Published online Aug 7, 2019. doi: 10.3748/wjg.v25.i29.3985
Peer-review started: March 26, 2019
First decision: May 24, 2019
Revised: June 13, 2019
Accepted: July 5, 2019
Article in press: July 3, 2019
Published online: August 7, 2019
Processing time: 134 Days and 6.4 Hours
Hepatitis B is a major public health problem in China. Accurate liver injury assessment is essential for clinical evidence-based treatment. Liver biopsy is considered the gold standard method to stage liver disease, but it is not widely used in resource-limited settings. Therefore, non-invasive liquid biopsy tests are needed.
To assess liver injury in hepatitis B patients using quantified cell free DNA combined with other serum biomarker as a liquid biopsy-based method.
A cohort of 663 subjects including 313 hepatitis B patients and 350 healthy controls were enrolled. Ultrasound-guided liver biopsies followed by histopathological assessments were performed for the 263 chronic hepatitis B patients to determine the degree of liver injury. Cell-free DNA was quantified using a novel duplex real-time polymerase chain reaction assay.
Compared with healthy controls, patients with hepatitis B virus (HBV) infection had significantly higher plasma DNA, serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), bilirubin, and HBV DNA levels (P < 0.01). Serum ALT, AST, bilirubin, and plasma DNA levels of patients with marked-severe inflammation were significantly higher than those with mild-moderate inflammation (P < 0.01). There was a statistically significant correlation between hepatocyte inflammation severity and serum bilirubin (R2 = 0.673, P < 0.01) or plasma DNA (R2 = 0.597, P < 0.01) levels. The areas under the curves of serum ALT, bilirubin, plasma DNA, and their combination to distinguish between patients with mild–moderate and marked-severe inflammation were 0.8059, 0.7910, 0.7921, and 0.9564, respectively.
The combination of plasma DNA, serum ALT, and bilirubin could be a candidate liquid biopsy for non-invasive assessment of liver injury in hepatitis B patients.
Core tip: Our study used quantified cell free DNA combined with other serum biomarker as a liquid biopsy-based method to assess liver injury in hepatitis B patients. A cohort of 663 subjects including 313 hepatitis B patients and 350 healthy controls were enrolled. Ultrasound-guided liver biopsies followed by histopathological assessments were performed for the 263 chronic hepatitis B patients to determine the degree of liver injury. Cell-free DNA was quantified using a novel duplex real-time polymerase chain reaction assay. Our results demonstrated that the combination of plasma DNA, serum alanine aminotransferase, and bilirubin could be a candidate liquid biopsy for non-invasive assessment of liver injury in hepatitis B patients.
- Citation: Xia WY, Gao L, Dai EH, Chen D, Xie EF, Yang L, Zhang SC, Zhang BF, Xu J, Pan SY. Liquid biopsy for non-invasive assessment of liver injury in hepatitis B patients. World J Gastroenterol 2019; 25(29): 3985-3995
- URL: https://www.wjgnet.com/1007-9327/full/v25/i29/3985.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i29.3985
The liver, playing an important role in many bodily functions from protein production and blood clotting to cholesterol, glucose, and iron metabolism, is the most important detoxification organ in human body[1]. Because of this, the liver becomes the vulnerable organ to be injured in various kinds of diseases, including pathogen infection, inherited metabolic disease, alcoholic hepatitis, drug-induced liver disease, autoimmune liver disease, and fatty liver disease. In clinical practice, accurate liver injury assessment is necessary for the evidence-based treatment.
Liver biopsy, which is thought to be the gold standard method for the assessment of liver injury severity, has been used to ascertain the degree of necroinflammation and fibrosis[1]. Unfortunately, liver biopsy is not always an available option for many patients with liver disease. It is also impractical to monitor the disease by frequent biopsy. Blood biomarkers, such as liver aminotransferases and bilirubin, are the routine assessments as liquid biopsy to monitor patients and to guide clinical treatment[1]. However, it is widely known that the degree of liver injury is quite different in various types of liver diseases. It is also thought that serum alanine aminotransferase (ALT) or aspartate aminotransferase (AST) concentrations are not necessarily a reliable index of hepatocyte necrosis, and this is especially seen in massive hepatic necrosis[2]. To assess liver injury more accurately, more reliable biomarkers for liquid biopsy are required.
In this study, we focused on hepatitis B caused by the hepatitis B virus (HBV) which is a DNA virus that infects the liver. There are approximately two billion people exposed to HBV and more than 350 million chronically infected people worldwide[3,4]. Hepatitis B is a major public health problem worldwide, especially in China[5]. HBV infection can be either asymptomatic or symptomatic depending on the severity of liver injury, which is important for patients’ outcome and to help guide the choice of clinical therapy. The aim of the present study was to quantify cell released DNA, a sensitive biomarker for cell death assessment, in hepatitis B patients using the duplex real-time quantitative polymerase chain reaction (PCR) to explore its potential use as a liquid biopsy-based biomarker combined with other liver function tests for assessment of liver injury.
This study included a total of 313 HBV-infected patients (median age, 36 years; range, 12-78 years; 89 females) with chronic (n = 263) or acute (n = 50) infection. Chronic HBV infection was defined as a continuous positive hepatitis B surface antigen (HBsAg) test result for more than 6 months, while acute HBV infection was a positive HBsAg test result for less than 6 months. Clinical information including demographic characteristics was obtained from the patients’ medical records. Healthy volunteers (n = 350; median age, 36 years; range, 18-72 years; 100 females) who attended the First Affiliated Hospital of Nanjing Medical University for an annual health check-up were recruited as a control group.
Ultrasound-guided liver biopsies followed by histopathological assessment were performed for the 263 chronic HBV infected patients to determine the degrees of liver injury including inflammation (grade) and fibrosis (stage) according to the Grading and Staging System in China[6], which was established based on the Ishak system[7]. In this study, mild or moderate portal area inflammation and spotty or piecemeal necrosis without bridging and multi-acinar necrosis were categorized as mild–moderate inflammation, while marked portal area inflammation, marked spotty or piecemeal necrosis, and/or bridging and multi-acinar necrosis were categorized marked-severe inflammation. The fibrosis stages range from 0 to 4, where Stage 0 is for no fibrosis, Stage 1 for portal fibrosis without septa, Stage 2 for portal fibrosis with few septa, Stage 3 for numerous septa without cirrhosis, and Stage 4 for cirrhosis.
After informed consent was obtained and the research protocol was approved by the Ethics Committee of Nanjing Medical University, a 2-mL peripheral blood sample was collected into an ethylenediamine tetraacetic acid-containing tube from each participant at the time of preliminary diagnosis. All of the blood samples were centrifuged (1600 g, 10 min) within 2 h after collection and plasma was carefully transferred into EP tubes. After an additional centrifugation at 16000 g for 10 min, 200 µL of plasma supernatant without blood cells was collected and 5 × 104 copies of recombinant internal standard plasmid DNA were added as described previously[8]. The mixed samples were stored at -80 °C until further processing. Serum samples were recovered from serum-separator tubes following centrifugation of whole blood at 3000 g for 10 min at room temperature.
DNA was extracted from the 200 µL plasma samples containing the internal standard using the BILATEST DNA/RNA kit (BILATEC, Viernheim, Germany), following the manufacturer’s recommendations. After DNA extraction, plasma DNA was detected with plasma cell-free DNA quantitative detection kit (Code Biotech, Jiangsu, China) using duplex real-time PCR, which was performed for both the human β–actin gene and the internal control recombinant plasmid DNA in the same tube. Duplex amplification curves were analyzed with Sequence Detection System Software (Ver. 1.4, Applied Biosystems). Specific probes and primers were described in our previous report[8].
The routine clinical chemistry panel, including ALT, AST, bilirubin, and albumin was measured on an automatic biochemistry analyzer (AU5800, Beckman-Coulter, United States). Serum HBV DNA was quantified by real-time PCR assay (ABI7500, Applied Biosystem, United States). ALT and AST activities were measured using the recommended IFCC method. Bilirubin was measured using the vanadic acid oxidation method. Albumin was measured using the bromocresol green colorimetry method. Serum HBV DNA was extracted using an HBV nucleic acid quantitative detection kit (KHB, China) and quantified by real-time PCR assay (ABI7500, Applied Biosystem, United States).
Results are presented as the median and interquartile range. Data were analyzed using the Kruskal–Wallis rank test and Mann–Whitney U test. Spearman’s rank correlation was performed to estimate the correlation between the degree of liver injury and plasma DNA, serum ALT, or bilirubin, respectively. The area under receiver operator characteristic (ROC) curve (AUC) was calculated to evaluate the diagnostic efficacy. All of these data were analyzed using Stata 9.2 software (Stata Corporation, College Station, TX, United States). A P-value < 0.05 was considered statistically significant.
The concentrations of serum ALT, AST, bilirubin, albumin, total plasma DNA, and HBV DNA in healthy controls and patients with HBV infection are shown in Table 1. There were statistically significant differences in serum ALT, AST, and plasma DNA levels between healthy males and females (P < 0.01). Healthy controls with a history of drinking alcohol had higher serum ALT, AST, bilirubin, and plasma DNA levels than those who rarely drank alcohol (P < 0.01).
| Group | Number of cases | Serum ALT (U/L) | P | Bilirubin (μM) | P | Plasma DNA (ng/mL) | P | HBV DNA (IU/mL) | P | Serum AST (U/L) | P | Albumin(g/L) | P |
| Healthy controls | 350 | 19.1 (15.1) | 25.5 (17.6) | 0.0 (0.0) | 16.4 (14.3) | 46.0 (22.2) | |||||||
| Sex | < 0.01 | 0.865 | < 0.01 | --- | < 0.01 | 0.062 | |||||||
| Male | 250 | 22.3 (14.1) | 10.6 (5.8) | 30.0 (16.1) | 0.0 (0.0) | 19.6 (14.1) | 46.4 (20.8) | ||||||
| Female | 100 | 12.6 (7.6) | 10.2 (5.5) | 16.7 (8.1) | 0.0 (0.0) | 10.1 (5.5) | 45.5 (20.1) | ||||||
| Age (yr) | 0.819 | 0.833 | 0.056 | --- | 0.916 | 0.066 | |||||||
| < 36 | 175 | 18.9 (17.9) | 9.7 (5.4) | 23.6 (16.4) | 0.0 (0.0) | 16.0 (15.9) | 47.6 (20.3) | ||||||
| ≥ 36 | 175 | 19.8 (13.7) | 10.5 (5.7) | 26.8 (19.3) | 0.0 (0.0) | 16.7 (12.2) | 45.8 (23.4) | ||||||
| HBsAb | 0.263 | 0.439 | 0.060 | --- | 0.549 | 0.884 | |||||||
| Nega-tive | 166 | 18.7 (15.1) | 10.3 (5.6) | 27.3 (19.1) | 0.0 (0.0) | 15.8 (15.1) | 45.7 (22.5) | ||||||
| Positive | 184 | 19.1 (15.0) | 10.3 (5.6) | 24.2 (16.0) | 0.0 (0.0) | 16.5 (15.2) | 46.2 (21.6) | ||||||
| Drink-ing history | < 0.01 | 0.002 | 0.000 | --- | 0.000 | 0.711 | |||||||
| No | 278 | 18.6 (12.1) | 10.4 (5.7) | 22.8 (15.5) | 0.0 (0.0) | 15.8 (11.1) | 46.1 (20.9) | ||||||
| Yes | 72 | 34.1 (29.2) | 21.0 (19.2) | 38.3 (19.0) | 0.0 (0.0) | 33.5 (27.4) | 45.4 (23.7) | ||||||
| Hepati-tis B patients | 313 | 103.3 (118.5) | < 0.011 | 85.6 (77.2) | 0.0001 | 132.9 (253.7) | < 0.011 | 6.1 (2.9) | < 0.011 | 97.0 (101.6) | < 0.011 | 42.7 (31.2) | < 0.011 |
| Sex | 0.972 | 0.606 | 0.772 | 0.149 | 0.656 | 0.761 | |||||||
| Male | 224 | 101.4 (114.4) | 88.3 (91.6) | 134.2 (260.2) | 6.2 (2.7) | 95.3 (99.5) | 42.4 (32.7) | ||||||
| Female | 89 | 113.5 (143.8) | 82.9 (89.1) | 132.2 (170.7) | 5.6 (2.8) | 101.7 (116.6) | 43.0 (33.2) | ||||||
| Age (yr) | 0.011 | 0.221 | 0.312 | < 0.01 | 0.831 | 0.591 | |||||||
| < 36 | 155 | 89.2 (120.0) | 86.3 (90.4) | 135.9 (256.6) | 6.7 (3.0) | 96.5 (98.3) | 42.2 (30.4) | ||||||
| ≥ 36 | 158 | 115.9 (124.9) | 83.9 (91.5) | 115.8 (244.5) | 5.6 (2.6) | 97.8 (103.2) | 43.1 (29.7) | ||||||
| HBeAg | 0.236 | 0.173 | 0.833 | < 0.01 | 0.742 | 0.070 | |||||||
| Nega-tive | 159 | 109.7 (126.6) | 84.4 (89.4) | 126.3 (294.0) | 5.4 (3.1) | 97.7 (101.8) | 43.3 (32.5) | ||||||
| Positive | 154 | 101.4 (117.0) | 86.1 (90.7) | 135.7 (228.9) | 6.5 (2.3) | 97.4 (99.6) | 40.6 (34.6) | ||||||
| Drink-ing history | 0.487 | 0.577 | 0.574 | 0.200 | 0.433 | 0.654 | |||||||
| No | 187 | 107.0 (122.8) | 85.0 (87.3) | 126.3 (255.4) | 6.2 (2.7) | 95.8 (98.5) | 42.0 (30.8) | ||||||
| Yes | 126 | 100.6 (118.0) | 86.9 (90.3) | 142.8 (243.6) | 6.0 (3.0) | 98.3 (100.1) | 43.2 (30.7) |
Compared with healthy controls, patients with HBV infection had significantly higher plasma DNA, serum ALT, AST, bilirubin, HBV DNA levels, and lower serum albumin levels (P < 0.01). There were no statistically significant differences in the six blood-based biomarkers between patients with different sex or drinking history (P > 0.05). Although there were significant differences in serum ALT and HBV DNA levels between patients younger and older than 36 years, the Spearman’s rank correlation analysis showed that there were no statistical correlations between patient age and the blood-based biomarkers. Serum HBV DNA levels in HBeAg positive patients were significantly higher than those with negative HBeAg results (P < 0.01).
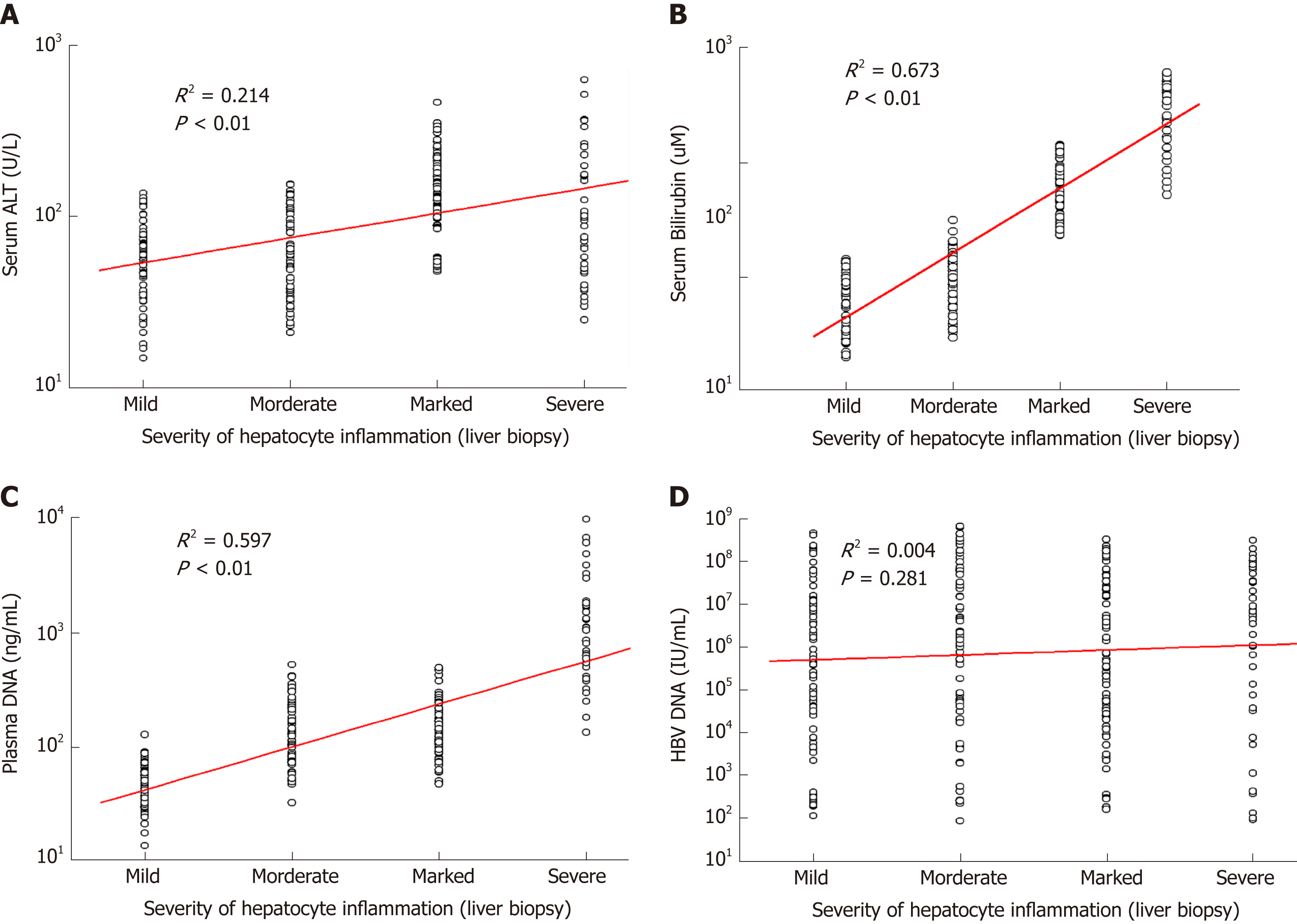
As shown in Table 2, serum ALT, AST, bilirubin, and plasma DNA levels of patients with marked-severe inflammation were significantly higher than those of patients with mild–moderate inflammation (P < 0.01). However, serum ALT or AST levels of 42 patients with severe liver injury were lower than those of patients with marked inflammation. There was no statistically significant difference in serum HBV DNA or albumin levels between patients with mild–moderate and marked-severe inflammation (P > 0.05). The positive correlation between serum ALT and inflammation severity was statistically significant but weak (R2 = 0.214, P < 0.01; Figure 1A) because the severe patients had low ALT levels. Inflammation severity determined by liver biopsy was correlated with serum bilirubin (R2 = 0.673, P < 0.01; Figure 1B) and plasma DNA (R2 = 0.597, P < 0.01; Figure 1C), but had no statistically significant correlation with HBV DNA (R2 = 0.004, P = 0.281; Figure 1D).
| Group | Number of cases | Serum ALT (U/L) | P | Bilirubin (μM) | P | Plasma DNA (ng/mL) | P | HBV DNA (IU/mL) | P | Serum AST (U/L) | P | Albumin(g/L) | P |
| Healthy controls | 350 | 19.1 (15.1) | 25.5 (17.6) | 0.0 (0.0) | 16.4 (14.3) | 46.0 (22.2) | |||||||
| Acute HBV infec-tion | 50 | 213.8 (355.9) | 121.6 (207.0) | 336.8 (179.5) | 5.5 (2.4) | 197.6 (312.5) | 45.4 (24.6) | ||||||
| Chronic HBV infec-tion | 263 | 88.3 (98.0) | < 0.011 | 73.9 (88.3) | < 0.011 | 106.4 (174.1) | < 0.011 | 6.2 (2.9) | 0.059* | 90.6 (93.7) | < 0.011 | 40.2 (41.3) | 0.0041 |
| Inflamma-tion | < 0.01 | < 0.01 | < 0.01 | 0.893 | < 0.01 | 0.444 | |||||||
| Mild | 68 | 59.9 (41.5) | 30.6 (31.4) | 46.9 (29.5) | 5.7 (2.5) | 51.2 (50.6) | 41.7 (28.6) | ||||||
| Modera-te | 70 | 62.8 (68.0) | 45.4 (50.4) | 106.1 (127.2) | 6.4 (3.1) | 61.3 (72.9) | 40.9 (30.4) | ||||||
| Marked | 83 | 152.8 (114.7) | 130 (125.5) | 141.1 (77.4) | 5.8 (2.8) | 153.2 (160.8) | 39.5 (32.6) | ||||||
| Severe | 42 | 99.9 (148.0) | 228 (254.9) | 862.5 (1213.6) | 6.8 (2.8) | 91.7 (135.1) | 39.8 (33.2) | ||||||
| Fibrosis | 0.552 | 0.015 | < 0.01 | 0.060 | 0.121 | 0.606 | |||||||
| No cirrhosis | 212 | 88.0 (106.4) | 65.2 (58.2) | 131.0 (235.8) | 6.3 (3.1) | 87.0 (93.0) | 40.8 (38.6) | ||||||
| Stage 0 | 59 | 83.5 (98.6) | 71.9 (70.3) | 111.8 (477.0) | 6.6 (3.0) | 87.7 (91.6) | 41.6 (38.5) | ||||||
| Stage 1 | 72 | 96.6 (81.2) | 60.2 (66.7) | 179.6 (235.3) | 6.5 (3.0) | 88.4 (90.3) | 40.1 (38.3) | ||||||
| Stage 2 | 46 | 87.4 (89.0) | 64.8 (79.6) | 98.8 (90.1) | 6.0 (2.7) | 79.1 (82.5) | 40.1 (38.7) | ||||||
| Stage 3 | 35 | 91.7 (110.3) | 77.7 (72.4) | 87.2 (77.7) | 6.0 (2.7) | 84.6 (88.0) | 40.2 (39.9) | ||||||
| Cirrho-sis (Stage 4) | 51 | 91.4 (73.5) | 92.0 (99.4) | 73.1 (44.8) | 5.7 (2.1) | 91.9 (94.5) | 39.9 (43.8) |
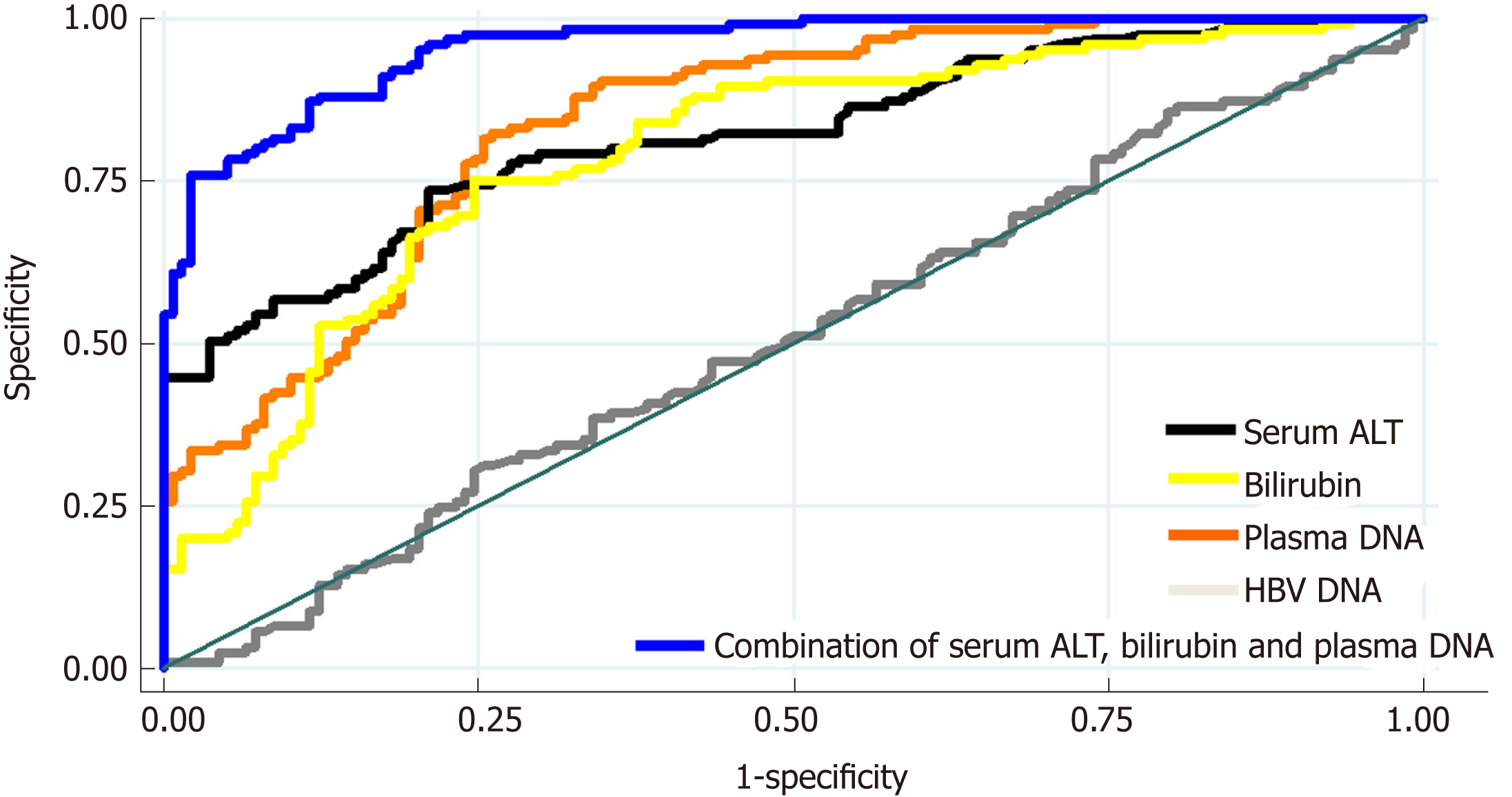
As shown in Figure 2, the AUC of using plasma DNA to distinguish between patients with mild–moderate and marked-severe inflammation was 0.7921, which was similar to the AUCs of using serum ALT and bilirubin (0.8059 and 0.7910). The AUCs of using serum AST, albumin, and HBV DNA to distinguish between patients with mild-moderate and marked-severe inflammation were 0.6530, 0.4877, and 0.4952, respectively. After the combination of serum ALT, bilirubin, and plasma DNA (blue), there was a statistically significant increase of AUC (0.9564). There was also a significant difference in plasma DNA levels or bilirubin between patients with and without cirrhosis (P < 0.01), but there was no difference in serum ALT, AST, albumin, or HBV DNA (P > 0.05; Table 2).
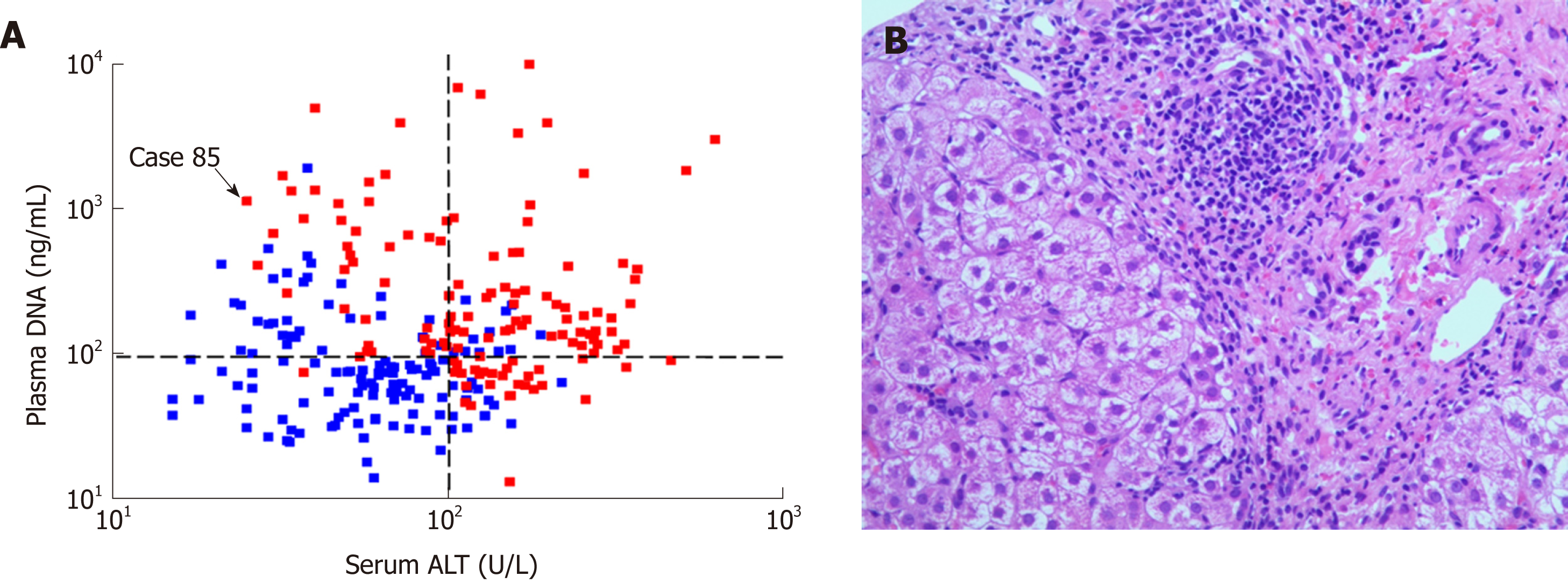
As shown in Figure 3A, there was no statistically significant correlation between plasma DNA and serum ALT (R2 = 0.012, P = 0.08). Most of the patients with high serum ALT levels (> 100.0 U/L) had marked-severe liver injury (red), suggesting a high specificity (89.3%) and positive predictive value (84.0%). However, there were still 33.5% of patients with low serum ALT levels (≤ 100.0 U/L) who had marked-severe liver injury, which suggests a low sensitivity (55.3%) of serum ALT. For example, patient 85 in Figure 3A was a 20-year-old man with chronic hepatitis B (CHB). Laboratory studies showed an HBV DNA level of 2.04 × 107 IU/mL and an ALT level of only 40 U/L. However, a needle biopsy of the liver showed severe piecemeal necrosis (marked portal inflammation, Grade 3; Figure 3B). These false negative patients could be further distinguished from patients with mild–moderate inflammation by plasma DNA with a sensitivity of 97.5% and specificity of 68.6% at the cutoff value of 95 ng/mL, as shown in Figure 3A. After the combination of serum ALT, bilirubin, and plasma DNA, there was a statistically significant increase in AUC (0.9564), with a maximum sensitivity and specificity of 88.64% and 80.15%, respectively (the blue line in Figure 2).
Serum ALT, AST, bilirubin, albumin, total plasma DNA, and HBV DNA levels in patients with chronic or acute HBV infection are shown in Table 2. There was no statistically significant difference in HBV DNA levels between patients with chronic and acute HBV infection (P > 0.05). However, total plasma DNA, bilirubin, serum ALT, AST, and albumin levels in patients with acute HBV infection were significantly higher than those of patients with chronic HBV infection (P < 0.01). ROC curve analysis showed that the AUC of using serum ALT levels to distinguish between patients with chronic and acute HBV infection was 0.8702, which was significantly higher than that of using plasma DNA (0.7800; P = 0.01).
Hepatitis B is a viral infectious disease caused by the HBV which primarily interferes with the functions of the liver by replicating in hepatocytes[5]. During HBV infection, the host immune response causes both viral clearance and hepatocellular damage[9]. In clinical practice, hepatitis B patients usually need to undergo a liver biopsy, which is the current gold standard method for liver injury assessment by direct cell morphological observation[1,10]. However, biopsy results are too dependent on the representation of the punctured sample and show significant variability which can lead to a wrong diagnosis[11]. Besides, liver biopsy requires a skilled expert and well-equipped hospital, and still has the risk of potentially lethal complications, such as pneumothorax and bleeding[12]. Therefore, liver biopsy is not always the best option for monitoring disease and some other more reliable techniques to assess liver injury, such as liquid biopsy focusing on biomarkers in body fluids, are required.
HBV infection can stimulate the body to produce various kinds of cell and humoral immunity responses to virus antigens and lead to persistent or massive hepatocellular apoptosis and necrosis[9,13,14]. As a result of cell damage, the components of liver cells including proteins and nucleic acids are released into the peripheral blood stream. This could increase the reference values and the quantitative detection of these substances released from damaged liver cells into body fluids may serve as a noninvasive liquid biopsy to evaluate and monitor hepatitis-related liver damage[15].
Liver-specific enzymes such as ALT and AST are the most common serum biomarkers for liver function assessment[16]. However, the point of using ALT or AST for liver cell injury assessment remains controversial. Desmet et al[17] found that the serum ALT level rose in almost all chronic liver diseases, yet this marker could not reliably reflect the degree of inflammatory injury. Kew suggested that serum ALT was not a reliable indicator in extensive hepatic necrosis in severe hepatitis, when decreasing serum ALT concentrations might signify a paucity of hepatocytes from which the enzymes could leak, rather than recovery[2]. Our current findings also demonstrate that serum ALT and AST levels are not always correlated with hepatocyte inflammation, especially in the patients with severe hepatocyte injury (Figure 1). The sensitivity of serum ALT or AST is less than 60%, which will lead to missed diagnosis and delay in clinical treatment.
Circulating plasma DNA, a kind of cell-free extracellular nucleic acid present in normal healthy individuals at low concentrations, is believed to derive primarily from apoptosis of normal cells[18]. The short half-life of plasma DNA in the circulation suggests a model of continuous release from apoptotic cells and rapid clearance[19]. In the context of various disease states characterized by abnormal cell death, such as cancer, trauma, and transplant rejection, a large amount of nucleic acids are released from necrotic cells into blood stream and significantly increase the level of plasma DNA[20-26]. Although plasma DNA quantification was proved to be a potential marker for cell damage, various preanalytical factors and lack of accurate and precise quantitative methods have become a considerable pitfall, hampering its application for liver injury assessment in clinical laboratories[30,31,32]. In our previous study, a duplex real-time PCR assay with a novel internal standard was developed for plasma DNA quantification and proved to be able to eliminate variations and allow for more sensitive, repeatable, accurate, and stable quantitative measurements of plasma DNA[8].
In the current study, we quantified total plasma DNA in 350 healthy controls and 313 HBV infected patients by using our novel assay, combined with several other serum biomarkers, to develop a novel liquid biopsy for non-invasive assessment of liver injury. Among healthy controls, higher serum ALT, AST, and plasma DNA levels were found in males and people with a history of drinking alcohol. While males have a higher basal metabolic rate than females, a higher activity rate of liver cells can lead to more ALT, AST, and genomic DNA released from apoptotic liver cells into the blood[27]. Drinking alcohol causes mild hepatocellular damage, which can lead to hepatocyte necrosis followed by ALT, AST, and genomic DNA release. We also demonstrated that HBV infected patients had statistically significantly higher serum ALT, AST, and total plasma DNA levels than healthy controls.
In 263 CHB patients with varying degrees of liver injury, we demonstrated statistically significantly higher serum ALT, AST, bilirubin, albumin, total plasma DNA, and HBV DNA levels than those of healthy controls. We then compared these blood-based biomarker levels in patients with different degrees of liver injury according to liver biopsy. While HBV is a noncytopathic virus and its replication does not directly damage the liver cells, the degree of hepatocyte injury has no direct correlation with the number of HBV DNA copies[28]. In this study, the HBV DNA level did not reflect the severity of liver injury in hepatitis B patients (Table 2). As to serum ALT, it had a high specificity (89.3%) but low sensitivity (55.3%) to discriminate between mild–moderate and marked-severe inflammation. Similar results were found for serum AST. This suggests that nearly half of patients, which were diagnosed with severe liver injury based solely on serum ALT or AST levels, may be misdiagnosed (e.g., patient 85 in Figure 3). By using our novel duplex real-time PCR assay with internal standard, it was demonstrated that plasma DNA concentration was more correlated with the severity of hepatocyte injury than serum ALT levels (Figure 1) and more sensitive to assess the severity of liver injury in patients with low serum ALT (≤ 100.0 U/L; Figure 3A). Cell-free plasma DNA, a superior indicator of cell death, was shown to be a good complement to serum aminotransferases to improve sensitivity. It has been estimated that when 1% of liver cells are damaged, enzymes such as ALT are released into the peripheral blood and this could increase the reference value about one-fold. Considering that 1% of genomic DNA out of the total 2500000000 liver cells is released into plasma, the concentration of plasma DNA would increase by approximate 66 ng/mL, which is 2.6-fold higher than the median value of healthy people. Here, we suggest that this might be an explanation for the effectiveness of plasma DNA in assessing the severity of liver injury in patients with low serum ALT. However, because of the low specificity (68.6%), plasma DNA alone is not sufficient to evaluate hepatic cell injury. After the combination of plasma DNA, serum ALT, and bilirubin, there was a significant improvement in AUC (Figure 2).
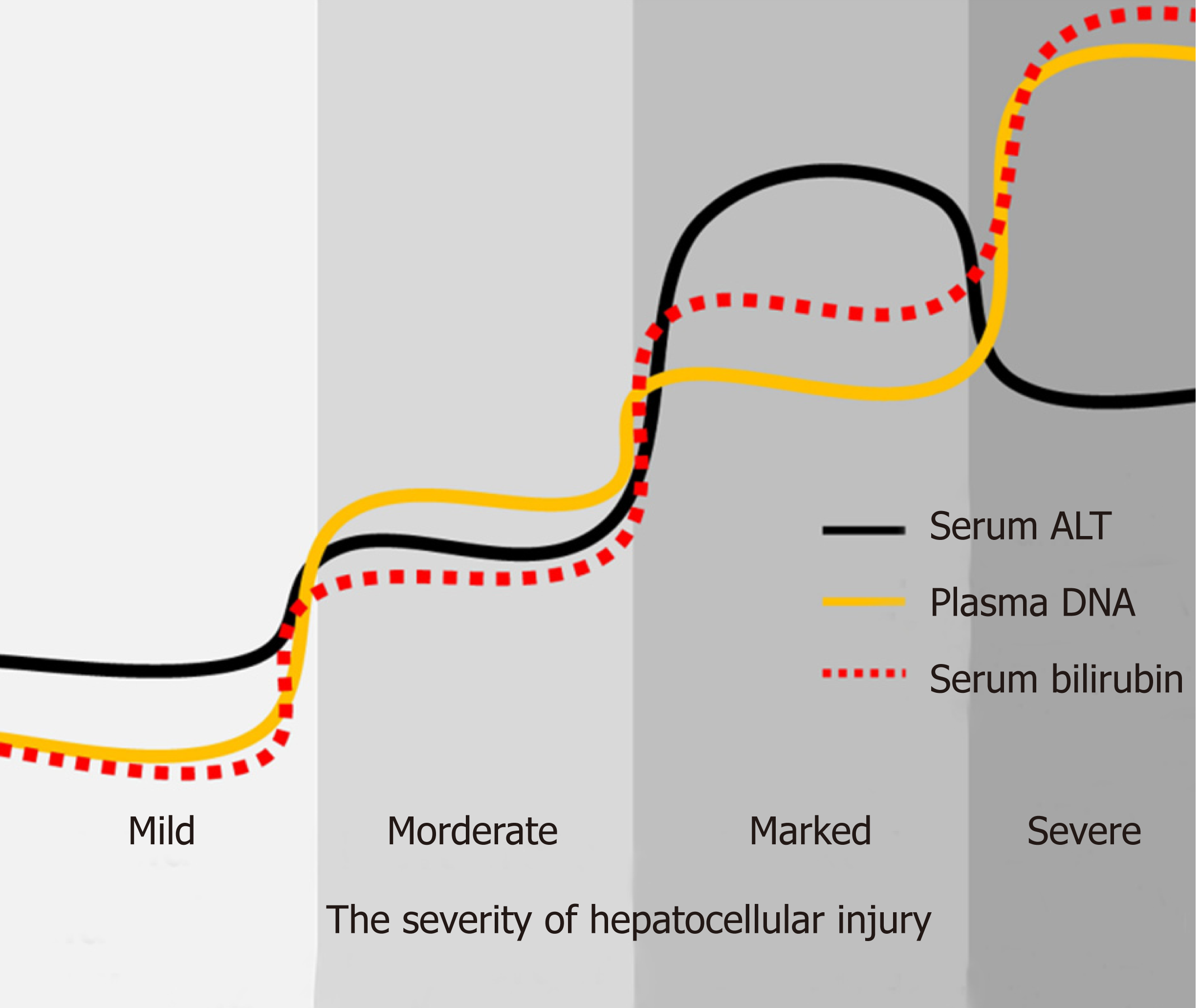
Because of the invasiveness and risk of complications, there are many limitations for liver biopsy in clinical practice. Certain conditions, including thrombocytopenia, bleeding diathesis, cirrhosis, ascites, and amyloidosis, are recognized relative or absolute contraindications to biopsy[29]. Therefore, noninvasive liquid biopsy is the necessary and useful substitution for patients who are not suitable to undertake liver biopsy. According to the WHO’s guidelines[1], there are several non-invasive tests (NITs) based on blood or serum now available and increasingly used for evaluating and staging liver fibrosis. However, except for serum ALT, AST, and bilirubin, few new NITs has been developed for assessment of liver injury, which can reduce the need for liver biopsy in persons with hepatitis B. In this study, by quantifying serum ALT and plasma DNA, clinicians can assess the severity of liver injury and evaluate the patient's condition to determine the best course of treatment. For example, as shown in Figure 4, low levels of serum ALT, bilirubin, and plasma DNA indicate that there is no significant liver injury, while persistent high level of plasma DNA combined with elevated bilirubin can indicate the severity of hepatocellular injury in the case of severe liver cell damage with the “enzyme bilirubin separate” phenomenon. Furthermore, liquid biopsy can be repeated and present a dynamic change throughout the clinical treatment. But as to liver biopsy, that is impossible. This liquid biopsy for non-invasive assessment of liver injury in hepatitis B patients may be an important supplement be written into the guideline in the future. It is also believed that this kind of non-invasive liquid biopsy can be applied for liver injury assessment in many other liver diseases besides hepatitis B.
This is believed to be the first study about combination of plasma DNA, serum ALT, and bilirubin as a sensitive and unique liquid biopsy for the noninvasive assessment of degree of liver injury. This novel liquid biopsy technique is expected to assist in making more precise diagnoses for hepatitis B patients.
Hepatitis B is a major public health problem in China. It is important that the severity of liver injury is evaluated accurately for clinical treatment. Liver biopsy is considered the gold standard method to stage liver disease. However, it is not widely used in resource-limited settings. Therefore, the methods of non-invasive liquid biopsy need to be explored for assessment of liver injury.
Plasma DNA quantification was proved to be a potential marker for cell damage, which may be a non-invasive method for evaluating the severity of liver injury. However, the application of plasma DNA quantification still needs to be investigated in patients with hepatitis B.
The aim of this study was to evaluate liver injury in hepatitis B patients using quantified cell free DNA combined with other serum biomarker as a liquid biopsy-based method.
A cohort of 663 subjects including 313 hepatitis B patients and 350 healthy controls were enrolled. Ultrasound-guided liver biopsies followed by histopathological assessments were performed for the 263 chronic hepatitis B patients to determine the degree of liver injury. Cell-free DNA was quantified using a novel duplex real-time polymerase chain reaction assay.
Compared with healthy controls, patients with hepatitis B virus (HBV) infection had significantly higher plasma DNA, serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), bilirubin, and HBV DNA levels (P < 0.01). Serum ALT, AST, bilirubin, and plasma DNA levels of patients with marked-severe inflammation were significantly higher than those of patients with mild-moderate inflammation (P < 0.01). There was a statistically significant correlation between hepatocyte inflammation severity and serum bilirubin (R2 = 0.673, P < 0.01) or plasma DNA (R2 = 0.597, P < 0.01) levels. The area under the curves of serum ALT, bilirubin, plasma DNA, and their combination to distinguish between patients with mild–moderate and marked-severe inflammation were 0.8059, 0.7910, 0.7921, and 0.9564, respectively.
The combination of plasma DNA, serum ALT, and bilirubin could be a candidate liquid biopsy for non-invasive assessment of liver injury in hepatitis B patients.
The combination of plasma DNA, serum ALT, and bilirubin as a novel liquid biopsy technique is expected to assist in making more precise diagnoses for hepatitis B patients, which will be validated in multiple clinical centers in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Demonacos C, Muench MO, Zapater P S-Editor: Yan JP L-Editor: Wang TQ E-Editor: Ma YJ
| 2. | Kew MC. Serum aminotransferase concentration as evidence of hepatocellular damage. Lancet. 2000;355:591-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 178] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 3. | Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1806] [Cited by in RCA: 1995] [Article Influence: 199.5] [Reference Citation Analysis (4)] |
| 4. | Ott JJ, Horn J, Krause G, Mikolajczyk RT. Time trends of chronic HBV infection over prior decades - A global analysis. J Hepatol. 2017;66:48-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 5. | Lao TT, Sahota DS, Law LW, Cheng YK, Leung TY. Age-specific prevalence of hepatitis B virus infection in young pregnant women, Hong Kong Special Administrative Region of China. Bull World Health Organ. 2014;92:782-789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Chinese Society of Hepatology and Chinese Society of Infectious Diseases, Chinese Medical Association. [The guideline of prevention and treatment for chronic hepatitis B (2010 version)]. Zhonghua Liu Xing Bing Xue Za Zhi. 2011;32:405-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 7. | Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, Desmet V, Korb G, MacSween RN. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3521] [Cited by in RCA: 3782] [Article Influence: 126.1] [Reference Citation Analysis (1)] |
| 8. | Chen D, Pan S, Xie E, Gao L, Xu H, Xia W, Xu T, Huang P. Development and Evaluation of a Duplex Real-Time PCR Assay With a Novel Internal Standard for Precise Quantification of Plasma DNA. Ann Lab Med. 2017;37:18-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Shin EC, Sung PS, Park SH. Immune responses and immunopathology in acute and chronic viral hepatitis. Nat Rev Immunol. 2016;16:509-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 259] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 10. | Mani H, Kleiner DE. Liver biopsy findings in chronic hepatitis B. Hepatology. 2009;49:S61-S71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Regev A, Berho M, Jeffers LJ, Milikowski C, Molina EG, Pyrsopoulos NT, Feng ZZ, Reddy KR, Schiff ER. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am J Gastroenterol. 2002;97:2614-2618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1504] [Cited by in RCA: 1569] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 12. | Lidbury JA. Getting the Most Out of Liver Biopsy. Vet Clin North Am Small Anim Pract. 2017;47:569-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Ganem D. Persistent infection of humans with hepatitis B virus: Mechanisms and consequences. Rev Infect Dis. 1982;4:1026-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1700] [Cited by in RCA: 1712] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 15. | Schütz E, Fischer A, Beck J, Harden M, Koch M, Wuensch T, Stockmann M, Nashan B, Kollmar O, Matthaei J, Kanzow P, Walson PD, Brockmöller J, Oellerich M. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PLoS Med. 2017;14:e1002286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 16. | Giannini EG, Testa R, Savarino V. Liver enzyme alteration: A guide for clinicians. CMAJ. 2005;172:367-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1178] [Cited by in RCA: 1175] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 17. | Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ. Classification of chronic hepatitis: Diagnosis, grading and staging. Hepatology. 1994;19:1513-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1506] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 18. | Chan AK, Chiu RW, Lo YM; Clinical Sciences Reviews Committee of the Association of Clinical Biochemists. Cell-free nucleic acids in plasma, serum and urine: A new tool in molecular diagnosis. Ann Clin Biochem. 2003;40:122-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64:218-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 812] [Cited by in RCA: 811] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 20. | Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, Roz E, Cirenei N, Bellomi M, Pelosi G, Pierotti MA, Pastorino U. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21:3902-3908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 417] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 21. | Jiang T, Zhai C, Su C, Ren S, Zhou C. The diagnostic value of circulating cell free DNA quantification in non-small cell lung cancer: A systematic review with meta-analysis. Lung Cancer. 2016;100:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Bedin C, Enzo MV, Del Bianco P, Pucciarelli S, Nitti D, Agostini M. Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patients. Int J Cancer. 2017;140:1888-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 23. | Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clin Chem. 2000;46:319-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Lam NY, Rainer TH, Chan LY, Joynt GM, Lo YM. Time course of early and late changes in plasma DNA in trauma patients. Clin Chem. 2003;49:1286-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 142] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 25. | Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011;108:6229-6234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 295] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 26. | Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem. 2002;48:421-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Dufour DR, Lott JA, Nolte FS, Gretch DR, Koff RS, Seeff LB. Diagnosis and monitoring of hepatic injury. I. Performance characteristics of laboratory tests. Clin Chem. 2000;46:2027-2049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 28. | Liu X, Chen JM, Lou JL, Huang YX, Yan Y, Sun GZ, Li N. Correlation between hepatitis B virus DNA levels and diagnostic tests for HBsAg, HBeAg, and PreS1-Ag in chronic hepatitis B. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 29. | Grant A, Neuberger J. Guidelines on the use of liver biopsy in clinical practice. British Society of Gastroenterology. Gut. 1999;45:IV1-IV11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 186] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 30. | Wang H. Zhang B, Chen D, Xia W, Zhang J, Wang F, Xu J, Zhang Y, Zhang M, Zhang L, Lu Y, Geng Y, Huang P, Huang P, Wang H, Pan S. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin Epigenetics. 2015;31:119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 186] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 31. | Gao L, Xie E, Yu T, Chen D, Zhang L, Zhang B, Wang F. Methylated APC and RASSF1A in multiple specimens contribute to the differential diagnosis of patients with undetermined solitary pulmonary nodules. J Thorac Dis. 2015;7:422-432. |
| 32. | Pan S, Xia W, Ding Q, Shu Y, Xu T, Geng Y, Lu Y. Can plasma DNA monitoring be employed in personalized chemotherapy for patients with advanced lung cancer? Biomed Pharmacother. 2012;66:131-137. |