cancer.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Guo K, Trovato GMM S-Editor: Ma YJ L-Editor: A E-Editor: Ma YJ
| 1. | Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM, Anderson RN, Ma J, Ly KN, Cronin KA, Penberthy L, Kohler BA. Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122:1312-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 633] [Cited by in RCA: 694] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 2. | Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1933] [Cited by in RCA: 1871] [Article Influence: 207.9] [Reference Citation Analysis (4)] |
| 3. | Siegel AB, Zhu AX. Metabolic syndrome and hepatocellular carcinoma: two growing epidemics with a potential link. Cancer. 2009;115:5651-5661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 302] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 4. | Diehl AM, Day C. Cause, Pathogenesis, and Treatment of Nonalcoholic Steatohepatitis. N Engl J Med. 2017;377:2063-2072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 918] [Article Influence: 114.8] [Reference Citation Analysis (0)] |
| 5. | Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 415] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 6. | Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ito T, Ohta M, Kera Y, Noda T, Igarashi K. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011;41:554-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 150] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 7. | Peng H, Li TW, Yang H, Moyer MP, Mato JM, Lu SC. Methionine adenosyltransferase 2B-GIT1 complex serves as a scaffold to regulate Ras/Raf/MEK1/2 activity in human liver and colon cancer cells. Am J Pathol. 2015;185:1135-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Yang H, Liu T, Wang J, Li TW, Fan W, Peng H, Krishnan A, Gores GJ, Mato JM, Lu SC. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans(‡). Hepatology. 2016;64:439-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 9. | Liu T, Yang H, Fan W, Tu J, Li TWH, Wang J, Shen H, Yang J, Xiong T, Steggerda J, Liu Z, Noureddin M, Maldonado SS, Annamalai A, Seki E, Mato JM, Lu SC. Mechanisms of MAFG Dysregulation in Cholestatic Liver Injury and Development of Liver Cancer. Gastroenterology. 2018;155:557-571.e14. |
| 10. | Gil B, Casado M, Pajares MA, Boscá L, Mato JM, Martín-Sanz P, Alvarez L. Differential expression pattern of S-adenosylmethionine synthetase isoenzymes during rat liver development. Hepatology. 1996;24:876-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 11. | Lu SC, Gukovsky I, Lugea A, Reyes CN, Huang ZZ, Chen L, Mato JM, Bottiglieri T, Pandol SJ. Role of S-adenosylmethionine in two experimental models of pancreatitis. FASEB J. 2003;17:56-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Murray B, Antonyuk SV, Marina A, Van Liempd SM, Lu SC, Mato JM, Hasnain SS, Rojas AL. Structure and function study of the complex that synthesizes S-adenosylmethionine. IUCrJ. 2014;1:240-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Quinlan CL, Kaiser SE, Bolaños B, Nowlin D, Grantner R, Karlicek-Bryant S, Feng JL, Jenkinson S, Freeman-Cook K, Dann SG, Wang X, Wells PA, Fantin VR, Stewart AE, Grant SK. Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A. Nat Chem Biol. 2017;13:785-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 14. | Yang H, Ara AI, Magilnick N, Xia M, Ramani K, Chen H, Lee TD, Mato JM, Lu SC. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134:281-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | González B, Garrido F, Ortega R, Martínez-Júlvez M, Revilla-Guarinos A, Pérez-Pertejo Y, Velázquez-Campoy A, Sanz-Aparicio J, Pajares MA. NADP+ binding to the regulatory subunit of methionine adenosyltransferase II increases intersubunit binding affinity in the hetero-trimer. PLoS One. 2012;7:e50329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Pajares MA, Alvarez L, Pérez-Sala D. How are mammalian methionine adenosyltransferases regulated in the liver? A focus on redox stress. FEBS Lett. 2013;587:1711-1716. |
| 17. | Reytor E, Pérez-Miguelsanz J, Alvarez L, Pérez-Sala D, Pajares MA. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 2009;23:3347-3360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | Xia M, Chen Y, Wang LC, Zandi E, Yang H, Bemanian S, Martínez-Chantar ML, Mato JM, Lu SC. Novel function and intracellular localization of methionine adenosyltransferase 2beta splicing variants. J Biol Chem. 2010;285:20015-20021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Murray B, Peng H, Barbier-Torres L, Robinson AE, Li TWH, Fan W, Tomasi ML, Gottlieb RA, Van Eyk J, Lu Z, Martínez-Chantar ML, Liangpunsakul S, Skill NJ, Mato JM, Lu SC. Methionine Adenosyltransferase α1 Is Targeted to the Mitochondrial Matrix and Interacts with Cytochrome P450 2E1 to Lower Its Expression. Hepatology. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 20. | González B, Pajares MA, Hermoso JA, Guillerm D, Guillerm G, Sanz-Aparicio J. Crystal structures of methionine adenosyltransferase complexed with substrates and products reveal the methionine-ATP recognition and give insights into the catalytic mechanism. J Mol Biol. 2003;331:407-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 21. | Komoto J, Yamada T, Takata Y, Markham GD, Takusagawa F. Crystal structure of the S-adenosylmethionine synthetase ternary complex: a novel catalytic mechanism of S-adenosylmethionine synthesis from ATP and Met. Biochemistry. 2004;43:1821-1831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Shafqat N, Muniz JR, Pilka ES, Papagrigoriou E, von Delft F, Oppermann U, Yue WW. Insight into S-adenosylmethionine biosynthesis from the crystal structures of the human methionine adenosyltransferase catalytic and regulatory subunits. Biochem J. 2013;452:27-36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 23. | Murray B, Antonyuk SV, Marina A, Lu SC, Mato JM, Hasnain SS, Rojas AL. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. Proc Natl Acad Sci U S A. 2016;113:2104-2109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 24. | Sánchez-Pérez GF, Bautista JM, Pajares MA. Methionine adenosyltransferase as a useful molecular systematics tool revealed by phylogenetic and structural analyses. J Mol Biol. 2004;335:693-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Panmanee J, Bradley-Clarke J, Mato JM, O'Neill PM, Antonyuk SV, Hasnain SS. Control and regulation of S-Adenosylmethionine biosynthesis by the regulatory β subunit and quinolone-based compounds. FEBS J. 2019;286:2135-2154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Kotb M, Mudd SH, Mato JM, Geller AM, Kredich NM, Chou JY, Cantoni GL. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet. 1997;13:51-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 177] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Kotb M, Kredich NM. S-Adenosylmethionine synthetase from human lymphocytes. Purification and characterization. J Biol Chem. 1985;260:3923-3930. [PubMed] |
| 28. | Parry RJ, Minta A. Studies of enzyme stereochemistry. Elucidation of the stereochemistry of S-adenosylmethionine formation by yeast methionine adenosyltransferase. J Am Chem Soc. 1982;104:871-872. [RCA] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Markham GD, Parkin DW, Mentch F, Schramm VL. A kinetic isotope effect study and transition state analysis of the S-adenosylmethionine synthetase reaction. J Biol Chem. 1987;262:5609-5615. [PubMed] |
| 30. | Markham GD, Pajares MA. Structure-function relationships in methionine adenosyltransferases. Cell Mol Life Sci. 2009;66:636-648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 108] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 31. | Okada G, Teraoka H, Tsukada K. Multiple species of mammalian S-adenosylmethionine synthetase. Partial purification and characterization. Biochemistry. 1981;20:934-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 100] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Pajares MA, Durán C, Corrales F, Pliego MM, Mato JM. Modulation of rat liver S-adenosylmethionine synthetase activity by glutathione. J Biol Chem. 1992;267:17598-17605. [PubMed] |
| 33. | Sullivan DM, Hoffman JL. Fractionation and kinetic properties of rat liver and kidney methionine adenosyltransferase isozymes. Biochemistry. 1983;22:1636-1641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 116] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, García-Trevijano ER, Huang ZZ, Chen L, Kanel G, Avila MA, Mato JM, Lu SC. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 219] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 35. | Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1:228-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 942] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 36. | Cai J, Mao Z, Hwang JJ, Lu SC. Differential expression of methionine adenosyltransferase genes influences the rate of growth of human hepatocellular carcinoma cells. Cancer Res. 1998;58:1444-1450. [PubMed] |
| 37. | Halim AB, LeGros L, Geller A, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J Biol Chem. 1999;274:29720-29725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 38. | Nordgren KK, Peng Y, Pelleymounter LL, Moon I, Abo R, Feng Q, Eckloff B, Yee VC, Wieben E, Weinshilboum RM. Methionine adenosyltransferase 2A/2B and methylation: gene sequence variation and functional genomics. Drug Metab Dispos. 2011;39:2135-2147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 39. | Avila MA, Berasain C, Torres L, Martín-Duce A, Corrales FJ, Yang H, Prieto J, Lu SC, Caballería J, Rodés J, Mato JM. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 274] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 40. | Lee TD, Sadda MR, Mendler MH, Bottiglieri T, Kanel G, Mato JM, Lu SC. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin Exp Res. 2004;28:173-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 116] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 41. | Lu SC, Mato JM. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. J Gastroenterol Hepatol. 2008;23 Suppl 1:S73-S77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 150] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 42. | Cai J, Sun WM, Hwang JJ, Stain SC, Lu SC. Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology. 1996;24:1090-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 104] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 43. | Martínez-Chantar ML, García-Trevijano ER, Latasa MU, Martín-Duce A, Fortes P, Caballería J, Avila MA, Mato JM. Methionine adenosyltransferase II beta subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 44. | Huang ZZ, Mao Z, Cai J, Lu SC. Changes in methionine adenosyltransferase during liver regeneration in the rat. Am J Physiol. 1998;275:G14-G21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 45. | Lu SC, Huang ZZ, Yang H, Mato JM, Avila MA, Tsukamoto H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;279:G178-G185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 123] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 46. | Huang ZZ, Mato JM, Kanel G, Lu SC. Differential effect of thioacetamide on hepatic methionine adenosyltransferase expression in the rat. Hepatology. 1999;29:1471-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 47. | Frau M, Tomasi ML, Simile MM, Demartis MI, Salis F, Latte G, Calvisi DF, Seddaiu MA, Daino L, Feo CF, Brozzetti S, Solinas G, Yamashita S, Ushijima T, Feo F, Pascale RM. Role of transcriptional and posttranscriptional regulation of methionine adenosyltransferases in liver cancer progression. Hepatology. 2012;56:165-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 72] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 48. | Zeng Z, Huang ZZ, Chen C, Yang H, Mao Z, Lu SC. Cloning and functional characterization of the 5'-flanking region of human methionine adenosyltransferase 1A gene. Biochem J. 2000;346 Pt 2:475-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 49. | Ikeda R, Nishida T, Watanabe F, Shimizu-Saito K, Asahina K, Horikawa S, Teraoka H. Involvement of CCAAT/enhancer binding protein-beta (C/EBPbeta) in epigenetic regulation of mouse methionine adenosyltransferase 1A gene expression. Int J Biochem Cell Biol. 2008;40:1956-1969. |
| 50. | Fan W, Yang H, Liu T, Wang J, Li TW, Mavila N, Tang Y, Yang J, Peng H, Tu J, Annamalai A, Noureddin M, Krishnan A, Gores GJ, Martínez-Chantar ML, Mato JM, Lu SC. Prohibitin 1 suppresses liver cancer tumorigenesis in mice and human hepatocellular and cholangiocarcinoma cells. Hepatology. 2017;65:1249-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Torres L, Avila MA, Carretero MV, Latasa MU, Caballería J, López-Rodas G, Boukaba A, Lu SC, Franco L, Mato JM. Liver-specific methionine adenosyltransferase MAT1A gene expression is associated with a specific pattern of promoter methylation and histone acetylation: implications for MAT1A silencing during transformation. FASEB J. 2000;14:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 52. | Tomasi ML, Li TW, Li M, Mato JM, Lu SC. Inhibition of human methionine adenosyltransferase 1A transcription by coding region methylation. J Cell Physiol. 2012;227:1583-1591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 53. | Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076-1087. |
| 54. | Vázquez-Chantada M, Fernández-Ramos D, Embade N, Martínez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnell RH, Caballería J, Laird-Offringa IA, Gorospe M, Lu SC, Mato JM, Martínez-Chantar ML. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943-1953. |
| 55. | Koturbash I, Melnyk S, James SJ, Beland FA, Pogribny IP. Role of epigenetic and miR-22 and miR-29b alterations in the downregulation of Mat1a and Mthfr genes in early preneoplastic livers in rats induced by 2-acetylaminofluorene. Mol Carcinog. 2013;52:318-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 56. | Yang H, Cho ME, Li TW, Peng H, Ko KS, Mato JM, Lu SC. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest. 2013;123:285-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 57. | Yang H, Huang ZZ, Wang J, Lu SC. The role of c-Myb and Sp1 in the up-regulation of methionine adenosyltransferase 2A gene expression in human hepatocellular carcinoma. FASEB J. 2001;15:1507-1516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 58. | Yang H, Huang ZZ, Zeng Z, Chen C, Selby RR, Lu SC. Role of promoter methylation in increased methionine adenosyltransferase 2A expression in human liver cancer. Am J Physiol Gastrointest Liver Physiol. 2001;280:G184-G190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 59. | Rodríguez JL, Boukaba A, Sandoval J, Georgieva EI, Latasa MU, García-Trevijano ER, Serviddio G, Nakamura T, Avila MA, Sastre J, Torres L, Mato JM, López-Rodas G. Transcription of the MAT2A gene, coding for methionine adenosyltransferase, is up-regulated by E2F and Sp1 at a chromatin level during proliferation of liver cells. Int J Biochem Cell Biol. 2007;39:842-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Yang H, Sadda MR, Yu V, Zeng Y, Lee TD, Ou X, Chen L, Lu SC. Induction of human methionine adenosyltransferase 2A expression by tumor necrosis factor alpha. Role of NF-kappa B and AP-1. J Biol Chem. 2003;278:50887-50896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 61. | Wang K, Fang S, Liu Q, Gao J, Wang X, Zhu H, Zhu Z, Ji F, Wu J, Ma Y, Hu L, Shen X, Gao D, Zhu J, Liu P, Zhou H. TGF-β1/p65/MAT2A pathway regulates liver fibrogenesis via intracellular SAM. EBioMedicine. 2019;42:458-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 62. | Ramani K, Tomasi ML. Transcriptional regulation of methionine adenosyltransferase 2A by peroxisome proliferator-activated receptors in rat hepatic stellate cells. Hepatology. 2012;55:1942-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 63. | Guo YT, Leng XS, Li T, Peng JR, Song SH, Xiong LF, Qin ZZ. Effect of ligand of peroxisome proliferator-activated receptor gamma on the biological characters of hepatic stellate cells. World J Gastroenterol. 2005;11:4735-4739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 64. | Hellemans K, Michalik L, Dittie A, Knorr A, Rombouts K, De Jong J, Heirman C, Quartier E, Schuit F, Wahli W, Geerts A. Peroxisome proliferator-activated receptor-beta signaling contributes to enhanced proliferation of hepatic stellate cells. Gastroenterology. 2003;124:184-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 108] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 65. | Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1α and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10:1113-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 66. | Liu Q, Chen J, Liu L, Zhang J, Wang D, Ma L, He Y, Liu Y, Liu Z, Wu J. The X protein of hepatitis B virus inhibits apoptosis in hepatoma cells through enhancing the methionine adenosyltransferase 2A gene expression and reducing S-adenosylmethionine production. J Biol Chem. 2011;286:17168-17180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 67. | Yang H, Zheng Y, Li TW, Peng H, Fernandez-Ramos D, Martínez-Chantar ML, Rojas AL, Mato JM, Lu SC. Methionine adenosyltransferase 2B, HuR, and sirtuin 1 protein cross-talk impacts on the effect of resveratrol on apoptosis and growth in liver cancer cells. J Biol Chem. 2013;288:23161-23170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 68. | Ramani K, Yang H, Xia M, Ara AI, Mato JM, Lu SC. Leptin's mitogenic effect in human liver cancer cells requires induction of both methionine adenosyltransferase 2A and 2beta. Hepatology. 2008;47:521-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 69. | Lo TF, Tsai WC, Chen ST. MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One. 2013;8:e75628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 70. | Tomasi ML, Cossu C, Spissu Y, Floris A, Ryoo M, Iglesias-Ara A, Wang Q, Pandol SJ, Bhowmick NA, Seki E, Posadas EM, Lu SC. S-adenosylmethionine and methylthioadenosine inhibit cancer metastasis by targeting microRNA 34a/b-methionine adenosyltransferase 2A/2B axis. Oncotarget. 2017;8:78851-78869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 71. | Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m<sup>6</sup>A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824-835.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 816] [Article Influence: 102.0] [Reference Citation Analysis (0)] |
| 72. | Shima H, Matsumoto M, Ishigami Y, Ebina M, Muto A, Sato Y, Kumagai S, Ochiai K, Suzuki T, Igarashi K. S-Adenosylmethionine Synthesis Is Regulated by Selective N<sup>6</sup>-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 2017;21:3354-3363. |
| 73. | Avila MA, Corrales FJ, Ruiz F, Sánchez-Góngora E, Mingorance J, Carretero MV, Mato IM. Specific interaction of methionine adenosyltransferase with free radicals. Biofactors. 1998;8:27-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 74. | Corrales F, Ochoa P, Rivas C, Martin-Lomas M, Mato JM, Pajares MA. Inhibition of glutathione synthesis in the liver leads to S-adenosyl-L-methionine synthetase reduction. Hepatology. 1991;14:528-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 5] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 75. | Pajares MA, Durán C, Corrales F, Mato JM. Protein kinase C phosphorylation of rat liver S-adenosylmethionine synthetase: dissociation and production of an active monomer. Biochem J. 1994;303:949-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 76. | Ramani K, Donoyan S, Tomasi ML, Park S. Role of methionine adenosyltransferase α2 and β phosphorylation and stabilization in human hepatic stellate cell trans-differentiation. J Cell Physiol. 2015;230:1075-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 77. | Ghioni P, D'Alessandra Y, Mansueto G, Jaffray E, Hay RT, La Mantia G, Guerrini L. The protein stability and transcriptional activity of p63alpha are regulated by SUMO-1 conjugation. Cell Cycle. 2005;4:183-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 78. | Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861-871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 947] [Cited by in RCA: 922] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 79. | Tomasi ML, Ryoo M, Ramani K, Tomasi I, Giordano P, Mato JM, Lu SC. Methionine adenosyltransferase α2 sumoylation positively regulate Bcl-2 expression in human colon and liver cancer cells. Oncotarget. 2015;6:37706-37723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 80. | Tomasi ML, Tomasi I, Ramani K, Pascale RM, Xu J, Giordano P, Mato JM, Lu SC. S-adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology. 2012;56:982-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 81. | Yang HB, Xu YY, Zhao XN, Zou SW, Zhang Y, Zhang M, Li JT, Ren F, Wang LY, Lei QY. Acetylation of MAT IIα represses tumour cell growth and is decreased in human hepatocellular cancer. Nat Commun. 2015;6:6973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 82. | García-Trevijano ER, Latasa MU, Carretero MV, Berasain C, Mato JM, Avila MA. S-adenosylmethionine regulates MAT1A and MAT2A gene expression in cultured rat hepatocytes: a new role for S-adenosylmethionine in the maintenance of the differentiated status of the liver. FASEB J. 2000;14:2511-2518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 91] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 83. | Martínez-Chantar ML, Latasa MU, Varela-Rey M, Lu SC, García-Trevijano ER, Mato JM, Avila MA. L-methionine availability regulates expression of the methionine adenosyltransferase 2A gene in human hepatocarcinoma cells: role of S-adenosylmethionine. J Biol Chem. 2003;278:19885-19890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 84. | Pérez C, Pérez-Zúñiga FJ, Garrido F, Reytor E, Portillo F, Pajares MA. The Oncogene PDRG1 Is an Interaction Target of Methionine Adenosyltransferases. PLoS One. 2016;11:e0161672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 85. | Jiang L, Luo X, Shi J, Sun H, Sun Q, Sheikh MS, Huang Y. PDRG1, a novel tumor marker for multiple malignancies that is selectively regulated by genotoxic stress. Cancer Biol Ther. 2011;11:567-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 86. | Peng H, Dara L, Li TW, Zheng Y, Yang H, Tomasi ML, Tomasi I, Giordano P, Mato JM, Lu SC. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology. 2013;57:2299-2313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 87. | Mato JM, Corrales FJ, Lu SC, Avila MA. S-Adenosylmethionine: a control switch that regulates liver function. FASEB J. 2002;16:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 315] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 88. | Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, Avila MA, Kanel G, Mato JM. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98:5560-5565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 353] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 89. | Santamaria E, Avila MA, Latasa MU, Rubio A, Martin-Duce A, Lu SC, Mato JM, Corrales FJ. Functional proteomics of nonalcoholic steatohepatitis: mitochondrial proteins as targets of S-adenosylmethionine. Proc Natl Acad Sci USA. 2003;100:3065-3070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 134] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 90. | Rao CV, Asch AS, Yamada HY. Emerging links among Chromosome Instability (CIN), cancer, and aging. Mol Carcinog. 2017;56:791-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 91. | Coleman WB, Tsongalis GJ. Multiple mechanisms account for genomic instability and molecular mutation in neoplastic transformation. Clin Chem. 1995;41:644-657. [PubMed] |
| 92. | Ferguson LR, Chen H, Collins AR, Connell M, Damia G, Dasgupta S, Malhotra M, Meeker AK, Amedei A, Amin A, Ashraf SS, Aquilano K, Azmi AS, Bhakta D, Bilsland A, Boosani CS, Chen S, Ciriolo MR, Fujii H, Guha G, Halicka D, Helferich WG, Keith WN, Mohammed SI, Niccolai E, Yang X, Honoki K, Parslow VR, Prakash S, Rezazadeh S, Shackelford RE, Sidransky D, Tran PT, Yang ES, Maxwell CA. Genomic instability in human cancer: Molecular insights and opportunities for therapeutic attack and prevention through diet and nutrition. Semin Cancer Biol. 2015;35 Suppl:S5-S24. |
| 93. | Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713-2722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 320] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 94. | Tomasi ML, Iglesias-Ara A, Yang H, Ramani K, Feo F, Pascale MR, Martínez-Chantar ML, Mato JM, Lu SC. S-adenosylmethionine regulates apurinic/apyrimidinic endonuclease 1 stability: implication in hepatocarcinogenesis. Gastroenterology. 2009;136:1025-1036. |
| 95. | Hsu CC, Tseng LM, Lee HC. Role of mitochondrial dysfunction in cancer progression. Exp Biol Med (Maywood). 2016;241:1281-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 223] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 97. | Ko KS, Tomasi ML, Iglesias-Ara A, French BA, French SW, Ramani K, Lozano JJ, Oh P, He L, Stiles BL, Li TW, Yang H, Martínez-Chantar ML, Mato JM, Lu SC. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology. 2010;52:2096-2108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 98. | Ramani K, Mavila N, Ko KS, Mato JM, Lu SC. Prohibitin 1 Regulates the H19-Igf2 Axis and Proliferation in Hepatocytes. J Biol Chem. 2016;291:24148-24159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 99. | Yang JW, Murray B, Barbier-Torres L, Liu T, Liu Z, Yang H, Fan W, Wang J, Li Y, Seki E, Mato JM, Lu SC. The mitochondrial chaperone Prohibitin 1 negatively regulates interleukin-8 in human liver cancers. J Biol Chem. 2019;294:1984-1996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 100. | Mavila N, Tang Y, Berlind J, Ramani K, Wang J, Mato JM, Lu SC. Prohibitin 1 Acts As a Negative Regulator of Wingless/Integrated-Beta-Catenin Signaling in Murine Liver and Human Liver Cancer Cells. Hepatol Commun. 2018;2:1583-1600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 101. | Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853-47861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 270] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 102. | Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu Rev Pharmacol Toxicol. 2004;44:27-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 399] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 103. | Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 2008;44:723-738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 611] [Cited by in RCA: 594] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 104. | Chagoya de Sánchez V, Chávez E, Velasco-Loyden G, Guadalupe Lozano-Rosas M, Rusbel Aparicio-Cadena A. Interaction of Mitochondrial and Epigenetic Regulation in Hepatocellular Carcinoma. Liver Cancer. Ahmed Lasfar: IntechOpen 2018; . [DOI] [Full Text] |
| 105. | Alison MR. Liver stem cells: implications for hepatocarcinogenesis. Stem Cell Rev. 2005;1:253-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 124] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 106. | Jelnes P, Santoni-Rugiu E, Rasmussen M, Friis SL, Nielsen JH, Tygstrup N, Bisgaard HC. Remarkable heterogeneity displayed by oval cells in rat and mouse models of stem cell-mediated liver regeneration. Hepatology. 2007;45:1462-1470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 84] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 107. | Dumble ML, Croager EJ, Yeoh GC, Quail EA. Generation and characterization of p53 null transformed hepatic progenitor cells: oval cells give rise to hepatocellular carcinoma. Carcinogenesis. 2002;23:435-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 172] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 108. | Rountree CB, Senadheera S, Mato JM, Crooks GM, Lu SC. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology. 2008;47:1288-1297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 109. | Ding W, Mouzaki M, You H, Laird JC, Mato J, Lu SC, Rountree CB. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49:1277-1286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 110. | Nguyen LN, Furuya MH, Wolfraim LA, Nguyen AP, Holdren MS, Campbell JS, Knight B, Yeoh GC, Fausto N, Parks WT. Transforming growth factor-beta differentially regulates oval cell and hepatocyte proliferation. Hepatology. 2007;45:31-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 111. | Tomasi ML, Ramani K, Lopitz-Otsoa F, Rodríguez MS, Li TW, Ko K, Yang H, Bardag-Gorce F, Iglesias-Ara A, Feo F, Pascale MR, Mato JM, Lu SC. S-adenosylmethionine regulates dual-specificity mitogen-activated protein kinase phosphatase expression in mouse and human hepatocytes. Hepatology. 2010;51:2152-2161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 112. | Pañeda C, Gorospe I, Herrera B, Nakamura T, Fabregat I, Varela-Nieto I. Liver cell proliferation requires methionine adenosyltransferase 2A mRNA up-regulation. Hepatology. 2002;35:1381-1391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 113. | Ramani K, Yang H, Kuhlenkamp J, Tomasi L, Tsukamoto H, Mato JM, Lu SC. Changes in the expression of methionine adenosyltransferase genes and S-adenosylmethionine homeostasis during hepatic stellate cell activation. Hepatology. 2010;51:986-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 114. | Vázquez-Chantada M, Ariz U, Varela-Rey M, Embade N, Martínez-Lopez N, Fernández-Ramos D, Gómez-Santos L, Lamas S, Lu SC, Martínez-Chantar ML, Mato JM. Evidence for LKB1/AMP-activated protein kinase/ endothelial nitric oxide synthase cascade regulated by hepatocyte growth factor, S-adenosylmethionine, and nitric oxide in hepatocyte proliferation. Hepatology. 2009;49:608-617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 115. | Personeni N, Rimassa L, Pressiani T, Destro A, Ligorio C, Tronconi MC, Bozzarelli S, Carnaghi C, Di Tommaso L, Giordano L, Roncalli M, Santoro A. Molecular determinants of outcome in sorafenib-treated patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2013;139:1179-1187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 116. | Negri FV, Dal Bello B, Porta C, Campanini N, Rossi S, Tinelli C, Poggi G, Missale G, Fanello S, Salvagni S, Ardizzoni A, Maria SE. Expression of pERK and VEGFR-2 in advanced hepatocellular carcinoma and resistance to sorafenib treatment. Liver Int. 2015;35:2001-2008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 117. | Sun H, Charles CH, Lau LF, Tonks NK. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 900] [Cited by in RCA: 939] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 118. | Calvisi DF, Pinna F, Meloni F, Ladu S, Pellegrino R, Sini M, Daino L, Simile MM, De Miglio MR, Virdis P, Frau M, Tomasi ML, Seddaiu MA, Muroni MR, Feo F, Pascale RM. Dual-specificity phosphatase 1 ubiquitination in extracellular signal-regulated kinase-mediated control of growth in human hepatocellular carcinoma. Cancer Res. 2008;68:4192-4200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 119. | Lin YW, Yang JL. Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling. J Biol Chem. 2006;281:915-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 120. | Korsse SE, Peppelenbosch MP, van Veelen W. Targeting LKB1 signaling in cancer. Biochim Biophys Acta. 2013;1835:194-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 121. | Huang YH, Chen ZK, Huang KT, Li P, He B, Guo X, Zhong JQ, Zhang QY, Shi HQ, Song QT, Yu ZP, Shan YF. Decreased expression of LKB1 correlates with poor prognosis in hepatocellular carcinoma patients undergoing hepatectomy. Asian Pac J Cancer Prev. 2013;14:1985-1988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 122. | Martínez-López N, García-Rodríguez JL, Varela-Rey M, Gutiérrez V, Fernández-Ramos D, Beraza N, Aransay AM, Schlangen K, Lozano JJ, Aspichueta P, Luka Z, Wagner C, Evert M, Calvisi DF, Lu SC, Mato JM, Martínez-Chantar ML. Hepatoma cells from mice deficient in glycine N-methyltransferase have increased RAS signaling and activation of liver kinase B1. Gastroenterology. 2012;143:787-798.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 123. | Barbier-Torres L, Delgado TC, García-Rodríguez JL, Zubiete-Franco I, Fernández-Ramos D, Buqué X, Cano A, Gutiérrez-de Juan V, Fernández-Domínguez I, Lopitz-Otsoa F, Fernández-Tussy P, Boix L, Bruix J, Villa E, Castro A, Lu SC, Aspichueta P, Xirodimas D, Varela-Rey M, Mato JM, Beraza N, Martínez-Chantar ML. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget. 2015;6:2509-2523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 124. | Tan X, Liao Z, Liang H, Chen X, Zhang B, Chu L. Upregulation of liver kinase B1 predicts poor prognosis in hepatocellular carcinoma. Int J Oncol. 2018;53:1913-1926. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 125. | Zubiete-Franco I, García-Rodríguez JL, Lopitz-Otsoa F, Serrano-Macia M, Simon J, Fernández-Tussy P, Barbier-Torres L, Fernández-Ramos D, Gutiérrez-de-Juan V, López de Davalillo S, Carlevaris O, Beguiristain Gómez A, Villa E, Calvisi D, Martín C, Berra E, Aspichueta P, Beraza N, Varela-Rey M, Ávila M, Rodríguez MS, Mato JM, Díaz-Moreno I, Díaz-Quintana A, Delgado TC, Martínez-Chantar ML. SUMOylation regulates LKB1 localization and its oncogenic activity in liver cancer. EBioMedicine. 2019;40:406-421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 126. | Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563-575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1555] [Cited by in RCA: 1520] [Article Influence: 95.0] [Reference Citation Analysis (0)] |
| 127. | Chen L, Zeng Y, Yang H, Lee TD, French SW, Corrales FJ, García-Trevijano ER, Avila MA, Mato JM, Lu SC. Impaired liver regeneration in mice lacking methionine adenosyltransferase 1A. FASEB J. 2004;18:914-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 128. | Avila MA, Mingorance J, Martínez-Chantar ML, Casado M, Martin-Sanz P, Boscá L, Mato JM. Regulation of rat liver S-adenosylmethionine synthetase during septic shock: role of nitric oxide. Hepatology. 1997;25:391-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 129. | Ruiz F, Corrales FJ, Miqueo C, Mato JM. Nitric oxide inactivates rat hepatic methionine adenosyltransferase In vivo by S-nitrosylation. Hepatology. 1998;28:1051-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 130. | Martínez-Chantar ML, Vázquez-Chantada M, Garnacho M, Latasa MU, Varela-Rey M, Dotor J, Santamaria M, Martínez-Cruz LA, Parada LA, Lu SC, Mato JM. S-adenosylmethionine regulates cytoplasmic HuR via AMP-activated kinase. Gastroenterology. 2006;131:223-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 131. | Martínez-López N, Varela-Rey M, Fernández-Ramos D, Woodhoo A, Vázquez-Chantada M, Embade N, Espinosa-Hevia L, Bustamante FJ, Parada LA, Rodriguez MS, Lu SC, Mato JM, Martínez-Chantar ML. Activation of LKB1-Akt pathway independent of phosphoinositide 3-kinase plays a critical role in the proliferation of hepatocellular carcinoma from nonalcoholic steatohepatitis. Hepatology. 2010;52:1621-1631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 132. | Mato JM, Cámara J, Fernández de Paz J, Caballería L, Coll S, Caballero A, García-Buey L, Beltrán J, Benita V, Caballería J, Solà R, Moreno-Otero R, Barrao F, Martín-Duce A, Correa JA, Parés A, Barrao E, García-Magaz I, Puerta JL, Moreno J, Boissard G, Ortiz P, Rodés J. S-adenosylmethionine in alcoholic liver cirrhosis: a randomized, placebo-controlled, double-blind, multicenter clinical trial. J Hepatol. 1999;30:1081-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 300] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 133. | Liu GY, Wang W, Jia WD, Xu GL, Ma JL, Ge YS, Yu JH, Sun QK, Meng FL. Protective effect of S-adenosylmethionine on hepatic ischemia-reperfusion injury during hepatectomy in HCC patients with chronic HBV infection. World J Surg Oncol. 2014;12:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 134. | Rambaldi A, Gluud C, Rambaldi A. S-adenosyl-L-methionine for alcoholic liver diseases. Rambaldi A. Cochrane Database of Systematic Reviews. Chichester, United Kingdom: John Wiley & Sons, Ltd; 2001; CD002235. [RCA] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 135. | Nieto N, Cederbaum AI. S-adenosylmethionine blocks collagen I production by preventing transforming growth factor-beta induction of the COL1A2 promoter. J Biol Chem. 2005;280:30963-30974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 136. | Ansorena E, García-Trevijano ER, Martínez-Chantar ML, Huang ZZ, Chen L, Mato JM, Iraburu M, Lu SC, Avila MA. S-adenosylmethionine and methylthioadenosine are antiapoptotic in cultured rat hepatocytes but proapoptotic in human hepatoma cells. Hepatology. 2002;35:274-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 115] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 137. | Yang H, Sadda MR, Li M, Zeng Y, Chen L, Bae W, Ou X, Runnegar MT, Mato JM, Lu SC. S-adenosylmethionine and its metabolite induce apoptosis in HepG2 cells: Role of protein phosphatase 1 and Bcl-x(S). Hepatology. 2004;40:221-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 138. | Lu SC, Ramani K, Ou X, Lin M, Yu V, Ko K, Park R, Bottiglieri T, Tsukamoto H, Kanel G, French SW, Mato JM, Moats R, Grant E. S-adenosylmethionine in the chemoprevention and treatment of hepatocellular carcinoma in a rat model. Hepatology. 2009;50:462-471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 139. | Chen YM, Shiu JY, Tzeng SJ, Shih LS, Chen YJ, Lui WY, Chen PH. Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma. Int J Cancer. 1998;75:787-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 140. | Li J, Ramani K, Sun Z, Zee C, Grant EG, Yang H, Xia M, Oh P, Ko K, Mato JM, Lu SC. Forced expression of methionine adenosyltransferase 1A in human hepatoma cells suppresses in vivo tumorigenicity in mice. Am J Pathol. 2010;176:2456-2466. |
| 141. | Lombardini JB, Coulter AW, Talalay P. Analogues of methionine as substrates and inhibitors of the methionine adenosyltransferase reaction. Deductions concerning the conformation of methionine. Mol Pharmacol. 1970;6:481-499. [PubMed] |
| 142. | Sviripa VM, Zhang W, Balia AG, Tsodikov OV, Nickell JR, Gizard F, Yu T, Lee EY, Dwoskin LP, Liu C, Watt DS. 2',6'-Dihalostyrylanilines, pyridines, and pyrimidines for the inhibition of the catalytic subunit of methionine S-adenosyltransferase-2. J Med Chem. 2014;57:6083-6091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 143. | Anwar SL, Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J Gastroenterol. 2014;20:7894-7913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 65] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 144. | Frau M, Feo F, Pascale RM. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J Hepatol. 2013;59:830-841. |
| 145. | An J, Na SK, Shim JH, Park YS, Jun MJ, Lee JH, Song GW, Lee HC, Yu E. Histological expression of methionine adenosyl transferase (MAT) 2A as a post-surgical prognostic surrogate in patients with hepatocellular carcinoma. J Surg Oncol. 2018;117:892-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 146. | Wang R, Jin Y, Yao XH, Fan W, Zhang J, Cao Y, Li J. A novel mechanism of the M1-M2 methionine adenosyltransferase switch-mediated hepatocellular carcinoma metastasis. Mol Carcinog. 2018;57:1201-1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |