Published online Jun 21, 2019. doi: 10.3748/wjg.v25.i23.2846
Peer-review started: March 20, 2019
First decision: April 5, 2019
Revised: April 26, 2019
Accepted: May 8, 2019
Article in press: May 8, 2019
Published online: June 21, 2019
Processing time: 93 Days and 9.7 Hours
Cytochromes P450s (CYPs) are terminal enzymes in CYP dependent monooxygenases, which constitute a superfamily of enzymes catalysing the metabolism of both endogenous and exogenous substances. One of their main tasks is to facilitate the excretion of these substances and eliminate their toxicities in most phase 1 reactions. Endogenous substrates of CYPs include steroids, bile acids, eicosanoids, cholesterol, vitamin D and neurotransmitters. About 80% of currently used drugs and environmental chemicals comprise exogenous substrates for CYPs. Genetic polymorphisms of CYPs may affect the enzyme functions and have been reported to be associated with various diseases and adverse drug reactions among different populations. In this review, we discuss the role of some critical CYP isoforms (CYP1A1, CYP2D6, CYP2J2, CYP2R1, CYP3A5, CYP3A7, CYP4F3, CYP24A1, CYP26B1 and CYP27B1) in the pathogenesis or aetiology of ulcerative colitis concerning gene polymorphisms. In addition, their significance in metabolism concerning ulcerative colitis in patients is also discussed showing a clear underestimation in genetic studies performed so far.
Core tip: The role of cytochrome P450s (CYPs) genes in the pathogenesis of ulcerative colitis (UC). Extrahepatic and extrarenal CYPs (e.g., macrophages and dendritic cells of colonic mucosa) have a critical role in UC development. Polymorphisms discussed can result in dysregulation of these enzymes in favour of alternative pathways producing more reactive metabolites. Production of reactive metabolites is favouring more severe disease states. Pharmacogenetics might facilitate individualized medicine for UC in the future although actually available data is limited.
- Citation: Sen A, Stark H. Role of cytochrome P450 polymorphisms and functions in development of ulcerative colitis. World J Gastroenterol 2019; 25(23): 2846-2862
- URL: https://www.wjgnet.com/1007-9327/full/v25/i23/2846.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i23.2846
Cytochrome P450s (CYPs) are a superfamily of the integral membrane, heme-thiolate proteins entailed in the synthesis and breakdown of various molecules and chemicals within cells[1-3]. CYPs play a role in the metabolism of many endogenous substances including steroids, bile acids, eicosanoids, cholesterol, vitamin D and neuro-transmitters, steroid hormones, cholesterol, fatty acids, bile acids[4-6]. Additional CYPs metabolise xenobiotics, such as drugs and endogenous molecules such as toxins that are shaped inside cells[7-11]. The human CYP isoenzymes superfamily is composed of 57 CYP genes and 58 pseudogenes arranged into 18 families and 43 subfamilies[12]. They are located either at endoplasmic reticulum or mitochondria of liver cells, but are also situated in other cells throughout the body[13-16]. Mitochondrial CYPs are commonly engaged in phase I reactions with anabolic and catabolic transformations of endogenous substances, while CYPs in the endoplasmic reticulum generally process xenobiotics. CYPs are gathered into families and subfamilies as indicated by the similarity index of the amino sequence. Every CYP is given a number relating a particular family inside the gene group, a letter exemplifying the subfamily, and a number allocated to the distinct gene inside the subgroup, e.g., the CYP gene that is in family 1, subgroup A, gene 1, is written as CYP1A1[12,17-20].
CYPs show intra- or interethnic and intra- or interindividual genetic variations. These variations or polymorphisms in CYP genes can largely alter the function of the enzymes. We continue to learn about the properties of these enzymes in humans and their roles in different diseases. As with many other genes and proteins associated with a critical life function, specific polymorphisms or variability in these CYPs and, hence, the gene product will result in pathology and lead to a severe human di-sease[21-24].
Ulcerative colitis (UC) is an idiopathic chronic inflammation condition with multifactorial determinants[25]. Populace-based careful surveys have shown that the frequency of UC worldwide has expanded in recent years. As opposed to the de-veloped communities of North America and Western Europe, where the prevalence of UC has levelled or even lessened, publications demonstrate that incidences have elevated in developing countries, for example, those in Latin America, Asia and Eastern Europe leaving an urgent medical need[26-28]. The progression of UC requires a hereditary predilection, dysregulated immune reactions and an environmental incites. Candidate genes comprise those that govern innate immunity and epithelial boundary function[29-32]. Consequently, the interplay among hereditary and environ-mental components will cast the gut epithelial-inborn immunity interface and lead to unique phenotypes in patients with Inflammatory Bowel Disease.
Several studies have demonstrated that the CYP gene polymorphisms have been associated with the susceptibility to UC, but this is, to the best of our knowledge the first systematic review on the role of the CYPs polymorphism and function in the vulnerability and the development of UC. Each CYP will be described and discussed in alphabetical order, rather than in its importance to UC.
CYP1A1 (EC 1.14.14.1), a notable aryl hydrocarbon hydroxylase, is expressed in the liver at exceptionally low quantities, and is mostly translated in human extrahepatic tissues, including digestive tract[8,33-37]. In humans, since the CYP1A1 is an extrahepatic protein, it is considered to assume a secondary function in the removal of medications in vivo, and consequently, the polymorphism of the CYP1A1 gene may have a minor impact on their metabolic clearance. CYP1A1 assume essential tasks in the bio-activation of a collection of cancer-causing polycyclic aromatic hydrocarbons [PAHs; e.g., benzo[a]pyrene (B[a]P)], aromatic amines and amides, and mycotoxins ascertained in certain grains, e.g., aflatoxin B1[10,38-41]. CYP1A1 is also committed in the metabolism of endogenous compounds such as the pineal hormone melatonin and 17 β-estradiol and estrone[42-44]. It has been acknowledged as the principal CYP responsible for the C2- or C4-hydroxylation of 17 β-estradiol and estrone in ex-trahepatic tissues[45].
The CYP1A1 is positioned on chromosome 15 close to the mannose phosphate isomerase (MPI) locus at 15q22-24[46]. To date, there are 2092 SNPs depicted for CYP1A1 in NCBI dbSNP (https://http://www.ncbi.nlm.nih.gov/snp, access date: February 23, 2019). There are just a couple of investigations on the relationship of CYP1A1 polymorphism and function with UC.
CYP1A1 is the least studied isoform on the genotyping and phenotyping in gastroenterological practice, on the assumption that it is not involved in the meta-bolism of the intestine and inadequacy of clinical response. Klotz and colleagues[47] have tested the hypothesis that the appearance, arrangement, and activity of drug-metabolising enzymes in the gut may produce one or more reactive metabolites and create UC. For this purpose, they have evaluated the terminal ileum and different regions of the colon biopsies from 37 patients with UC by staining immu-nohistochemically for CYP1A1 isozyme. All proteins aside from CYP1A1 were displayed with comparable recurrence in both control and UC patients. CYP1A1 staining was definite substantially more frequently in patients with UC (39.4%) than in control (irritable colon, no clinical or histological indications of inflammations; 19.2%). These results may confirm the involvement of this protein in the aetiology or pathogenesis of inflammation in this tissue.
On the other hand, the more frequent appearance of CYP1A1 could be due to secondarily to the appearance of inflammation. Other recent studies have further supported the latter suggestion by demonstrating that the CYP1A1 regulates immune responses in the intestine and confers protection against intestinal inflammation[48-49]. On the other hand, Plewka and colleagues[50] have pointed out that the expression of CYP1A1 in enterocytes from UC patients was lower than in control, equivalent to 80% of the latter. Therefore, CYP1A1 needs to be further studied to resolve its role in UC.
Furthermore, our study included 161 Turkish patients with ulcerative colitis (94 males, 67 females; all Caucasian) consulting the outpatient clinic of the Department of Gastroenterology, Ege University, Turkey[51,52]. A group of 198 healthy Turkish Caucasians adjusted for age and sex (115 males and 83 females) were utilised as controls.
DNA preparations from the Turkish population were subjected to genotype analysis of CYP1A1*2B (rs4646903, T>C, 3′-flanking region, linked with increased enzyme activity). Our results showed that the CYP1A1*2A alleles correlate with an increased predisposition to UC, a piece of further supporting evidence that the increased CYP1A1 activity might cause the abundant accumulation of reactive metabolites, which advances an irregular intestinal immune response, causing irreversible harm to the colonic mucosa and eventually UC. Since there is limited and conflicting literature available on CYP1A1 polymorphism and function in UC, more studies are required to clarify the role of CYP1A1 in UC.
The CYP2D6, also known as debrisoquine hydroxylase, presents a small percentage of all hepatic cytochrome P450s[53]. It is encoded by the CYP2D6 gene that is localised on chromosome 22q13.1 and participates in the biotransformation of about 20-25% of the clinically used drugs[54,55]. The endogenous substrates of CYP2D6 include neuro-transmitters and neurosteroids, pinoline, progesterone and lipids[55,56]. It is highly polymorphic in the human population, and marked inter-racial variation observed.
Individuals are identified as ultra-rapid, extensive, intermediate or poor metabolizer, according to the number of functional alleles. To date, there are 3257 SNPs described for CYP2D6 in NCBI dbSNP (https://http://www.ncbi.nlm.nih.gov/snp, access date: February 23, 2019). CYP2D6 is another isoform that genotyping and phenotyping are not usually performed for gastroenterological inflammation, on the assumption that the irrelativity of the medications to intestinal digestion or generally little metabolic limit of CYP2D6 in the small digestive tract[57,58]. CYP2D6 is another isoform on which genotyping and phenotyping are not usually performed for gastroenterological inflammation, on the belief that there is non-relativity of the medications to intestinal metabolism or the generally little metabolic limit of CYP2D6 in the small intestine[57,58].
Dudarewicz et al[59] have analysed CYP2D6 genotypes *1 (wild type), *3 and *4 using the PCR-RFLP method in 258 people from central Poland; 65 patients with UC and 50 with Crohn’s disease (CD); and 143 healthy controls. They announced that despite the fact that the odds ratio (OR) was higher in carriers of the CYP2D6*1 /CYP2D6*3 genotype [extensive metabolizer (EM)], there was no factually significant difference in the recurrence of the CYP2D6 alleles in patients with UC and CD. Similarly, Trzcinski et al[60] have shown that the increased OR for inflammatory bowel disease (IBD) was without statistical significance. These researches suggest that there were no measurably significant variations in the appearance of the CYP2D6 alleles in IBD patients (UC or CD). However, the EM genotype might be the risk factor for IBD. Future investigations applied to a larger group of patients are needed to confirm presumptions.
Handersson et al[61] reported a real-world clinical case portraying the drug-drug interactions in a UC patient. The patient was on tamoxifen therapy for breast cancer and was prescribed rifampin for worsening UC. The patient is known to be a CYP2D6 intermediate metabolizer, and rifampin significantly lowered the endoxifen level. It constitutes a substantial risk of sub-therapeutic efficacy in tamoxifen patients, which may be of distinct concern among high-risk patients. This study illustrates the clinical value of CYPs genotyping in UC patients for therapeutic efficiencies of combinative therapies; exceptional consideration must be practised when required.
Le and Bae[62] carried out a meta-analysis probing the association between functional CYP2D6 polymorphisms (*3 and *4) and susceptibility to autoimmune diseases, including IBD. This meta-analysis demonstrated the association and susceptibility of the functional CYP2D6*3 and *4 polymorphisms with the autoimmune disease in Caucasians. Therefore, the CYP2D6 gene plays a role in the aetiology and the development of autoimmune diseases.
More direct evidence was very recently reported relating the CYP2D6*4 allele (PM) with susceptibility to UC[63]. The researchers studied CYP2D6*4 polymorphisms in 215 unrelated UC patients and 212 separate healthy controls by PCR-RFLP in a Kurdish population from Iran. A significantly higher frequency of CYP2D6*4 A allele in UC patients (12.6%) compared to healthy subjects (8.5%, P = 0.046) was reported. Also, the presence of A allele significantly increased the risk of UC by odds ratio (OR) = 1.56-fold (P = 0.047). This report is consistent with the Dudarewicz report showing a higher frequency of the CYP2D6*4 A allele in patients from Poland. Thus, this study suggests that CYP2D6*4 polymorphisms may be risk factors for UC susceptibility.
Moreover, a short while ago we reported the role of CYP2D6 in the metabolism of 5-aminosalicylic acid (5ASA), which is an anti-inflammatory drug used to treat ulcerative colitis. It is known that 5ASA is mainly metabolised to N-acetyl-5-ASA by N-acetyltransferases (NAT). However, no information is available on the oxidation of 5ASA by CYPs. Also, scarce pharmacogenetic analysis has focused directly on 5-ASA metabolism. Our study presented compelling evidence indicating that the 5-ASA is a substrate for CYP2D6[64,65]. Therefore, knowledge of CYP2D6 allelic variants is required for the better response of UC patients to this specific medication.
All these studies have strongly suggested that further studies are required to clarify the role of the CYP2D6 gene in aetiology, development and pathogenesis and the treatment of UC.
CYP2J2 is the sole member of human CYP2J subfamily and the prominent arachidonic acid (AA) epoxygenases. It is localised on chromosome 1q32.1 and participates in the metabolism of AA to all four cis-eicosatrienoic acid epoxides (EETs) as 5,6-EET, 8,9-EET, 11,12-EET and 14,15-EET, some of them exhibits signalling properties in anti-inflammatory pathways[66]. It is shown mainly in the cardiovascular system, yet is also displayed in the intestines, stomach, and other tissues[67-70].
Bystrom et al[71] demonstrated that CYP2J2 is an inflammation-induced enzyme displaying anti-inflammatory activities and raises phagocytosis of both gram-positive and negative bacteria. It was noted that the deficiency of CYP2J2 in IBD (CD) macrophages in response to bacterial infection may participate in the pathogenesis of the disease. Boosting the epoxygenase metabolites or the use of 11,12-EET mimetics may present useful therapeutic approaches for the treatment of IBD[66,71]. Therefore, the proper function of CYP2J2 is vital for bacterial clearance in IBD, and a deficit in the CYP2J2 pathway may advance bacterial pathogenesis resulting in the development of IBD. Likewise, Qiu et al[72] assessed the EETs and the expression of CYP2J2 in colon tissue biopsies collected from UC patients along with adjacent unaffected tissues to study the role of CYP2J2 in UC. It was found that the quantities of EETs were significantly higher in UC tissues matched with adjacent unaffected tissues (1.91 ± 0.98 ng/mg vs 0.96 ± 0.77 ng/mg, mean ± SD, P < 0.01). Also, the expression of CYP2J2 rose significantly in UC tissues (P < 0.05). Thus, the increment in EET levels may be part of a defence mechanism in UC and CYP2J2 could be a key target for drug therapy for UC.
In addition, it was reported that endocannabinoids such as arachidonoyl ethanolamide (AEA) are significantly increased during intestinal inflammation and the CYP2J2 produced metabolites are related to pathologic conditions of the gastrointestinal tract[66,73-76]. CYP2J2 converts EAE to 20-hydroxyeicosatetraenoic acid ethanolamide (20-HETE-EA) and some other EET-EA metabolites which bind to cannabinoid-1 receptors (CB1-Rs) on neurons and cannabinoid-2 receptors (CB-2Rs) on immune and epithelial cells of the gut reducing digestion and represses the liberation of inflammatory mediators[77,78]. Also, UC patients display raised histamine levels which increase pathogenic neutrophil invasion into the colonic mucosa, intensifying the symptoms of colitis[79-82]. CYP2J2 has appeared to assume a fundamental role in the intestinal metabolism of antihistamines such as astemizole and ebastine. Thus, the knowledge of pharmacogenetics of CYP2J2 is essential and contribute to useful therapeutic approaches for the treatment of IBD.
There have been 7214 SNPs in human CYP2J2 gene in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp, access date: 24 February 2019). A portion of these SNPs has been appeared to have a potential association with certain diseases, particularly cancer and heart diseases[83]. However, there is only a single report in the literature focusing on the relationship of CYP2J2 variation to UC. Otte and collaborators[84] have screened the polymorphism at position -50 (G-50T) in the promoter region of CYP2J2 (CYP2J2*7) in 146 UC and 147 CD patients matched to 357 healthy German people. CYP2J2*7 has a G>T replacement in the regulatory region at -50 position at the transcriptional start, causing diminished translation because of the loss of the Sp1 binding site[85,86]. Thus, this creates a smaller amounts of the CYP2J2 protein causing decreased CYP2J2 epoxygenase metabolites in vivo. The -50T allele was identified in 19.9% of subjects with UC 14.3% of subjects with CD, and 10.9% of the control group (P < 0.05). Additionally, a noteworthy higher recurrence of this allele was distinguished in patients with UC in contrast to the CD group. Their outcomes unequivocally support the relationship of UC with the promoter polymorphism in the CYP2J2 gene showing a critical function of epoxyeicosatrienoic acids in the pathophysiology of IBD. Further examinations are expected to depict the actions of CYP2J2 in pathology and the treatment of UC.
The CYP2R1 gene produces an enzyme called vitamin D-25-hydroxylase (EC 1.14.14.24), showing a 25-hydroxylase action on both types of vitamin D, vitamin D2 and D3. It catalyses the initial reaction leading to the production of 1,25-dihydroxy vitamin D3, also called calcitriol[87]. CYP2R1 is located on chromosome 11p15.2 and converts vitamin D into 25-hydroxyvitamin D (calcidiol), which is the essential circulatory form of vitamin D.
There have been 4247 SNPs in human CYP2R1 gene in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp, access date: 25 February 2019). However, there is no one study on the association of any of these SNPs with IBD, both CD and UC. Moreover, most studies focused on its role in vitamin D and related clinically significant diseases such as vitamin D deficiency[88]. However, few studies have examined its role in some autoimmune diseases such as multiple sclerosis, T1D, Hashimoto’s disease and Grave’s disease[89-94].
There is a convincing amount of evidence that vitamin D deficiency is one of a designated set of factors proposed to intervene in the contemplated relationship between environmental exposures and IBD, CD and UC[95-98]. It would be expected that the enzyme responsible for the production of active vitamin D, i.e., CYP2R1, has some role in IBD. However, since it has been proposed that there is redundancy in the enzymes responsible for the 25-hydroxylation step, the identification of affected individuals with CYP2R1 polymorphisms has been difficult.
The rs10741657, located near the CYP2R1 gene has been linked by several studies to the increased risk of vitamin D insufficiency[99]. Moreover, an immense connection between the CYP2R1 gene transcript and 1,25(OH)2D3 plasma levels in the helper T cells was affirmed[99]. Ramos-Lopes et al[89] genotyped 203 simplex type 1 German diabetes families for the rs10741657 polymorphism. They examined the 25(OH)D3 quantities comparing to CYP2R1 polymorphisms to its mRNA transcripts from peripheral blood mononuclear cells (PBMCs) in 133 T1D patients. The G variant of the rs10741657 polymorphism was more regularly transmitted to influenced children and was likewise more prevalent in cases than in the controls (46.1% vs 35.7%, P = 0.03). Patients conveying the genotype “GG” or “GA” of the rs10741657 polymorphism had, by and large, lower amounts of 25(OH)D3 in contrast to those with the genotype “AA”. They showed an association of CYP2R1 polymorphisms with 25(OH)D3 levels in T1D patients. Coper et al[90] and Hussein et al[91] likewise detailed a similar association. Also, the relationship between the variant CYP2R1 alleles and MS risk suggested being dependency on the presence of the HLA-DR15 risk allele and other factors such as gender[89,94].
In conclusion, taking all of these studies into account, we have to remain sceptical regarding any association between CYP2R1 and IBD, both CD and UC, and em-phasise that research efforts must be accelerated in oder to generate the answers. It would be not unexpected that CYP2R1 had a role on which we can only speculate with given data so far.
CYP3A4 (EC 1.14.13.97) is one of the most widely studied enzymes among CYPs since it metabolises approximately 60% of prescribed drugs, is localised mainly in the liver and in the intestine and is induced by glucocorticoids and some pharmacological agents[53,100]. In grown-ups, they are the main CYP3A subfamily members expressed in the liver and the intestine. CYP3A4 is the portion of a cluster of CYP genes on chromosome 7q22.1. It is called niphedipine oxidase but has many other aliases for its activity such as cholesterol 25-hydroxylase, taurochenodeoxycholate 6-alpha-hydroxylase and 1,8-cineole 2-exo-monooxygenase[101-103]. CYP3A4 also metabolises arachidonic acid to EETs or 20-HETE, expressing both epoxygenase and mono-oxygenase activities[104,105].
On the other hand, CYP3A5, a highly polymorphic enzyme, has been reported to range from undetectable amounts to amounts comparable to those of CYP3A4 in the human liver[106]. It is also part of a cluster of CYP genes on chromosome 7q21.1 and exhibits similar enzymatic activities[54,107].
5521 and 6608 SNPs have been found in human CYP3A4 and CYP3A5 genes, respectively in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp, access date: 26 February 2019). However, there is no study on the association of any of these SNPs with IBD, both CD and UC. Yet, the role of variants in the transformation of the drugs used for the treatment of UC was studied in detail, particularly tacrolimus[108-110].
Tacrolimus is an immunosuppressive drug, utilised in the treatment of UC patients who have no response to 5-ASA or corticosteroids[111]. Tacrolimus is a substrate of CYP3A4 and CYP3A5, and they are in charge of the metabolism of tacrolimus[112]. CYP3A5’s ancestral polymorphisms influence tacrolimus metabolism, for the most part. An SNP in the CYP3A5 gene including an A>G change at position 6986 inside intron 3 (rs776746) was determined to be strongly associated with CYP3A5 protein synthesis. CYP3A5*3/*3 genotypes are viewed as CYP3A5 non-expressers, while CYP3A5 expressers convey at least one CYP3A5*1 allele[113,114].
Many studies have shown that tacrolimus dose requirements are influenced by CYP3A4/5 genetic polymorphisms and the adverse events especially nephrotoxicity were frequently observed in CYP3A4/5 expressers. In addition, CYP3A4/5 expressers require that particular attention should be paid to the onset of nephrotoxicity. Thus, genotyping for CYP3A5 variants allows individualised care to be practised[57,108,110,115,116].
Plewka et al[50] reported that the CYP3A4 level was slightly higher in UC as compared to the control level. Additionally, in this case, the expression of CYP3A4 was restrained particularly in epithelial cells of the mucosa in the colon. The expression level of the fetal form of CYP3A, i.e., CYP3A7, was increased 3-fold in the colonic tissues of UC patients[117]. In addition, it was reported that CYP3A4 were induced by mesalazine (5-ASA), a drug used to treat colitis, in a concentration-dependent manner, both in cultured hepatocytes and human cryopreserved hepatocyte from UC[118]. Similarly, we have also determined that 5-ASA is both inducer and substrates for CYP3A4[64,65]. Accordingly, alteration in CYP3A4 activity in disease states may be an underappreciated determinant of difference in the aetiology of UC.
CYP4F3 known as leukotriene-B4 omega-hydroxylase encodes two distinct enzymes and is part of a cluster of CYP genes on chromosome 19. CYP4F3 encodes two splice-variants, CYP4F3A and CYP4F3B, and their expression is tissue-specific with CYP3F3A being expressed mostly in leukocytes and CYP4F3B chiefly in the liver[119,120]. They metabolise leukotriene B4 and very likely 5-hydroxyeicosatetraenoic acid by an omega oxidation reaction, leading to the inactivation and degradation of well-known mediators of inflammation[121]. Thus, CYP4F has underlain the proposed roles of cytochromes in depressing inflammatory responses, and CYP4F3 is associated with IBD[122,123]. CYP4F3A/B also omega oxidise arachidonic acid to 20-HETE as well as EETs[124].
Five thousand five hundred and seventy-five SNPs have been found in human CYP4F3 in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp, access date: 27 February 2019). Ananthakrishnan and colleagues[125] have genotyped CYP4F3 in 101 CD and 139 UC patients matched to 495 controls. They revealed that that high consumption of a high amount of n3:n6 PUFA (above median) exhibited a re-lationship to a decreased risk of UC (OR = 0.71, 95%CI: 0.47-1.09, P = 0.11). In addition, high n3:n6 PUFA intake was related with a decreased risk of UC in people with the GG/AG genotype (rs4646904) in CYP4F3 (OR = 0.57, 95%CI: 0.32-0.99) compared to those with the AA genotype (OR = 0.95, 95%CI: 0.47-1.93) (P-connection = 0.049). Despite the fact that the rs4646904 (converged into rs1805042) CYP4F3 variant is synonymous, it is proposed that it changes the splicing efficiency and gene expression level[126]. Hence, CYP4F3 variations are determinant of the relationship between dietary n3:n6 PUFA intake and the risk of UC. It is the only study present in the literature involving the CYP4F3 variants and an association between the UC. There is a definite need for additional studies examining the impact of other polymorphisms in exploring the association of specific CYP4F3 variants with UC as well as defining the diet-UC associations in patients.
CYP24A1 is a mitochondrial monooxygenase which assumes a crucial function in calcium homeostasis through controlling the level of vitamin D3[127]. It is characterised as vitamin D3 24-hydroxylase (EC 1.14.15.16; Entrez Gene: CYP24A1 cytochrome P450 family 24 subfamily A part 1 [Homo sapiens (human)] accessed 28 February 2019). It facilitates hydroxylation reactions leading to the degradation of 1,25-dihydroxy vitamin D3, the physiologically active class of vitamin D. Hydroxylation of the side chain of vitamin D3 produces calcitroic acid and various metabolites which are discharged in bile. Inactivation of vitamin D is achieved by the mitochondrial catalyst, 25-hydroxyvitamin D3-24-hydroxylase, first portrayed in the mid-1970s and at first accepted to be included exclusively in the renal 24-hydroxylation of 25-OH-D3. Work performed over the ensuing 35 years has demonstrated that 24-hydroxylase is the aftereffect of CYP24A1[128,129]. CYP24A1 catalyses the transformation of both 25-OH-D3 and 1,25-(OH)2D3 into a series of 24- and 23-hydroxylated compounds directed to well-known pathways ending in the water-soluble biliary metabolite such as calcitroic acid and 26,23-lactone[130].
Besides being a self-induction of CYP24A1 by the 1,25-(OH)2D3 itself, the enzyme is controlled by crucial determinants such as the parathyroid hormone (PTH) and the fibroblast growth factor (FGF). 1,25-(OH)2D3-mediated PTH significantly reduces induction of the CYP24A1 expression because of destabilisation and enhanced degradation of CYP24A1 mRNA[130-132]. The translation of CYP24A1 is enhanced via PI3K-Akt-facilitated IRES within 5'UTR-dependent manner in response to the inflammatory condition by shifting from monosomal to polysomal fractions[133]. CYP24A1 is located on chromosome 20q13.2, and 6746 SNPs have been found in human CYP24A1 in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp, access date: 28 February 2019).
CYP24A1 is correlated with idiopathic infantile hypercalcemia and is associated with the risk of cell-mediated immune mechanisms in MS and IBD[98,134,135]. The increased expression of CYP24A1 accompanied by CYP27B1 in inflamed colonic tissues in IBD patients results in the loss of 25(OH)D thereby exerting downward pressure on the vitamin D status. Therefore, it is essential in the regulation of inflammations.
Recently Chen et al[136] have genotyped rs4809957, rs6068816, rs6091822 and rs8124792 SNPs in 44 ulcerative colitis patients along with a control group composed of 504 East Asians enrolled in the 1000 Genomes Project. CYP24A1 polymorphisms rs4809957 A/G and rs6068816 C/T demonstrated a statistically noteworthy association with the UC when both the genotypes and allele frequencies were considered together. Regarding rs6091822 G/T, UC was identified with risk allele bearers (GT + TT) vs wild-type (GG). However, the relationship between the allele frequencies and the disease were not significant. These alleles were additionally observed to be related to the risk of colonic polyps and colon cancer, and low-dose aspirin-related small intestine bleedings[137,138]. Rs4809957 situated in the 3′ untranslated vicinity neighbouring the polyA microsatellite repeat influences the stability of CYP24A1 mRNA and is proposed to be a coupling site for the retinoic acid-responsive element. Contrarily, rs4809957 may influence methylation of 3'UTR. It was accounted for that the 3'UTR of mRNA was the objective of miRNA to hasten degradation[139,140]. Rs6068816 likewise demonstrated a factually strong relationship with the risk of UC. Then OR for rs6068816 C vs T was again high (OR = 18.260, 95%CI: 8.350-39.932). Consequently, these discoveries demonstrate that rs6068816 T is a strong risk factor for UC as well. Changes in rs6068816 would not influence the amino acid residue of the CYP24A1 protein yet may influence intron splicing[141].
Interestingly, several SNPs in CYP24A1 and the other genes involved in vitamin D metabolism and signalling seem to exhibit susceptibility to UC in Asians, yet do not have a statistically significant effect on IBD risk in Europeans[136,142-145]. It is proposed that SNPs in CYP24A1 take part in the initiation or development of UC, and are not merely the result of ulcerative colitis-related malfunctions[136]. Despite the fact that the mechanisms are indistinct, it might be identified with vitamin D metabolism and signalling since both in vivo and in vitro investigations have shown the function of vitamin D in immune intervened diseases[98,145,146]. It is realised that vitamin D insufficiency cause diminished colonic bacterial clearance, decreased expression of tight junctions in the intestinal epithelium, and raised Th1 cell-driven inflammation at the gut level[97,127,145]. Therefore, CYPs in vitamin D metabolism are associated with the UC and deserve further detailed examinations.
CYP26B1 is defined on chromosome 2p13.2, and the protein product is localised on the endoplasmic reticulum. It works as a crucial switch of all-trans retinoic acid (RA) levels. It inactivates all-trans retinoic acid to hydroxylated forms, including 4-OH-RA, 4-oxo-RA, and 18-OH-RA[147]. Mutations in this gene are related to radiohumeral fusions and other skeletal and craniofacial abnormalities, and an increased level of the protein is associated with atherosclerotic lesions. Alternative splicing results in multiple transcripts.
Kang et al[148] demonstrated that both high and low vitamin A levels brought about ameliorated intestinal inflammation and differentially activated subsets of FoxP3+ cells in SAMP1/YP mice. Likewise, Takeuchi et al[149] concluded that CYP26B1 expression was stimulated by all trans-RA in T-cells of the mesenteric lymph nodes (MLN) and Peyer's patches (PP) and changed the expression of CYP26B1 altering T cell dealing and separation in the gut. As of lately, Chenery et al[150] have demonstrated that CYP26B1 can restrict the differentiation of iTreg and Th17 cells and is di-fferentially expressed by these cells to tweak RA responsiveness. In this manner, CYP26B1 in T cells is associated with the pathogenesis of T cell-mediated chronic inflammation in the colon, possibly by controlling T cell effector activity in the intestinal tissue.
Five thousand three hundred and thirty-nine SNPs have been found in human CYP26B1 in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp, access date: 28 February 2019). There is only one study reporting the SNPs in CYP26B1 and the risk of IBD (UC and CD). Fransen et al[151] investigated the association of rs2241057 polymorphism with the threat of CD and UC, knowing that rs2241057 has an elevated catabolic function of retinoic acid. DNA from 1378 IBD patients (871 CD and 507 UC) and 1205 healthy controls gathered at Orebro University Hospital, and Karolinska University Hospital was genotyped for the CYP26B1 rs2241057 polymorphism. They detailed that a higher recurrence of patients homozygous for the dominant (T) allele was associated with the CD though not UC compared to the recurrence found in healthy controls. Hence, CYP26B1 polymorphism rs2241057 may have an increased risk for the progression of CD, which conceivably might be because of raised levels of RA. It was criticised that the lower number of enrolled patients with UC and the lack of standardisation in terms of ethnicity might be the reason for the absence of significant associations in UC. Therefore, their study requires further replication efforts and mechanistic studies to confirm the reported associations. In conclusion, further studies exploring vitamin A and CYP26B1 in the pathogenesis of both CD and UC are needed.
The CYP27B1 gene supplies information to produce a mitochondrial protein called 25-dihydroxy vitamin D3 1-α-hydroxylase (EC 1.14.13.13). This catalyst performs the second of two consecutive reactions to transform vitamin D to its active structure, 1,25-dihydroxy vitamin D3 [1,25-(OH)2-D3], also called calcitriol. Vitamin D can be obtained from the diet or can be made on the body surface with the assistance of sunlight exposure. Whenever active, this vitamin is associated with retaining up the best possible balance of a few minerals in the body, including calcium and phosphate, which are fundamental for the regular composition of bones and teeth. Vitamin D is likewise engaged with a few other processes unrelated to bone and tooth or-ganisation. It is currently kown that the protein exists in non-renal tissues to help increase the production of cellular 1,25-(OH)2-D3, a paracrine/autocrine system[129,152]. It affects immunomodulatory activities by repressing human leukocyte antigen (HLA) class II expression on endocrine cells, T cell proliferation and secretion of in-flammatory cytokines that are thought to function in autoimmune tissue dis-ruption[153-159]. In this way, the expression of CYP27B1 in cells of the colon, breast, prostate and monocyte/macrophage are so fundamental to the ordinary functioning of these tissues.
The cytogenetic location of the CYP27B1 is the long arm of chromosome 12 at position 14.1 (12q14.1). 1929 SNPs have been found in human CYP27B1 in NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp, access date: 5 March 2019). Diseases associated with CYP27B1 include Type 1A and hypocalcemic vitamin D-dependent rickets and autoimmune diseases such as Addison, Hashimoto, Grave’s, T1D and MS[158,160-164].
Du et al[159] surveyed the expressions of colonic CYP27B1 in UC patients. They gathered colon mucosal biopsies from the inflamed lesions and adjacent normal tissues from a cohort of patients with active UC. They revealed that CYP27B1 originating from colon epithelium, were notably induced in the lesions contrasting to the adjacent normal tissues in these patients. It was additionally bolstered by the observation that colon mucosal CYP27B1 was likewise induced significantly in an experimental colitis mouse model, and this nearby CYP27B1 induction and colonic inflammation required the community of commensal microscopic organisms. Therefore, in this context, it is vital to keep a regular vitamin D status with the goal that adequate substrate can be provided to CYP27B1, which produces local 1,25(OH)2D3 to protect the mucosal barrier and decrease colonic inflammation.
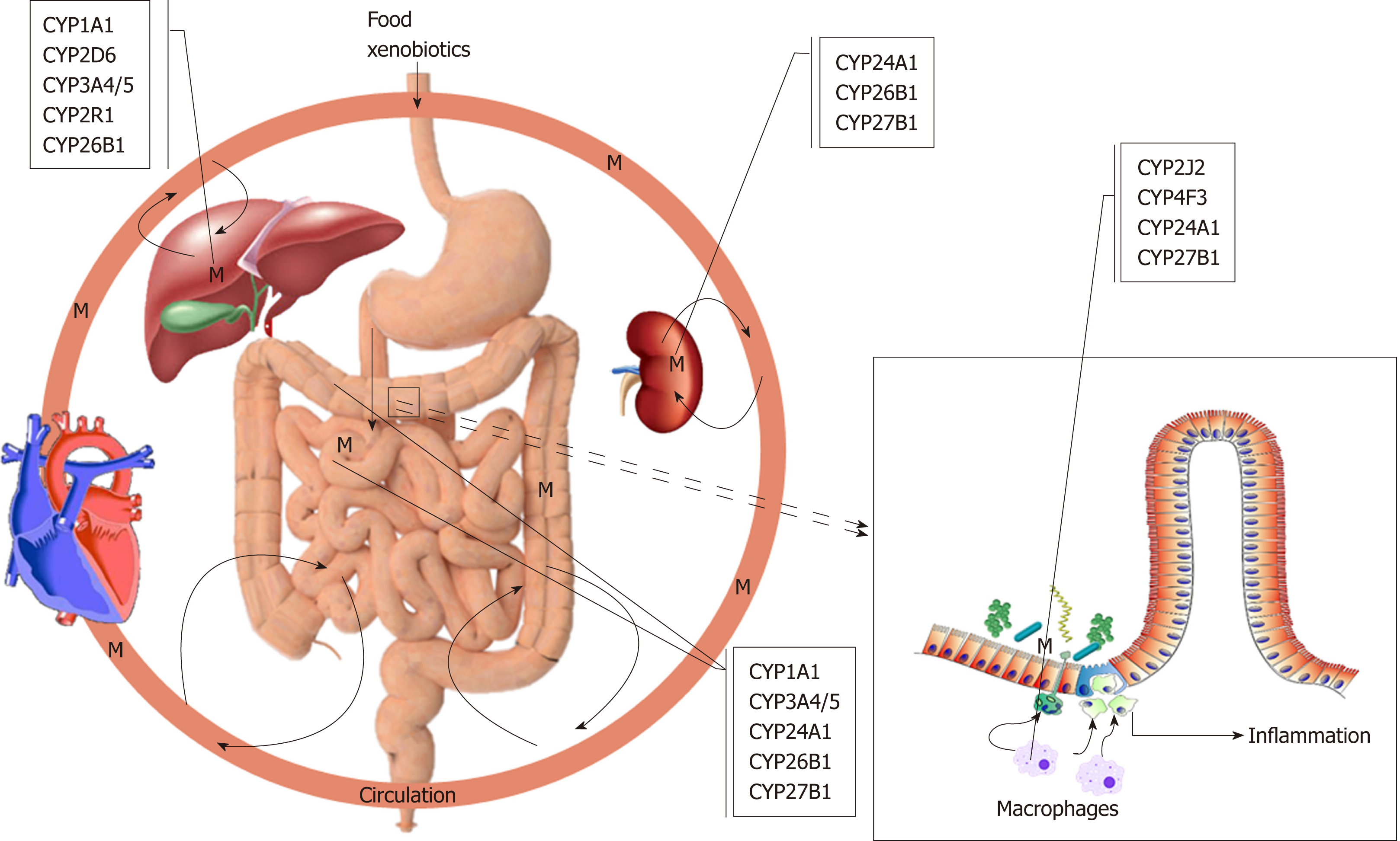
It is a widely accepted hypothesis that UC is caused by a reactive xenobiotic me-tabolite, which is conjugated before excretion. As was pointed out throughout this review, the function and the polymorphism of CYP1A1, 2D6, 2J2, 2R1, 3A4/5, 4F3, 24A1, 26B1 and 27B1 genes are undoubtedly important in the pathogenesis and clinical interest of UC, possibly producing reactive metabolites (Figure 1). These CYPs are somewhat involved in the metabolism of endogenous substrates, most notably vitamin D. It is evident that more of these polymorphisms are either loss-of-function mutations changing the amount of reactive metabolite produced and inhibiting or inducing the enzymes catalysing alternative pathways with the possibility of more severe conditions leading to disease states. However, some polymorphisms result in dysregulation of these enzymes leading to disease state. Most importantly, the exact role of the extrahepatic and extrarenal CYPs such as macrophages and dendritic cells of colonic mucosa is more critical to the development of UC and should require more thorough examinations to clarify their exact roles. The release of these metabolites resulted from polymorphic CYPs functions damage the colonic epithelial barrier and expose the mucosal immune system to luminal contents, thereby initiating an inflammatory response. It is undoubtedly an exciting moment to be involved in the study role of CYPs in autoimmune diseases like UC. An additional benefit is for rationalising the use of current therapeutics, i.e., administering the right drug at the right time and place to the right person. In addition to the polymorphism of CYPs, epigenetic regulation of UC cannot be excluded[165,166].
Early determination of disease-specific genes at the phase when their tolerance is still preserved would be the most critical since the essential treatment can be appropriately begun before auto-animosity happens. The candidate genes for genetic variant analysis was usually chosen from the genes associated with the pathogenesis of UC. Albeit a few SNPs have been identified to be related to drug response in patients with UC, most of these discoveries are as yet uncertain. Since not a single gene but rather different genes are engaged with the pathogenesis of UC just as drug responses are, a genome-wide association studies (GWAS) could be a progressively powerful methodology for distinguishing candidate genes to incorporate into these pharmacogenomic models. However, since the GWAS do not characterise the association between a gene and phenotype of the disease, discoveries from GWAS as well as the biological and clinical associations between the particular loci and diseases ought to be additionally investigated by conventional candidate gene examinations, for example, allelic separation by real-time PCR.
In future, more thorough investigations are necessary to elucidate the act of CYPs in the pathogenesis of UC. Furthermore, better learning of the role of genetic po-lymorphisms and haplotypes in CYPs expression and function will add to an excellent comprehension of inter-individual, ethnic and inter-ethnic variations in drug metabolism and impacts.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Turkey
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chiba T, Demonacos C S-Editor: Ma RY L-Editor: A E-Editor: Ma YJ
| 1. | Omura T, Sato R. A new cytochrome in liver microsomes. J Biol Chem. 1962;237:1375-1376. [PubMed] |
| 2. | Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 982] [Cited by in RCA: 940] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 3. | Ortiz de Montellano PR. Hydrocarbon hydroxylation by cytochrome P450 enzymes. Chem Rev. 2010;110:932-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 945] [Cited by in RCA: 858] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 5. | Rendic S, Guengerich FP. Survey of Human Oxidoreductases and Cytochrome P450 Enzymes Involved in the Metabolism of Xenobiotic and Natural Chemicals. Chem Res Toxicol. 2015;28:38-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 324] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 6. | Guengerich FP. Intersection of the Roles of Cytochrome P450 Enzymes with Xenobiotic and Endogenous Substrates: Relevance to Toxicity and Drug Interactions. Chem Res Toxicol. 2017;30:2-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 7. | Yoshimoto FK, Guengerich FP. Mechanism of the third oxidative step in the conversion of androgens to estrogens by cytochrome P450 19A1 steroid aromatase. J Am Chem Soc. 2014;136:15016-15025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 8. | Shimada T, Yamazaki H, Mimura M, Wakamiya N, Ueng YF, Guengerich FP, Inui Y. Characterization of microsomal cytochrome P450 enzymes involved in the oxidation of xenobiotic chemicals in human fetal liver and adult lungs. Drug Metab Dispos. 1996;24:515-522. [PubMed] |
| 9. | Arinç E, Sen A. In vivo effects of the anesthetic, benzocaine, on liver microsomal cytochrome P450 and mixed-function oxidase activities of gilthead seabream (Sparus aurata). Comp Biochem Physiol Pharmacol Toxicol Endocrinol. 1994;107:399-404. [PubMed] |
| 10. | Sen A, Arinç E. Further immunochemical and biocatalytic characterization of CYP1A1 from feral leaping mullet liver (Liza saliens) microsomes. Comp Biochem Physiol C Toxicol Pharmacol. 2000;126:235-244. [PubMed] |
| 11. | Guengerich FP, Waterman MR, Egli M. Recent Structural Insights into Cytochrome P450 Function. Trends Pharmacol Sci. 2016;37:625-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 254] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 12. | Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1-18. [PubMed] |
| 13. | Porter TD, Coon MJ. Cytochrome P-450. Multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J Biol Chem. 1991;266:13469-13472. [PubMed] |
| 14. | Thelen K, Dressman JB. Cytochrome P450-mediated metabolism in the human gut wall. J Pharm Pharmacol. 2009;61:541-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 15. | Neve EP, Ingelman-Sundberg M. Cytochrome P450 proteins: retention and distribution from the endoplasmic reticulum. Curr Opin Drug Discov Devel. 2010;13:78-85. [PubMed] |
| 16. | Renaud HJ, Cui JY, Khan M, Klaassen CD. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci. 2011;124:261-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 17. | Nebert DW, Adesnik M, Coon MJ, Estabrook RW, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, Kemper B, Levin W. The P450 gene superfamily: recommended nomenclature. DNA. 1987;6:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 522] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 18. | Elfaki I, Mir R, Almutairi FM, Duhier FMA. Cytochrome P450: Polymorphisms and Roles in Cancer, Diabetes and Atherosclerosis. Asian Pac J Cancer Prev. 2018;19:2057-2070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 93] [Reference Citation Analysis (0)] |
| 19. | Sim SC, Ingelman-Sundberg M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Hum Genomics. 2010;4:278-281. [PubMed] |
| 20. | Sim SC, Ingelman-Sundberg M. Update on allele nomenclature for human cytochromes P450 and the Human Cytochrome P450 Allele (CYP-allele) Nomenclature Database. Methods Mol Biol. 2013;987:251-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Makita K, Falck JR, Capdevila JH. Cytochrome P450, the arachidonic acid cascade, and hypertension: new vistas for an old enzyme system. FASEB J. 1996;10:1456-1463. [PubMed] |
| 22. | Bell JC, Strobel HW. Regulation of cytochrome P450 4F11 by nuclear transcription factor-κB. Drug Metab Dispos. 2012;40:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 23. | Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 361] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 24. | Pikuleva IA, Waterman MR. Cytochromes p450: roles in diseases. J Biol Chem. 2013;288:17091-17098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 25. | Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2199] [Cited by in RCA: 2487] [Article Influence: 310.9] [Reference Citation Analysis (2)] |
| 26. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2677] [Cited by in RCA: 4108] [Article Influence: 513.5] [Reference Citation Analysis (110)] |
| 27. | Sýkora J, Pomahačová R, Kreslová M, Cvalínová D, Štych P, Schwarz J. Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J Gastroenterol. 2018;24:2741-2763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 271] [Cited by in RCA: 262] [Article Influence: 37.4] [Reference Citation Analysis (6)] |
| 28. | da Silva BC, Lyra AC, Rocha R, Santana GO. Epidemiology, demographic characteristics and prognostic predictors of ulcerative colitis. World J Gastroenterol. 2014;20:9458-9467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 219] [Cited by in RCA: 206] [Article Influence: 18.7] [Reference Citation Analysis (3)] |
| 29. | Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2894] [Cited by in RCA: 3353] [Article Influence: 186.3] [Reference Citation Analysis (11)] |
| 30. | Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1967] [Cited by in RCA: 2202] [Article Influence: 137.6] [Reference Citation Analysis (6)] |
| 31. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2019] [Cited by in RCA: 1883] [Article Influence: 134.5] [Reference Citation Analysis (2)] |
| 32. | Luo Y, de Lange KM, Jostins L, Moutsianas L, Randall J, Kennedy NA, Lamb CA, McCarthy S, Ahmad T, Edwards C, Serra EG, Hart A, Hawkey C, Mansfield JC, Mowat C, Newman WG, Nichols S, Pollard M, Satsangi J, Simmons A, Tremelling M, Uhlig H, Wilson DC, Lee JC, Prescott NJ, Lees CW, Mathew CG, Parkes M, Barrett JC, Anderson CA. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat Genet. 2017;49:186-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 33. | Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, D'Amato M, Deffontaine V, Dmitrieva J, Docampo E, Elansary M, Farh KK, Franke A, Gori AS, Goyette P, Halfvarson J, Haritunians T, Knight J, Lawrance IC, Lees CW, Louis E, Mariman R, Meuwissen T, Mni M, Momozawa Y, Parkes M, Spain SL, Théâtre E, Trynka G, Satsangi J, van Sommeren S, Vermeire S, Xavier RJ; International Inflammatory Bowel Disease Genetics Consortium, Weersma RK, Duerr RH, Mathew CG, Rioux JD, McGovern DPB, Cho JH, Georges M, Daly MJ, Barrett JC. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 428] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 34. | Mhandire D, Lacerda M, Castel S, Mhandire K, Zhou D, Swart M, Shamu T, Smith P, Musingwini T, Wiesner L, Stray-Pedersen B, Dandara C. Effects of CYP2B6 and CYP1A2 Genetic Variation on Nevirapine Plasma Concentration and Pharmacodynamics as Measured by CD4 Cell Count in Zimbabwean HIV-Infected Patients. OMICS. 2015;19:553-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Paine MF, Schmiedlin-Ren P, Watkins PB. Cytochrome P-450 1A1 expression in human small bowel: interindividual variation and inhibition by ketoconazole. Drug Metab Dispos. 1999;27:360-364. [PubMed] |
| 36. | Prueksaritanont T, Gorham LM, Hochman JH, Tran LO, Vyas KP. Comparative studies of drug-metabolizing enzymes in dog, monkey, and human small intestines, and in Caco-2 cells. Drug Metab Dispos. 1996;24:634-642. [PubMed] |
| 37. | Willey JC, Coy EL, Frampton MW, Torres A, Apostolakos MJ, Hoehn G, Schuermann WH, Thilly WG, Olson DE, Hammersley JR, Crespi CL, Utell MJ. Quantitative RT-PCR measurement of cytochromes p450 1A1, 1B1, and 2B7, microsomal epoxide hydrolase, and NADPH oxidoreductase expression in lung cells of smokers and nonsmokers. Am J Respir Cell Mol Biol. 1997;17:114-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 95] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Dey A, Parmar D, Dayal M, Dhawan A, Seth PK. Cytochrome P450 1A1 (CYP1A1) in blood lymphocytes evidence for catalytic activity and mRNA expression. Life Sci. 2001;69:383-393. [PubMed] |
| 39. | Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mutat Res. 1998;400:201-213. [PubMed] |
| 40. | Oda Y, Aryal P, Terashita T, Gillam EM, Guengerich FP, Shimada T. Metabolic activation of heterocyclic amines and other procarcinogens in Salmonella typhimurium umu tester strains expressing human cytochrome P4501A1, 1A2, 1B1, 2C9, 2D6, 2E1, and 3A4 and human NADPH-P450 reductase and bacterial O-acetyltransferase. Mutat Res. 2001;492:81-90. [PubMed] |
| 41. | Eshkoor SA, Ismail P, Rahman SA. Does CYP1A1 gene polymorphism affect cell damage biomarkers and ageing? Turk J Biol. 2014;38:219-225. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 42. | Spink DC, Hayes CL, Young NR, Christou M, Sutter TR, Jefcoate CR, Gierthy JF. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on estrogen metabolism in MCF-7 breast cancer cells: evidence for induction of a novel 17 beta-estradiol 4-hydroxylase. J Steroid Biochem Mol Biol. 1994;51:251-258. [PubMed] |
| 43. | Schwarz D, Kisselev P, Schunck WH, Chernogolov A, Boidol W, Cascorbi I, Roots I. Allelic variants of human cytochrome P450 1A1 (CYP1A1): effect of T461N and I462V substitutions on steroid hydroxylase specificity. Pharmacogenetics. 2000;10:519-530. [PubMed] |
| 44. | Ma X, Idle JR, Krausz KW, Gonzalez FJ. Metabolism of melatonin by human cytochromes p450. Drug Metab Dispos. 2005;33:489-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 233] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 45. | Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT. Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology. 2003;144:3382-3398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 365] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 46. | Hildebrand CE, Gonzalez FJ, McBride OW, Nebert DW. Assignment of the human 2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible cytochrome P1-450 gene to chromosome 15. Nucleic Acids Res. 1985;13:2009-2016. [PubMed] |
| 47. | Klotz U, Hoensch H, Schütz T, Beaune P, Zanger U, Bode JC, Fritz P. Expression of intestinal drug-metabolizing enzymes in patients with chronic inflammatory bowel disease. Curr Ther Res. 1998;59:556–563. [RCA] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 48. | Arsenescu R, Arsenescu V, Zhong J, Nasser M, Melinte R, Dingle RW, Swanson H, de Villiers WJ. Role of the xenobiotic receptor in inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:1149-1162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 49. | Manzella C, Singhal M, Alrefai WA, Saksena S, Dudeja PK, Gill RK. Serotonin is an endogenous regulator of intestinal CYP1A1 via AhR. Sci Rep. 2018;8:6103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 50. | Plewka D, Plewka A, Szczepanik T, Morek M, Bogunia E, Wittek P, Kijonka C. Expression of selected cytochrome P450 isoforms and of cooperating enzymes in colorectal tissues in selected pathological conditions. Pathol Res Pract. 2014;210:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 51. | Ozkarsli M, Buyukgoze O, Sabah S, Osmanoglu N, Sen A. Genetic polymorphisms of biotransformation enzymes CYP1A1 P4502A6 and microsomal epoxide hydrolase. FEBS Journal. 2008;275:110. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 52. | Buyukgoze O, Osmanoglu N, Arslan S, Sen A. Association of the CYP1A1*2A, GSTT1 null, GSTM1 null, mEPHX*3, and XRCC1-399 genetic polymorphisms with ulcerative colitis. Int J Colorectal Dis. 2013;28:593-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 53. | Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 767] [Cited by in RCA: 764] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 54. | Givens RC, Watkins PB. Pharmacogenetics and clinical gastroenterology. Gastroenterology. 2003;125:240-248. [RCA] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 55. | Teh LK, Bertilsson L. Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance. Drug Metab Pharmacokinet. 2012;27:55-67. [PubMed] |
| 56. | Wang X, Li J, Dong G, Yue J. The endogenous substrates of brain CYP2D. Eur J Pharmacol. 2014;724:211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 57. | Camilleri M. The role of pharmacogenetics in nonmalignant gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. 2012;9:173-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 58. | Kawakami M, Takenoshita-Nakaya S, Takeba Y, Nishimura Y, Oda M, Watanabe M, Ohta Y, Kobayashi S, Ohtsubo T, Kobayashi S, Uchida N, Matsumoto N. Evaluation of CYP2D6 Protein Expression and Activity in the Small Intestine to Determine Its Metabolic Capability in the Japanese Population. Biol Pharm Bull. 2017;40:1344-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 59. | Dudarewicz M, Rychlik-Sych M, Barańska M, Wojtczak A, Trzciński R, Dziki A, Skrętkowicz J. Significance of the genetic polymorphism of CYP2D6 and NAT2 in patients with inflammatory bowel diseases. Pharmacol Rep. 2014;66:686-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 60. | Trzcinski R, Skretkowicz J, Dziki A, Rychlik-Sych M, Baranska M. Genetic polymorphisms of CYP2D6 oxidation in patients with inflammatory bowel disease. Dig Dis Sci. 2010;55:1037-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 61. | Henderson SL, Teft WA, Kim RB. Profound reduction in tamoxifen active metabolite endoxifen in a breast cancer patient treated with rifampin prior to initiation of an anti-TNFα biologic for ulcerative colitis: a case report. BMC Cancer. 2016;16:304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 62. | Lee YH, Bae SC. Association between Functional CYP2D6 Polymorphisms and Susceptibility to Autoimmune Diseases: A Meta-Analysis. Immunol Invest. 2017;46:109-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 63. | Lotfi F, Bahrehmand F, Vaisi-Raygani A, Khodarahmi R, Tanhapour M, Kiani A, Rahimi Z, Pourmotabbed T. Cytochrome P450 (CYP450,2D6*A), N-Acetyltransferase-2 (NAT2*7, A) and Multidrug Resistance 1 (MDR1 3435 T) Alleles Collectively Increase Risk of Ulcerative Colitis. Arch Iran Med. 2018;21:530-535. [PubMed] |
| 64. | Kale E, Sen A. Identification of the cytochrome P4502D6 in the metabolism of 5-aminosalicylic acid: in vitro investigations of potential co-prescription interactions. The 3rd International Symposium on EuroAsian Biodiversity, Minsk, Belarus, 5-8 July, 2017. Abstract Book 484-484. Available from: http://elib.bsu.by/bitstream/123456789/180188/1/p200.pdf. |
| 65. | Kale E, Sen A. Characterization of Human CYP450 Isozymes Responsible For The In Vitro Oxidative Metabolism of Mesalamine Used For Colitis. Innovations in Science and Education. 2019;7:In Press. |
| 66. | Walker VJ, Griffin AP, Hammar DK, Hollenberg PF. Metabolism of Anandamide by Human Cytochrome P450 2J2 in the Reconstituted System and Human Intestinal Microsomes. J Pharmacol Exp Ther. 2016;357:537-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 67. | Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J Biol Chem. 1996;271:3460-3468. [PubMed] |
| 68. | Enayetallah AE, French RA, Thibodeau MS, Grant DF. Distribution of soluble epoxide hydrolase and of cytochrome P450 2C8, 2C9, and 2J2 in human tissues. J Histochem Cytochem. 2004;52:447-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 204] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 69. | Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC. The human intestinal cytochrome P450 "pie". Drug Metab Dispos. 2006;34:880-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 727] [Cited by in RCA: 615] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 70. | Karkhanis A, Hong Y, Chan ECY. Inhibition and inactivation of human CYP2J2: Implications in cardiac pathophysiology and opportunities in cancer therapy. Biochem Pharmacol. 2017;135:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 71. | Bystrom J, Thomson SJ, Johansson J, Edin ML, Zeldin DC, Gilroy DW, Smith AM, Bishop-Bailey D. Inducible CYP2J2 and its product 11,12-EET promotes bacterial phagocytosis: a role for CYP2J2 deficiency in the pathogenesis of Crohn's disease? PLoS One. 2013;8:e75107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 72. | Qiu YE, Qin J, Luo Y, Qin SL, Mu YF, Cun R, Jiang HL, Chen JJ, Yu MH, Zhong M. Increased epoxyeicosatrienoic acids may be part of a protective mechanism in human ulcerative colitis, with increased CYP2J2 and reduced soluble epoxide hydrolase expression. Prostaglandins Other Lipid Mediat. 2018;136:9-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 73. | Izzo AA, Fezza F, Capasso R, Bisogno T, Pinto L, Iuvone T, Esposito G, Mascolo N, Di Marzo V, Capasso F. Cannabinoid CB1-receptor mediated regulation of gastrointestinal motility in mice in a model of intestinal inflammation. Br J Pharmacol. 2001;134:563-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 197] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 74. | D'Argenio G, Valenti M, Scaglione G, Cosenza V, Sorrentini I, Di Marzo V. Up-regulation of anandamide levels as an endogenous mechanism and a pharmacological strategy to limit colon inflammation. FASEB J. 2006;20:568-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 174] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 75. | Di Marzo V, Izzo AA. Endocannabinoid overactivity and intestinal inflammation. Gut. 2006;55:1373-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 76. | Askari AA, Thomson S, Edin ML, Lih FB, Zeldin DC, Bishop-Bailey D. Basal and inducible anti-inflammatory epoxygenase activity in endothelial cells. Biochem Biophys Res Commun. 2014;446:633-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Izzo AA. Cannabinoids and intestinal motility: welcome to CB2 receptors. Br J Pharmacol. 2004;142:1201-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 78. | Mathison R, Ho W, Pittman QJ, Davison JS, Sharkey KA. Effects of cannabinoid receptor-2 activation on accelerated gastrointestinal transit in lipopolysaccharide-treated rats. Br J Pharmacol. 2004;142:1247-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 110] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 79. | Nolte H, Spjeldnaes N, Kruse A, Windelborg B. Histamine release from gut mast cells from patients with inflammatory bowel diseases. Gut. 1990;31:791-794. [PubMed] |
| 80. | King T, Biddle W, Bhatia P, Moore J, Miner PB. Colonic mucosal mast cell distribution at line of demarcation of active ulcerative colitis. Dig Dis Sci. 1992;37:490-495. [PubMed] |
| 81. | Lennon EM, Borst LB, Edwards LL, Moeser AJ. Mast Cells Exert Anti-Inflammatory Effects in an IL10<sup>-/-</sup> Model of Spontaneous Colitis. Mediators Inflamm. 2018;2018:7817360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 82. | Wechsler JB, Szabo A, Hsu CL, Krier-Burris RA, Schroeder HA, Wang MY, Carter RG, Velez TE, Aguiniga LM, Brown JB, Miller ML, Wershil BK, Barrett TA, Bryce PJ. Histamine drives severity of innate inflammation via histamine 4 receptor in murine experimental colitis. Mucosal Immunol. 2018;11:861-870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 83. | Xu M, Ju W, Hao H, Wang G, Li P. Cytochrome P450 2J2: distribution, function, regulation, genetic polymorphisms and clinical significance. Drug Metab Rev. 2013;45:311-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 84. | Otte JM, Spiecker M, Jagiello P, Brand S, Felderbauer P, Griga T, Epplen JT, Schmitz F, Schmidt WE. Crohn's disease and ulcerative colitis are associated with a promoter polymorphism of the cytochrome P450 epoxygenase CYP2J2 gene. Gastroenterology. 2005;128:A446-A447. [DOI] [Full Text] |
| 85. | King LM, Ma J, Srettabunjong S, Graves J, Bradbury JA, Li L, Spiecker M, Liao JK, Mohrenweiser H, Zeldin DC. Cloning of CYP2J2 gene and identification of functional polymorphisms. Mol Pharmacol. 2002;61:840-852. [PubMed] |
| 86. | Spiecker M, Darius H, Hankeln T, Soufi M, Sattler AM, Schaefer JR, Node K, Börgel J, Mügge A, Lindpaintner K, Huesing A, Maisch B, Zeldin DC, Liao JK. Risk of coronary artery disease associated with polymorphism of the cytochrome P450 epoxygenase CYP2J2. Circulation. 2004;110:2132-2136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 195] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 87. | Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci U S A. 2004;101:7711-7715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 489] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 88. | Slater NA, Rager ML, Havrda DE, Harralson AF. Genetic Variation in CYP2R1 and GC Genes Associated With Vitamin D Deficiency Status. J Pharm Pract. 2017;30:31-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 89. | Ramos-Lopez E, Brück P, Jansen T, Herwig J, Badenhoop K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab Res Rev. 2007;23:631-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 129] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 90. | Cooper JD, Smyth DJ, Walker NM, Stevens H, Burren OS, Wallace C, Greissl C, Ramos-Lopez E, Hyppönen E, Dunger DB, Spector TD, Ouwehand WH, Wang TJ, Badenhoop K, Todd JA. Inherited variation in vitamin D genes is associated with predisposition to autoimmune disease type 1 diabetes. Diabetes. 2011;60:1624-1631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 201] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 91. | Hussein AG, Mohamed RH, Alghobashy AA. Synergism of CYP2R1 and CYP27B1 polymorphisms and susceptibility to type 1 diabetes in Egyptian children. Cell Immunol. 2012;279:42-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 92. | Inoue N, Watanabe M, Ishido N, Katsumata Y, Kagawa T, Hidaka Y, Iwatani Y. The functional polymorphisms of VDR, GC and CYP2R1 are involved in the pathogenesis of autoimmune thyroid diseases. Clin Exp Immunol. 2014;178:262-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 93. | Al-Barry MA, Albalawi AM, Sayf MA, Badawi A, Afzal S, Latif M, Samman MI, Basit S. Sequence analysis of four vitamin D family genes (VDR, CYP24A1, CYP27B1 and CYP2R1) in Vogt-Koyanagi-Harada (VKH) patients: identification of a potentially pathogenic variant in CYP2R1. BMC Ophthalmol. 2016;16:172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 94. | Scazzone C, Agnello L, Ragonese P, Lo Sasso B, Bellia C, Bivona G, Schillaci R, Salemi G, Ciaccio M. Association of CYP2R1 rs10766197 with MS risk and disease progression. J Neurosci Res. 2018;96:297-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 95. | El-Matary W, Sikora S, Spady D. Bone mineral density, vitamin D, and disease activity in children newly diagnosed with inflammatory bowel disease. Dig Dis Sci. 2011;56:825-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 96. | Ulitsky A, Ananthakrishnan AN, Naik A, Skaros S, Zadvornova Y, Binion DG, Issa M. Vitamin D deficiency in patients with inflammatory bowel disease: association with disease activity and quality of life. JPEN J Parenter Enteral Nutr. 2011;35:308-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 240] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 97. | Palmer MT, Weaver CT. Linking vitamin d deficiency to inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:2245-2256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 98. | Del Pinto R, Pietropaoli D, Chandar AK, Ferri C, Cominelli F. Association Between Inflammatory Bowel Disease and Vitamin D Deficiency: A Systematic Review and Meta-analysis. Inflamm Bowel Dis. 2015;21:2708-2717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 188] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 99. | Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, Cooper JD, O'Reilly PF, Houston DK, Glazer NL, Vandenput L, Peacock M, Shi J, Rivadeneira F, McCarthy MI, Anneli P, de Boer IH, Mangino M, Kato B, Smyth DJ, Booth SL, Jacques PF, Burke GL, Goodarzi M, Cheung CL, Wolf M, Rice K, Goltzman D, Hidiroglou N, Ladouceur M, Wareham NJ, Hocking LJ, Hart D, Arden NK, Cooper C, Malik S, Fraser WD, Hartikainen AL, Zhai G, Macdonald HM, Forouhi NG, Loos RJ, Reid DM, Hakim A, Dennison E, Liu Y, Power C, Stevens HE, Jaana L, Vasan RS, Soranzo N, Bojunga J, Psaty BM, Lorentzon M, Foroud T, Harris TB, Hofman A, Jansson JO, Cauley JA, Uitterlinden AG, Gibson Q, Järvelin MR, Karasik D, Siscovick DS, Econs MJ, Kritchevsky SB, Florez JC, Todd JA, Dupuis J, Hyppönen E, Spector TD. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1279] [Cited by in RCA: 1202] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 100. | Morán-Auth Y, Penna-Martinez M, Shoghi F, Ramos-Lopez E, Badenhoop K. Vitamin D status and gene transcription in immune cells. J Steroid Biochem Mol Biol. 2013;136:83-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 101. | Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2251] [Cited by in RCA: 2764] [Article Influence: 230.3] [Reference Citation Analysis (0)] |
| 102. | Ashida R, Okamura Y, Ohshima K, Kakuda Y, Uesaka K, Sugiura T, Ito T, Yamamoto Y, Sugino T, Urakami K, Kusuhara M, Yamaguchi K. <i>CYP3A4</i> Gene Is a Novel Biomarker for Predicting a Poor Prognosis in Hepatocellular Carcinoma. Cancer Genomics Proteomics. 2017;14:445-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 103. | Gupta RP, He YA, Patrick KS, Halpert JR, Bell NH. CYP3A4 is a vitamin D-24- and 25-hydroxylase: analysis of structure function by site-directed mutagenesis. J Clin Endocrinol Metab. 2005;90:1210-1219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 104. | Miyata N, Taniguchi K, Seki T, Ishimoto T, Sato-Watanabe M, Yasuda Y, Doi M, Kametani S, Tomishima Y, Ueki T, Sato M, Kameo K. HET0016, a potent and selective inhibitor of 20-HETE synthesizing enzyme. Br J Pharmacol. 2001;133:325-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 168] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 105. | Bishop-Bailey D, Thomson S, Askari A, Faulkner A, Wheeler-Jones C. Lipid-metabolizing CYPs in the regulation and dysregulation of metabolism. Annu Rev Nutr. 2014;34:261-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 106. | Lin YS, Dowling AL, Quigley SD, Farin FM, Zhang J, Lamba J, Schuetz EG, Thummel KE. Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol. 2002;62:162-172. [PubMed] |
| 107. | Williams JA, Ring BJ, Cantrell VE, Jones DR, Eckstein J, Ruterbories K, Hamman MA, Hall SD, Wrighton SA. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos. 2002;30:883-891. [PubMed] |
| 108. | Onodera M, Endo K, Kakuta Y, Kuroha M, Kimura T, Hiramoto K, Kanazawa Y, Negoro K, Shiga H, Kinouchi Y, Shimosegawa T. ATP-binding cassette subfamily B member 1 1236C/T polymorphism significantly affects the therapeutic outcome of tacrolimus in patients with refractory ulcerative colitis. J Gastroenterol Hepatol. 2017;32:1562-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 109. | Onodera M, Endo K, Naito T, Moroi R, Kuroha M, Kanazawa Y, Kimura T, Shiga H, Kakuta Y, Negoro K, Kinouchi Y, Shimosegawa T. Tacrolimus Dose Optimization Strategy for Refractory Ulcerative Colitis Based on the Cytochrome P450 3A5 Polymorphism Prediction Using Trough Concentration after 24 Hours. Digestion. 2018;97:90-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 110. | Hirai F, Takatsu N, Yano Y, Satou Y, Takahashi H, Ishikawa S, Tsurumi K, Hisabe T, Matsui T. Impact of CYP3A5 genetic polymorphisms on the pharmacokinetics and short-term remission in patients with ulcerative colitis treated with tacrolimus. J Gastroenterol Hepatol. 2014;29:60-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 111. | Naganuma M, Fujii T, Watanabe M. The use of traditional and newer calcineurin inhibitors in inflammatory bowel disease. J Gastroenterol. 2011;46:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 112. | Cervelli M, Russ G. Specialty practice series: intrapatient variability with tacrolimus. Australian J Pharmacy. 2012;93:83-86. |
| 113. | Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1583] [Cited by in RCA: 1597] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 114. | Hesselink DA, van Schaik RH, van der Heiden IP, van der Werf M, Gregoor PJ, Lindemans J, Weimar W, van Gelder T. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 529] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 115. | Herrlinger KR, Koc H, Winter S, Teml A, Stange EF, Fellermann K, Fritz P, Schwab M, Schaeffeler E. ABCB1 single-nucleotide polymorphisms determine tacrolimus response in patients with ulcerative colitis. Clin Pharmacol Ther. 2011;89:422-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 116. | Asada A, Bamba S, Morita Y, Takahashi K, Imaeda H, Nishida A, Inatomi O, Sugimoto M, Sasaki M, Andoh A. The effect of CYP3A5 genetic polymorphisms on adverse events in patients with ulcerative colitis treated with tacrolimus. Dig Liver Dis. 2017;49:24-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 117. | Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445-456. [PubMed] |
| 118. | Kim YH, Bae YJ, Kim HS, Cha HJ, Yun JS, Shin JS, Seong WK, Lee YM, Han KM. Measurement of Human Cytochrome P450 Enzyme Induction Based on Mesalazine and Mosapride Citrate Treatments Using a Luminescent Assay. Biomol Ther (Seoul). 2015;23:486-492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 119. | Kikuta Y, Kato M, Yamashita Y, Miyauchi Y, Tanaka K, Kamada N, Kusunose M. Human leukotriene B4 omega-hydroxylase (CYP4F3) gene: molecular cloning and chromosomal localization. DNA Cell Biol. 1998;17:221-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 120. | Corcos L, Lucas D, Le Jossic-Corcos C, Dréano Y, Simon B, Plée-Gautier E, Amet Y, Salaün JP. Human cytochrome P450 4F3: structure, functions, and prospects. Drug Metabol Drug Interact. 2012;27:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 121. | Powell WS, Rokach J. Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochim Biophys Acta. 2015;1851:340-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 239] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 122. | Costea I, Mack DR, Israel D, Morgan K, Krupoves A, Seidman E, Deslandres C, Lambrette P, Grimard G, Levy E, Amre DK. Genes involved in the metabolism of poly-unsaturated fatty-acids (PUFA) and risk for Crohn's disease in children & young adults. PLoS One. 2010;5:e15672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 123. | Scaioli E, Liverani E, Belluzzi A. The Imbalance between n-6/n-3 Polyunsaturated Fatty Acids and Inflammatory Bowel Disease: A Comprehensive Review and Future Therapeutic Perspectives. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 124. | Johnson AL, Edson KZ, Totah RA, Rettie AE. Cytochrome P450 ω-Hydroxylases in Inflammation and Cancer. Adv Pharmacol. 2015;74:223-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 125. | Ananthakrishnan AN, Khalili H, Song M, Higuchi LM, Lochhead P, Richter JM, Chan AT. Genetic Polymorphisms in Fatty Acid Metabolism Modify the Association Between Dietary n3: n6 Intake and Risk of Ulcerative Colitis: A Prospective Cohort Study. Inflamm Bowel Dis. 2017;23:1898-1904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 126. | Yin J, Liu H, Liu Z, Owzar K, Han Y, Su L, Wei Y, Hung RJ, Brhane Y, McLaughlin J, Brennan P, Bickeboeller H, Rosenberger A, Houlston RS, Caporaso N, Landi MT, Heinrich J, Risch A, Christiani DC, Amos CI, Wei Q. Pathway-analysis of published genome-wide association studies of lung cancer: A potential role for the CYP4F3 locus. Mol Carcinog. 2017;56:1663-1672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 127. | Jones G, Prosser DE, Kaufmann M. Cytochrome P450-mediated metabolism of vitamin D. J Lipid Res. 2014;55:13-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 340] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 128. | Jones G, Strugnell SA, DeLuca HF. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78:1193-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 902] [Cited by in RCA: 858] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 129. | Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29:664-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 458] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 130. | Shinki T, Jin CH, Nishimura A, Nagai Y, Ohyama Y, Noshiro M, Okuda K, Suda T. Parathyroid hormone inhibits 25-hydroxyvitamin D3-24-hydroxylase mRNA expression stimulated by 1 alpha,25-dihydroxyvitamin D3 in rat kidney but not in intestine. J Biol Chem. 1992;267:13757-13762. [PubMed] |
| 131. | Shimada T, Yamazaki Y, Takahashi M, Hasegawa H, Urakawa I, Oshima T, Ono K, Kakitani M, Tomizuka K, Fujita T, Fukumoto S, Yamashita T. Vitamin D receptor-independent FGF23 actions in regulating phosphate and vitamin D metabolism. Am J Physiol Renal Physiol. 2005;289:F1088-F1095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 258] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 132. | Ramon I, Kleynen P, Body JJ, Karmali R. Fibroblast growth factor 23 and its role in phosphate homeostasis. Eur J Endocrinol. 2010;162:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 133. | Rübsamen D, Kunze MM, Buderus V, Brauß TF, Bajer MM, Brüne B, Schmid T. Inflammatory conditions induce IRES-dependent translation of cyp24a1. PLoS One. 2014;9:e85314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 134. | Schlingmann KP, Kaufmann M, Weber S, Irwin A, Goos C, John U, Misselwitz J, Klaus G, Kuwertz-Bröking E, Fehrenbach H, Wingen AM, Güran T, Hoenderop JG, Bindels RJ, Prosser DE, Jones G, Konrad M. Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med. 2011;365:410-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 409] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 135. | Gong C, Long Z, Yu Y, Zhu L, Tian J, Li S, Li J, Yu H, Chi Q, Piao D, Wang F, Zhao Y, Cui B. Dietary factors and polymorphisms in vitamin D metabolism genes: the risk and prognosis of colorectal cancer in northeast China. Sci Rep. 2017;7:8827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 136. | Chen XQ, Mao JY, Li WB, Li J, Yang H, Qian JM, Li JN. Association between CYP24A1 polymorphisms and the risk of colonic polyps and colon cancer in a Chinese population. World J Gastroenterol. 2017;23:5179-5186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 137. | Dong J, Hu Z, Wu C, Guo H, Zhou B, Lv J, Lu D, Chen K, Shi Y, Chu M, Wang C, Zhang R, Dai J, Jiang Y, Cao S, Qin Z, Yu D, Ma H, Jin G, Gong J, Sun C, Zhao X, Yin Z, Yang L, Li Z, Deng Q, Wang J, Wu W, Zheng H, Zhou G, Chen H, Guan P, Peng Z, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Amos CI, Chen F, Tan W, Jin L, Wu T, Lin D, Shen H. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat Genet. 2012;44:895-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 138. | Shiotani A, Murao T, Fujita Y, Fujimura Y, Sakakibara T, Nishio K, Haruma K. Novel single nucleotide polymorphism markers for low dose aspirin-associated small bowel bleeding. PLoS One. 2013;8:e84244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 139. | Kong J, Xu F, Qu J, Wang Y, Gao M, Yu H, Qian B. Genetic polymorphisms in the vitamin D pathway in relation to lung cancer risk and survival. Oncotarget. 2015;6:2573-2582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 140. | Zheng B, Jeong S, Zhu Y, Chen L, Xia Q. miRNA and lncRNA as biomarkers in cholangiocarcinoma(CCA). Oncotarget. 2017;8:100819-100830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 141. | Yu S, Li X, Wang Y, Mao Z, Wang C, Ba Y, Li W. Transmission disequilibrium of rs4809957 in type 2 diabetes mellitus families and its association with vitamin D deficiency: A family-based case-control study. J Diabetes Complications. 2018;32:406-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 142. | Pei FH, Wang YJ, Gao SL, Liu BR, DU YJ, Liu W, Yu HY, Zhao LX, Chi BR. Vitamin D receptor gene polymorphism and ulcerative colitis susceptibility in Han Chinese. J Dig Dis. 2011;12:90-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 143. | Hughes DJ, McManus R, Neary P, O'morain C, O'sullivan M. Common variation in the vitamin D receptor gene and risk of inflammatory bowel disease in an Irish case-control study. Eur J Gastroenterol Hepatol. 2011;23:807-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 144. | Xue LN, Xu KQ, Zhang W, Wang Q, Wu J, Wang XY. Associations between vitamin D receptor polymorphisms and susceptibility to ulcerative colitis and Crohn's disease: a meta-analysis. Inflamm Bowel Dis. 2013;19:54-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 145. | Cantorna MT, Munsick C, Bemiss C, Mahon BD. 1,25-Dihydroxycholecalciferol prevents and ameliorates symptoms of experimental murine inflammatory bowel disease. J Nutr. 2000;130:2648-2652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 349] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 146. | Di Rosa M, Malaguarnera G, De Gregorio C, Palumbo M, Nunnari G, Malaguarnera L. Immuno-modulatory effects of vitamin D3 in human monocyte and macrophages. Cell Immunol. 2012;280:36-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 115] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 147. | Nelson DR. A second CYP26 P450 in humans and zebrafish: CYP26B1. Arch Biochem Biophys. 1999;371:345-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 148. | Kang SG, Wang C, Matsumoto S, Kim CH. High and low vitamin A therapies induce distinct FoxP3+ T-cell subsets and effectively control intestinal inflammation. Gastroenterology. 2009;137:1391-402.e1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 149. | Takeuchi H, Yokota A, Ohoka Y, Iwata M. Cyp26b1 regulates retinoic acid-dependent signals in T cells and its expression is inhibited by transforming growth factor-β. PLoS One. 2011;6:e16089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 150. | Chenery A, Burrows K, Antignano F, Underhill TM, Petkovich M, Zaph C. The retinoic acid-metabolizing enzyme Cyp26b1 regulates CD4 T cell differentiation and function. PLoS One. 2013;8:e72308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 151. | Fransén K, Franzén P, Magnuson A, Elmabsout AA, Nyhlin N, Wickbom A, Curman B, Törkvist L, D'Amato M, Bohr J, Tysk C, Sirsjö A, Halfvarson J. Polymorphism in the retinoic acid metabolizing enzyme CYP26B1 and the development of Crohn's Disease. PLoS One. 2013;8:e72739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 152. | Holick MF. Resurrection of vitamin D deficiency and rickets. J Clin Invest. 2006;116:2062-2072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 966] [Cited by in RCA: 888] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 153. | Aranow C. Vitamin D and the immune system. J Investig Med. 2011;59:881-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 851] [Cited by in RCA: 743] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 154. | Hahn HJ, Kuttler B, Mathieu C, Bouillon R. 1,25-Dihydroxyvitamin D3 reduces MHC antigen expression on pancreatic beta-cells in vitro. Transplant Proc. 1997;29:2156-2157. [PubMed] |
| 155. | Piemonti L, Monti P, Sironi M, Fraticelli P, Leone BE, Dal Cin E, Allavena P, Di Carlo V. Vitamin D3 affects differentiation, maturation, and function of human monocyte-derived dendritic cells. J Immunol. 2000;164:4443-4451. [PubMed] |
| 156. | Lopez ER, Zwermann O, Segni M, Meyer G, Reincke M, Seissler J, Herwig J, Usadel KH, Badenhoop K. A promoter polymorphism of the CYP27B1 gene is associated with Addison's disease, Hashimoto's thyroiditis, Graves' disease and type 1 diabetes mellitus in Germans. Eur J Endocrinol. 2004;151:193-197. [PubMed] |
| 157. | Hewison M, Burke F, Evans KN, Lammas DA, Sansom DM, Liu P, Modlin RL, Adams JS. Extra-renal 25-hydroxyvitamin D3-1alpha-hydroxylase in human health and disease. J Steroid Biochem Mol Biol. 2007;103:316-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 294] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 158. | Adams JS, Rafison B, Witzel S, Reyes RE, Shieh A, Chun R, Zavala K, Hewison M, Liu PT. Regulation of the extrarenal CYP27B1-hydroxylase. J Steroid Biochem Mol Biol. 2014;144 Pt A:22-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 159. | Du J, Wei X, Ge X, Chen Y, Li YC. Microbiota-Dependent Induction of Colonic Cyp27b1 Is Associated With Colonic Inflammation: Implications of Locally Produced 1,25-Dihydroxyvitamin D3 in Inflammatory Regulation in the Colon. Endocrinology. 2017;158:4064-4075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 160. | Alzahrani AS, Zou M, Baitei EY, Alshaikh OM, Al-Rijjal RA, Meyer BF, Shi Y. A novel G102E mutation of CYP27B1 in a large family with vitamin D-dependent rickets type 1. J Clin Endocrinol Metab. 2010;95:4176-4183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 161. | Fichna M, Zurawek M, Januszkiewicz-Lewandowska D, Gryczyñska M, Fichna P, Sowiñski J, Nowak J. Association of the CYP27B1 C(-1260)A polymorphism with autoimmune Addison's disease. Exp Clin Endocrinol Diabetes. 2010;118:544-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 162. | Ramagopalan SV, Dyment DA, Cader MZ, Morrison KM, Disanto G, Morahan JM, Berlanga-Taylor AJ, Handel A, De Luca GC, Sadovnick AD, Lepage P, Montpetit A, Ebers GC. Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Ann Neurol. 2011;70:881-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 163. | Ross JP, Bernales CQ, Lee JD, Sadovnick AD, Traboulsee AL, Vilariño-Güell C. Analysis of CYP27B1 in multiple sclerosis. J Neuroimmunol. 2014;266:64-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 164. | Demir K, Kattan WE, Zou M, Durmaz E, BinEssa H, Nalbantoğlu Ö, Al-Rijjal RA, Meyer B, Özkan B, Shi Y. Novel CYP27B1 Gene Mutations in Patients with Vitamin D-Dependent Rickets Type 1A. PLoS One. 2015;10:e0131376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 165. | Gould NJ, Davidson KL, Nwokolo CU, Arasaradnam RP. A systematic review of the role of DNA methylation on inflammatory genes in ulcerative colitis. Epigenomics. 2016;8:667-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 166. | Barnicle A, Seoighe C, Greally JM, Golden A, Egan LJ. Inflammation-associated DNA methylation patterns in epithelium of ulcerative colitis. Epigenetics. 2017;12:591-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |