Published online Mar 21, 2019. doi: 10.3748/wjg.v25.i11.1341
Peer-review started: November 20, 2018
First decision: January 30, 2019
Revised: February 19, 2019
Accepted: February 22, 2019
Article in press: February 22, 2019
Published online: March 21, 2019
Processing time: 123 Days and 6.6 Hours
Genomic profiling of tumors has contributed to the understanding of colorectal cancer (CRC), facilitating diagnosis, prognosis and selection of treatments, including targeted regimens. A report suggested that a 19-gene-based risk classifier (TCA19) was a prognostic tool for patients with stage III CRC. The survival outcomes in patients with stage IV CRC are still poor and appropriate selection of targeted therapies and immunotherapies is challenging.
To assess clinical implication of TCA19 in patients with stage IV CRC, and to identify TCA19 with involvement in immune-oncology.
A retrospective review of the medical records of 60 patients with stage IV CRC was conducted, assessing clinicopathological variables and progression-free survival (PFS). TCA19 gene expression was determined by quantitative polymerase chain reaction (qPCR) in matched normal and tumor tissues taken from the study cohort. Expression of potential immune-oncology regulatory proteins and targets was examined by immunohistochemistry (IHC), western blot, immunofluorescence staining in tissues from a validation cohort of 10 patients, and in CRC cell lines co-cultured with monocyte in vitro.
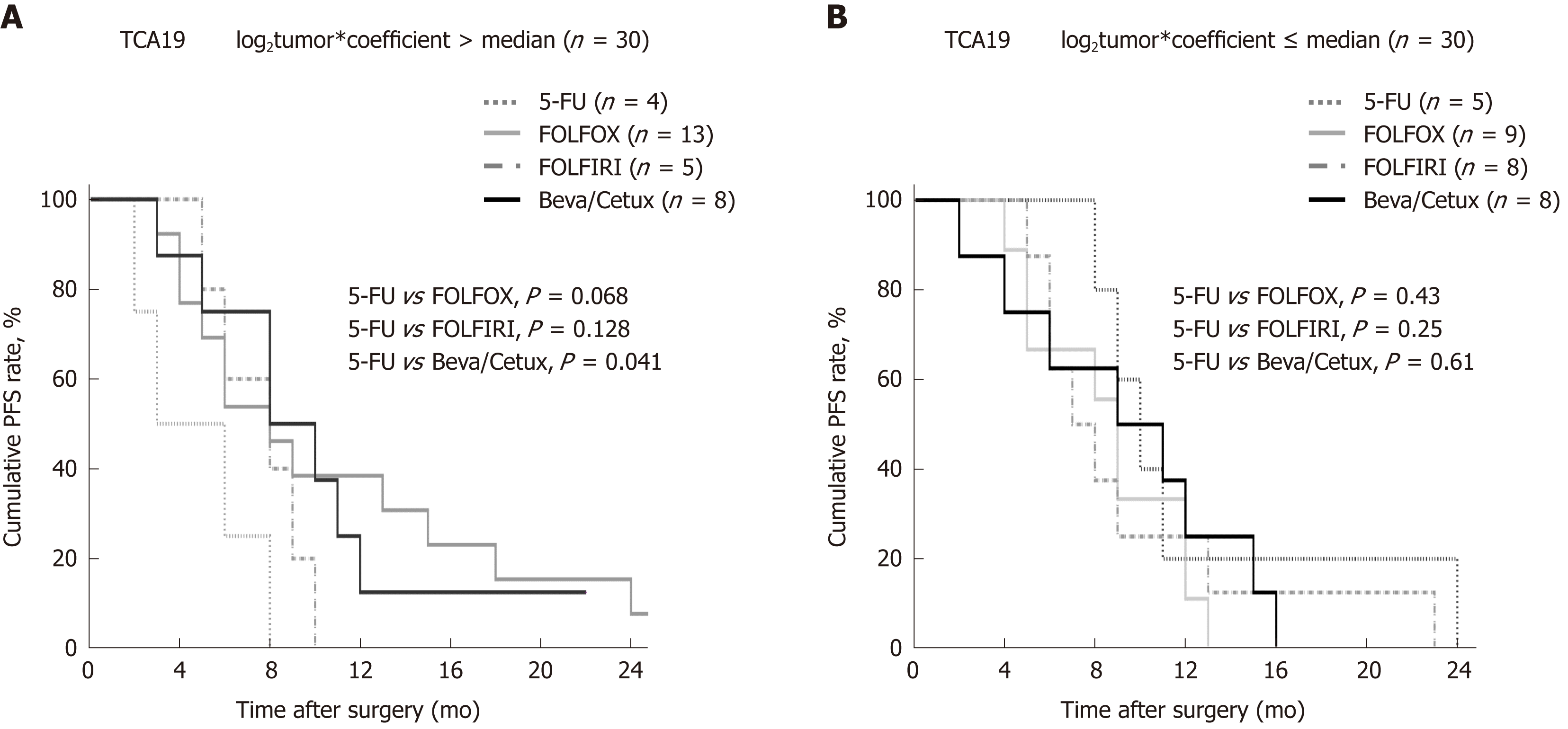
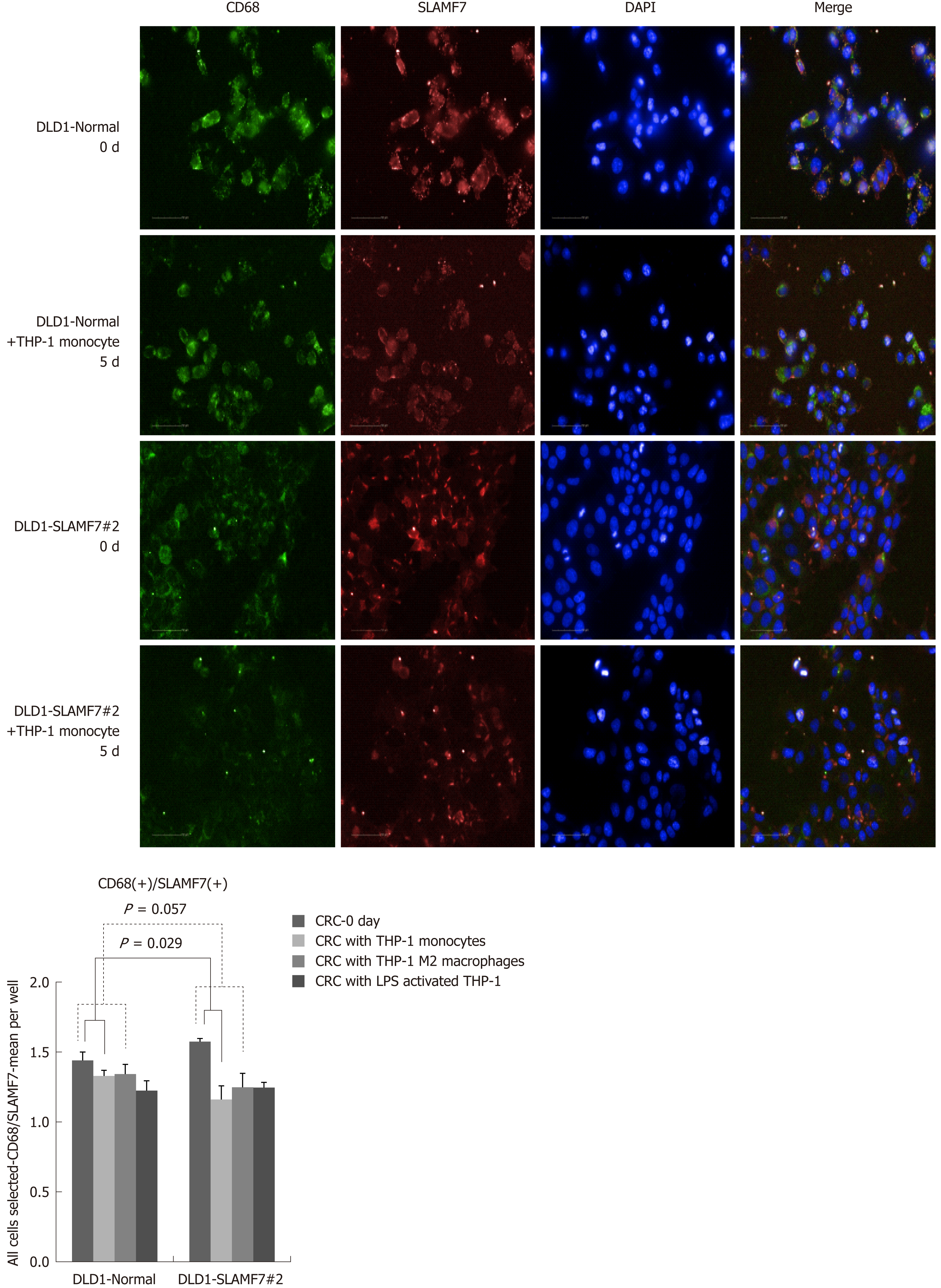
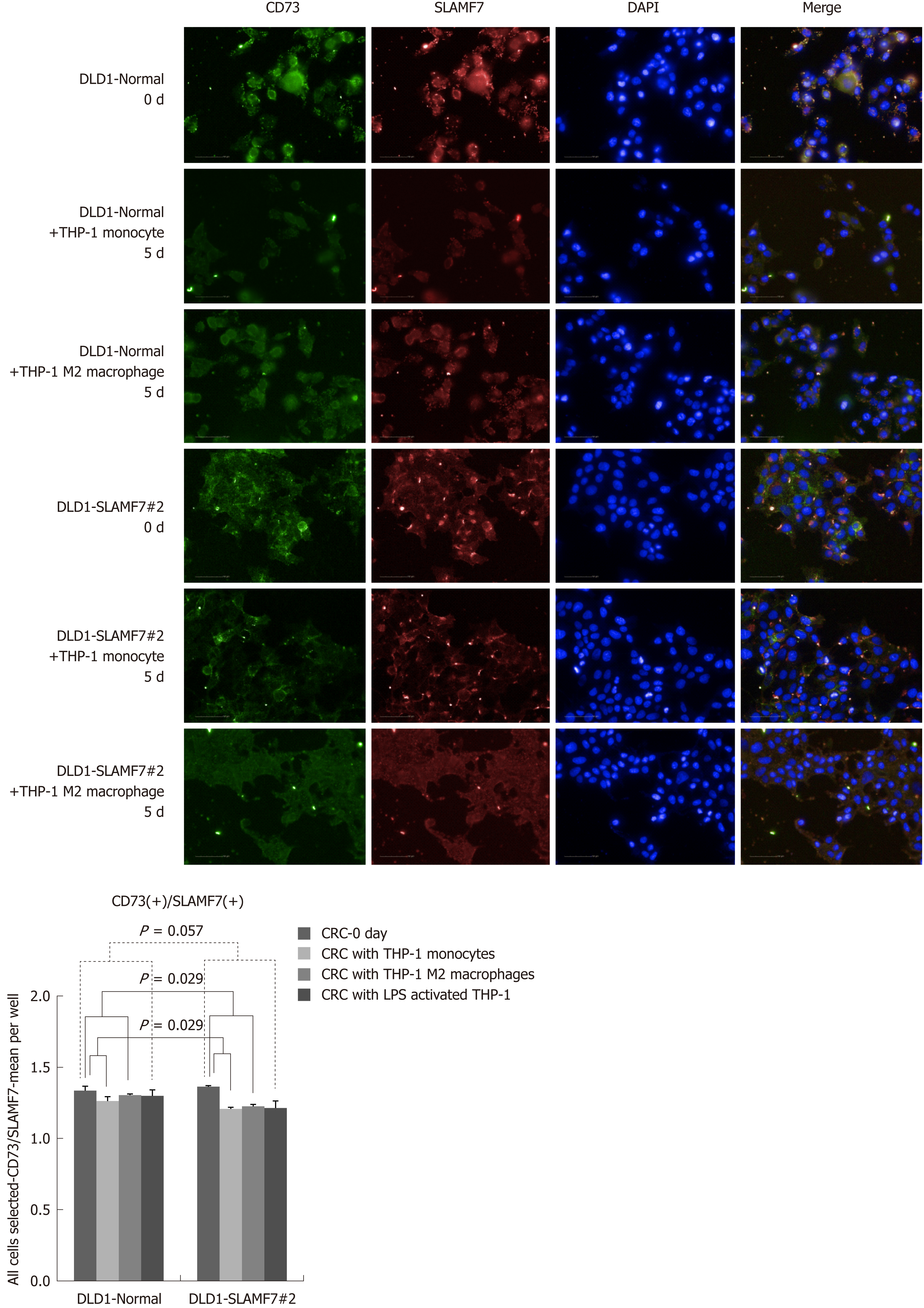
In the patients with TCA19 score higher than the median, the PFS rates of eight patients who received the targeted regimens were significantly higher than the PFS rates of four patients who received 5-fluorouracil-based regimen (P = 0.041). In multivariate analysis, expression of signaling lymphocytic activation molecule family, member 7 (SLAMF7) and triggering receptor expressed on myeloid cells 1 (TREM1) was associated with PFS in the 60-patient cohort. After checking another 10 validate set, the expression of the IHC, the level of real-time qPCR, and the level of western blot were lower for SLAMF7 and higher for TREM7 in primary and metastatic tumors than in normal tissues. In CRC cells expressing SLAMF7 that were co-cultured with a monocytic cell line, levels of CD 68 and CD 73 were significantly lower at day 5 of co-culture than at day 0.
The TCA19 score might be prognostic for target-regimen-specific PFS in stage IV CRC. Down-regulation of SLAMF7 and up-regulation of TREM1 occur in primary and metastatic tumor tissues.
Core tip: The current study showed that in the patients with stage IV colorectal cancer (CRC) and a higher 19-gene based risk classifier score, the target-regimen-specific progression-free survival (PFS) was significantly increased compared with the 5-fluorouracil-regimen-specific PFS. Using another 10 validate set, down-regulation of signaling lymphocytic activation molecule family, member 7 (SLAMF7) and up-regulation of triggering receptor expressed on myeloid cells 1 were identified in primary and metastatic tumors compared with normal tissue. In CRC cells expressing SLAMF7 that were co-cultured with a monocytic cell line, levels of CD68 and CD73 were significantly lower at day 5 of co-culture than at day 0.
- Citation: Lee JL, Roh SA, Kim CW, Kwon YH, Ha YJ, Kim SK, Kim SY, Cho DH, Kim YS, Kim JC. Clinical assessment and identification of immuno-oncology markers concerning the 19-gene based risk classifier in stage IV colorectal cancer. World J Gastroenterol 2019; 25(11): 1341-1354
- URL: https://www.wjgnet.com/1007-9327/full/v25/i11/1341.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i11.1341
According to the Korean National Cancer Information Center, colorectal cancer (CRC) is the third leading cause of cancer death in South Korea[1]. The survival rate for CRC in Korea has increased over the past two or three decades, with screening programs, new chemotherapy regimens (including targeted agents) and advances in surgical techniques all contributing to survival improvements[2-4]. Further improvements could be obtained by optimizing selection of treatment regimens for different patients. Pathological staging according to tumor burden (T), presence of cancer cells in lymph nodes (N) and evidence for metastasis (M) is a well-known standard for prognosis[5], and along with other clinicopathological characteristics is used for selection of patients for adjuvant chemotherapy. However, the response to chemotherapy varies because of heterogeneity among these patients[6-9].
Genomic profiling of tumors has contributed to our understanding of CRC, facilitating diagnosis, prognosis and selection of treatments, including targeted regimens[10]. Multi-gene prognostic classifiers developed in the past decade on the basis of next-generation sequencing have been used clinically to identify patients at risk of recurrence, or to select targeted therapies[11,12]. Previously, we developed a risk-score system based on 19 genes (TCA19) regulated by activation of triggering receptor expressed on myeloid cells 1 (TREM1) or connective tissue growth factor (CTGF)[8]. The genes were selected by their involvement in tumorigenesis and progression, and in providing benefit from adjuvant chemotherapy for stage III CRC[8]. Because survival outcomes in patients with stage IV CRC are poor, and appropriate selection of targeted therapies is challenging, we attempted in the present study to identify whether TCA19 scores predicted survival outcomes in patients with stage IV CRC undergoing different chemotherapy regimens.
The immune-checkpoint inhibitor, pembrolizumab has been shown to have a greater effect on disease-free survival in CRC with microsatellite instability than mincrosatellite-stable CRC[13]. The effects of pembrolizumab on survival in patients with stage IV CRC also depends on patterns of metastasis[14]. Tumor-assosicated macrophages (TAMs) of the M2 phenotype are present in the stroma of many tumors, and frequently associated with the progression of several types of cancer and CD68 acts as macrophage marker and TAMs marker[15]. TAMs infiltration at the invasive hotspot is associated with improvement in both hepatic metastasis and overall survival in CRC[16]. The function of CD73 in tumor is to facilitate escape from immune surveillance and to orchestrate the tumor-stroma interaction to promote cancer growth and metastasis[15]. High expression of CD73 was poor prognostic factor in CRC[17].
Here, we aim to determine whether the TCA19 system could be used as a prognostic indicator for stage IV CRC. We also attempt to identify possible target or marker genes associated with immune functions among the 19 genes, and examined the biologic behavior of the selected genes in association with immune function.
The study included matched tissues from 60 patients with stage IV CRC that were histologically identified as adenocarcinoma and normal colonic tissue (> 5 cm from the tumor border), as well as blood. Another 10 patients were also enrolled for investigation of candidate gene (Supplementary Figure 1). All patients received surgery at the Asan Medical Center between December 2008 and October 2014, and CRC tissue and blood samples were acquired at the time of surgery under the patient consent for tissue and blood sample donation and examination. The metastatic tissue samples were acquired during the resection of metastatic disease and if the resection was unavailable, tissue biopsy of metastatic disease during work-up or surgery was also accepted. The serum was used for extraction of DNA from the centrifuged blood sample. The study protocol was approved by the Institutional Review Board of Asan Medical Center (registration No. 2015-0581), in accord with the Declaration of Helsinki. Clinical and pathologic data were extracted from the medical records. Patients were staged according to the 8th American Joint Committee on Cancer (AJCC) staging system.
Total RNA was extracted from the matched normal and tumor tissues of the patients using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. The cDNA was synthesized from total RNA by amplification using random primers and SuperScript II RT (Invitrogen). Quantitative real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed on a LightCycler 96 using the SYBR Green I Master Mix (Roche, Mannheim, Germany), in a total volume of 20 μL with the following amplification steps: initial denaturation at 95 °C for 10 min, which was followed by 45 cycles of amplification (95 °C for 10 s, Tm for 10 s, and 72 °C for 20 s). Gene expression determined by RT-qPCR was normalized to human glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primers for TCA19 genes are listed in Supplementary Table 1. Relative levels of gene expression were determined using the ΔΔCt method in which ΔCt values between one of the TCA19 genes and the GAPDH control [ΔCt = (Ct)TCA19 genes - (Ct)GAPDH][18]. ΔΔCt was defined as a difference in the ΔCt values between a normal tissue and a tumor tissue of the same patient [ΔΔCt = (ΔCt)normal - (ΔCt)tumor] as previously reported.18 A 2-ΔΔCt value over 1-fold indicates upregulation of the tested TCA19 gene in a tumor tissue compared with a normal counterpart of the same patient.
| Variables (No. of patients) | Percentage of tumors with a log22-ΔΔCT value > 1 compared with matched normal tissue (P value) | |||||||||||||||||||
| GADD45B1 | S1PR3 | CDKN2B | EGR2 | CTGF | SERPINE1 | RGS16 | RHOU | TIMP1 | PHLDA1 | IL36RN | SLAMF7 | E2F7 | DTL | CFB | CDK1 | CXCL1 | CXCL3 | CKS2 | TREM1 | |
| Mean ± SEM of 2ΔΔCT | 0.78 ± 0.74 | 1.45 ± 4.23 | 1.64 ± 6.03 | 3.24 ± 10.88 | 1.23 ± 1.66 | 8.19 ± 22.49 | 10.39 ± 35.49 | 1.01 ± 1.59 | 2.90 ± 2.79 | 5.49 ± 14.95 | 4.40 ± 15.22 | 1.17 ± 5.10 | 9.84 ± 34.23 | 2.46 ± 4.68 | 3.29 ± 5.62 | 2.94 ± 6.51 | 22.76 ± 75.79 | 30.55 ± 76.03 | 2.65 ± 4.14 | 14.14 ± 71.65 |
| Sex | 25.9/24.2 | 25.9/27.3 | 22.2/15.2 | 33.3/39.4 | 37.0/36.4 | 74.1/81.8 | 81.5/69.7 | 33.3/24.2 | 74.1/75.8 | 63.0/66.7 | 33.3/42.4 | 3.7/18.2 | 85.2/84.8 | 44.4/60.6 | 70.4/69.7 | 51.9/60.6 | 92.6/78.8 | 96.3/84.8 | 55.6/75.8 | 29.6/42.4 |
| Female/male (27/33) | 0.56 | 0.57 | 0.35 | 0.42 | 0.58 | 0.34 | 0.23 | 0.31 | 0.56 | 0.49 | 0.33 | 0.89 | 0.63 | 0.16 | 0.59 | 0.34 | 0.13 | 0.15 | 0.08 | 0.23 |
| Age | 23.2/50.0 | 26.8/25.0 | 17.9/25.0 | 37.5/25.0 | 35.7/50.0 | 76.8/100 | 75.0/75.0 | 28.6/25.0 | 73.2/100 | 64.3/75.0 | 39.3/25.0 | 10.7/25.0 | 83.9/100 | 55.4/25.0 | 71.4/50.0 | 58.9/25.0 | 83.9/100 | 89.3/100 | 66.1/75.0 | 35.7/50.0 |
| ≤ 75 vs > 75 (56/4) | 0.26 | 0.71 | 0.57 | 0.53 | 0.47 | 0.37 | 0.69 | 0.68 | 0.31 | 0.56 | 0.5 | 0.4 | 0.51 | 0.26 | 0.35 | 0.21 | 0.51 | 0.65 | 0.59 | 0.47 |
| Comorbidity | 20.0/32.0 | 31.4/20.0 | 20.0/16.0 | 42.9/28.0 | 37.1/36.0 | 74.3/84.0 | 80.0/.68.0 | 34.3/20.0 | 68.6/84.0 | 62.9/68.0 | 45.7/28.0 | 8.6/16.0 | 80.0/92.0 | 60.0/44.0 | 71.4/68.0 | 68.6/40.0 | 82.9/88.0 | 88.6/92.0 | 68.6/64.0 | 37.1/36.0 |
| No/Yes (35/25) | 0.22 | 0.25 | 0.48 | 0.18 | 0.57 | 0.28 | 0.22 | 0.18 | 0.14 | 0.45 | 0.13 | 0.31 | 0.18 | 0.17 | 0.5 | 0.03 | 0.43 | 0.51 | 0.46 | 0.57 |
| Family history | 24.0/30.0 | 28.0/20.0 | 18.0/20.0 | 38.0/30.0 | 36.0/40.0 | 78.0/80.0 | 72.0/90.0 | 28.0/30.0 | 74.0/80.0 | 64.0/70.0 | 42.0/20.0 | 12.0/10.0 | 88.0/70.0 | 56.0/40.0 | 70.0/70.0 | 60.0/40.0 | 88.0/70.0 | 94.0/70.0 | 72.0/40.0 | 36.0/40.0 |
| No/Yes (50/10) | 0.48 | 0.46 | 0.59 | 0.46 | 0.54 | 0.63 | 0.22 | 0.59 | 0.52 | 0.51 | 0.17 | 0.67 | 0.16 | 0.28 | 0.63 | 0.21 | 0.16 | 0.05 | 0.06 | 0.54 |
| Synchronous Lm | 33.3/21.4 | 33.3/23.8 | 22.2/16.7 | 33.3/38.1 | 33.3/38.1 | 72.2/81.0 | 83.3/71.4 | 44.4/21.4 | 77.8/73.8 | 66.7/64.3 | 44.4/35.7 | 22.2/7.1 | 72.2/90.5 | 55.6/52.4 | 83.3/64.3 | 72.2/50.0 | 100/78.6 | 100/85.7 | 83.3/59.5 | 38.9/35.7 |
| No/Yes (18/42) | 0.25 | 0.32 | 0.43 | 0.48 | 0.48 | 0.33 | 0.26 | 0.07 | 0.51 | 0.55 | 0.36 | 0.11 | 0.08 | 0.52 | 0.12 | 0.09 | 0.03 | 0.1 | 0.06 | 0.52 |
| Synchronous Pm | 28.9/13.3 | 26.7/26.7 | 17.8/20.0 | 37.8/33.3 | 35.6/40.0 | 77.8/80.0 | 77.8/66.7 | 31.1/20.0 | 77.8/66.7 | 66.7/60.0 | 40.0/33.3 | 13.3/6.7 | 86.7/80.0 | 57.8/40.0 | 71.1/66.7 | 62.2/40.0 | 88.9/73.3 | 91.1/86.7 | 66.7/66.7 | 40.0/26.7 |
| No/Yes (45/15) | 0.2 | 0.64 | 0.56 | 0.51 | 0.49 | 0.58 | 0.3 | 0.32 | 0.3 | 0.43 | 0.44 | 0.43 | 0.4 | 0.18 | 0.49 | 0.11 | 0.15 | 0.47 | 0.62 | 0.27 |
| AJCC stage | 0/25.9 | 0/27.6 | 0/11 | 0/37.9 | 50/36.2 | 50/79.3 | 100/74.1 | 50/27.6 | 0/77.6 | 50/65.5 | 50/37.9 | 0/12.1 | 100/84.5 | 100/51.7 | 100/69.0 | 100/55.2 | 100/84.5 | 100/89.7 | 100/65.5 | 0/37.9 |
| III/IV (2/58) | 0.56 | 0.53 | 0.66 | 0.4 | 0.6 | 0.39 | 0.56 | 0.49 | 0.06 | 0.58 | 0.62 | 0.78 | 0.72 | 0.28 | 0.49 | 0.32 | 0.72 | 0.81 | 0.44 | 0.4 |
| LVI | 10.0/32.5 | 20.0/30.0 | 15.0/20.0 | 25.0/42.5 | 25.0/42.5 | 65.0/85.0 | 65.0/80.0 | 30.0/27.5 | 75.0/75.0 | 45.0/75.0 | 25.0/45.0 | 0/17.5 | 70.0/92.5 | 35.0/62.5 | 60.0/75.0 | 45.0/62.5 | 75.0/90.0 | 85.0/92.5 | 55.0/72.5 | 30.0/40.0 |
| No/Yes (20/40) | 0.05 | 0.31 | 0.47 | 0.15 | 0.15 | 0.08 | 0.17 | 0.53 | 0.63 | 0.02 | 0.11 | 0.05 | 0.03 | 0.04 | 0.18 | 0.16 | 0.13 | 0.31 | 0.14 | 0.32 |
| PNI | 12.0/34.3 | 32.0/22.9 | 16.0/20.0 | 40.0/34.3 | 32.0/40.0 | 76.0/80.0 | 60.0/85.7 | 36.0/22.9 | 72.0/77.1 | 56.0/71.4 | 40.0/37.1 | 8.0/14.3 | 76.0/91.4 | 52.0/54.3 | 68.0/71.4 | 56.0/57.1 | 76.0/91.4 | 84.0/94.3 | 64.0/68.6 | 24.0/45.7 |
| No/Yes (25/35) | 0.04 | 0.31 | 0.48 | 0.43 | 0.36 | 0.47 | 0.02 | 0.2 | 0.44 | 0.17 | 0.52 | 0.37 | 0.1 | 0.53 | 0.5 | 0.57 | 0.1 | 0.19 | 0.46 | 0.07 |
| CRM involvement | 22.4/36.4 | 26.5/27.3 | 18.4/18.2 | 34.7/45.5 | 38.8/27.3 | 79.6/72.7 | 73.5/81.8 | 30.6/18.2 | 73.5/81.8 | 63.3/72.7 | 36.7/45.5 | 6.1/36.4 | 83.7/90.9 | 51.0/63.6 | 69.4/72.7 | 53.1/72.7 | 85.7/81.8 | 89.8/90.9 | 61.2/90.9 | 30.6/63.6 |
| No/Yes (49/11) | 0.27 | 0.61 | 0.68 | 0.37 | 0.36 | 0.44 | 0.44 | 0.34 | 0.44 | 0.41 | 0.42 | 0.02 | 0.47 | 0.34 | 0.57 | 0.2 | 0.52 | 0.7 | 0.05 | 0.04 |
A risk score was developed via a previously reported strategy using the Cox regression coefficient for the TCA19 genes[8,19,20]. The risk score for each patient was calculated as the sum of each gene’s score, which was derived by multiplying the expression level of a gene by its corresponding coefficient using previously reported values (Supplementary Table 2) [8]. The patients were then divided into two groups, for high or low expression of TCA19 genes using the median of the risk score as the threshold.
| P value | Hazard ratio | CI lower | CI upper | |
| N-category | 0.02 | 0.569 | 0.349 | 0.927 |
| LVI | 0.004 | 2.559 | 1.347 | 4.862 |
| PNI | 0.538 | 0.837 | 0.476 | 1.473 |
| CRM | 0.004 | 2.931 | 1.398 | 6.143 |
| Bormann type | 0.3 | 0.731 | 0.402 | 1.327 |
| SLAMF7 | 0.003 | 0.206 | 0.072 | 0.589 |
| CKS2 | 0.018 | 0.484 | 0.266 | 0.883 |
| TREM1 | 0.017 | 0.471 | 0.254 | 0.875 |
Normal tissue, and primary and metastatic tumor tissues of 10 patients with metastatic CRC (separate from the initial study cohort of 60 patients), were used for immunohistochemistry (IHC) studies of the expression of signaling lymphocytic activation molecule family, member 7 (SLAMF7), TREM1, CD73 and CD68. Formalin fixed, paraffin-embedded tissue sections were immunohistochemically stained for protein expression of SLAMF7, TREM1, CD68, and CD73 with a BenchMark XT automatic immunostaining device (Ventana Medical Systems, Tucson, AZ, United States) with an OptiView DAB IHC Detection Kit (Ventana Medical Systems) according to the manufacturer's instructions.
Proteins were extracted from cultured cells using lysis buffer (Cells Signaling Technology, Danvers, MA, United States). Equal amounts of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Milipore, Billerica, MA, United States). The membranes were blocked with 5% nonfat milk diluted in Tris-buffer saline containing of Tween-20 (TBST) for 1h at room temperature before the addition of the appropriate primary antibody. The membranes were then washed with TBST and incubated with the appropriate HRP-conjugated secondary antibody (1:10000; Abcam, Cambridge, United Kingdom) for 1h at room temperature. Protein-antibody complexes were visualized using a chemiluminescence reagent (New England Nuclear, Boston, MA, United States). Antibodies used in IHC and western blotting were listed in Supplementary Table 3.
The 10 CRC cell lines (DLD-1, HCT116, HCT15, HT29, LoVo, LS174T, RKO, SW480, SW620 and WIDR) and two normal colonic cell lines (CCD-18Co and CCD841) were purchased from the American Type Tissue Culture Collection (ATCC, Manassas, VA, United States) and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). CRC cell lines (DLD-1, RKO, HCT116 and HT29) with minimal protein expression for SLAMF7 and TREM1 were selected for gene transfection (Supplementary Figure 2). SLAMF7 and TREM1 cDNA tagged with the peptide epitope Myc-DDK were purchased from OriGene (Rockville, MD, United States). Transient transfection was performed to establish each cell mixture using Lipofectamine 2000 (Invitrogen). Stable clones were selected by culturing with aminoglycoside antibiotic G418 for 10 d, and at least two different clones were generated for each cell line. Human monocytic THP-1 cells were purchased from ATCC and maintained in RPMI-1640 medium supplemented with 10% FBS. M2-polarized THP-1 cells were generated by treatment 50 ng/mL phorbol myristate acetate (PMA, Sigma-Aldrich, St. Louis, MO, United States) for 24 h followed by incubation with 25 ng/mL interleukin (IL)-4 and 25 ng/mL IL-13 for 18 h. For lipopolysaccharide (LPS)-mediated THP-1 activation, cells were plated at a concentration of 1 × 104 cells/ well in the presence of 1 μg/mL LPS for 18 h.
DLD-1, RKO, HCT116 and HT29 cells were co-cultured with THP-1 cells using Transwell inserts (Becton Dickinson, Franklin Lakes, NJ, United States) with a 0.4 μm porous membrane to create separate upper and lower chambers. CRC cells were cultured in the lower chamber at 1 × 104 cells/mL, and THP-1 cells were cultured in the upper chamber. CRC cells and THP-1 cells were collected 5 d after co-culture (Supplementary Figure 3).
Adherent CRC cells collected after 5 d of co-culture were seeded on 96-well plates coated with collagen type I (Greiner Bio-One #655956, Frickenhansen, Germany). Cells were fixed for 30 min with 4% paraformaldehyde in ice-cold phosphate-buffered saline (PBS), quenched for 5 min with 50 mmol/L NH4Cl, and incubated overnight at 4 °C with primary antibody diluted 1:100 in 5% normal goat serum (NGS) in PBS. Cells were washed three times with PBS and incubated for 1 h with secondary antibody at 1:500 dilution. Primary and secondary antibodies used in immunofluorescence staining (IFS) were listed in Supplementary Table 3. Cells were then washed three times with PBS, followed by nuclear counterstaining with 2 μg/mL DAPI for 1 min.
Stained 96-well plates were imaged with a high-content wide-field fluorescence imaging system coupled to Harmony software version 3.5 (Operetta: PerkinElmer, Waltham, MA, United States). Wells were imaged with a × 40 objective lens in a single focal plane across each plate. Twelve fields of views (each 510 μm × 675 μm) were imaged per well, with an identical pattern of fields used in every well.
Differential expression of individual TCA19 genes was compared with levels of clinico-pathological variables by the χ-square test, and the unpaired Student’s t-test or Mann-Whitney U-test, as appropriate. The Kaplan-Meier method was used to calculate the survival outcomes, and the difference in survival between two groups was assessed using log-rank tests. The prognostic association between the signature and potential risk factors was assessed using multivariate Cox proportional hazard regression models. Statistical significance was expressed as P < 0.05, and the analyses were performed using SPSS software version 21 (IBM Corporation, Armonk, NY, United States).
Of the study cohort, 33 were male and 27 were female, with a median age of 61 years (interquartile range 52-69 years). The cancer was located in the right colon (cecum-splenic flexure of transverse colon) in 16 patients, the left colon (splenic flexure of transverse colon-distal sigmoid colon) in 28 patients, and in the rectum in 16 patients. Curative surgery (R0 resection) was performed in 14 patients. All patients received chemotherapy after the surgery consisting of single-agent treatment with 5-fluorouracil (5-FU) or capecitabine (n = 9), a combination of 5-FU and oxaliplatin (n = 22), 5-FU and irinotecan (n = 13), or targeted agents (n = 16), whether the choice of all chemotherapy was according to the oncologist’s opinion. Nine patients received postoperative concurrent chemoradiotherapy (50.4 Gy in 28 fractions with concurrent 5-FU). The median follow-up period was 16 mo with an interquartile range (IQR) of 9-24 mo (Supplementary Table 4).
The study cohort was dichotomized on the basis of TCA19 risk scores. Progression-free survival (PFS) was not significantly different in the high (8 mo, IQR 6-10 mo) and low (9 months, IQR 8-10 mo) TCA19-score groups (P = 0.42). Among 30 patients with high TCA19 scores, the PFS rates were significantly different (P = 0.041) between four patients who received 5-FU regimen and eight patients who received the targeted regimens (bevacizumab or cetuximab). PFS rates among patients with low TCA19 scores did not differ significantly according to the chemotherapy regimens (P = 0.61) (Figure 1).
For each clinicopathological variable, the proportion of patients in each different condition with qPCR log22−ΔΔCt values greater than one (indicating a greater than one-fold increase in gene expression in the tumor relative to normal tissue) were compared (Table 1). For lymphovascular invasion (LVI), the proportions of patients with relative expression (log22−ΔΔCt greater than one) were significantly higher in the presence of invasion than in its absence for PHLDA1, E2F7, and DTL, and SLAMF7 (P = 0.02, 0.05, 0.03, 0.04, respectively). Similarly, for perineural invasion, the proportion of patients with relative expression was significantly higher in the presence of invasion than in its absence for RGS16 (P = 0.02). A lower proportion of patients had CXCL1 log22−ΔΔCt values greater than one in the presence of synchronous hepatic metastasis than its absence (P = 0.03). The proportion of patients with log22−ΔΔCt values greater than one was significantly higher in the presence than in the absence of involvement of circumferential resection margin (CRM) for SLAMF7, CKS2 and TREM1 (P = 0.02, 0.05, 0.04, respectively). These associations, as well as associations with the presence of comorbidity, family history, tumor location and the pattern of carcinoembryonic antigen, are shown in Table 1.
PFS was positively associated with relatively higher expression in tumor of SLAMF7, CXCL1, and CXCL3 and negatively associated with relatively higher expression of TREM1 (Supplementary Figure 4). Additionally, a multivariate Cox proportonal hazard regression model was used to analyze the associations of PFS with expression of individual TCA19 genes and with clinicopathological variables. This analysis showed that LVI, CRM involvement, and expression of SLAMF7, CKS2 and TREM1 were significantly associated with PFS (Table 2).
The association of expression of SLAMF7 and TREM1 with clinicopathological variables and PFS, as well as their immune-related gene functions, suggested that they were possible candidate genes. The relative mRNA expression of the SLAMF7 and TREM1 genes was assessed in the 60 CRC patients of the primary study group. In 53 patients, SLAMF7 expression was lower in tumor tissue than in normal tissue, giving a mean 2−ΔΔCt value of 0.2, compared with a mean 2−ΔΔCt value of 1.17 in 7 patients. In 22 patients, TREM1 expression was higher in tumor tissue than in normal tissue, giving a mean 2−ΔΔCt value of 37.95, compared with a mean 2−ΔΔCt value of 14.14 in 38 patients.
Additional validation for the pattern of mRNA expression of SLAMF7 and TREM1 was obtained through RT-qPCR, western blotting and IHC of normal and tumor tissue samples of 10 additional patients with stage IV CRC. The results showed similar pattern to those with the initial 60-patient cohort, with lower expression of SLAMF7 and higher expression of TREM1 mRNA and protein in tumor tissue than normal tissue (Supplementary Figure 5). In additional analysis of 27 patients (separate from the 60 study cohort and the 10 validation set), SLAMF7 expression in 7 patients was lower in tumor tissue than in normal tissue (mean 2−ΔΔCt value: 20.21) and TREM1 expression in 18 was higher in tumor tissue than in normal tissue (mean 2−ΔΔCt value: 37.21) (Supplementary Table 5).
The results of western blotting were consistent with those of the mRNA expression study, with lower expression of SLAF7 and higher expression of TREM1 in primary or metastatic tumor tissues than in normal colonic tissue (Supplementary Figure 6). However, in IHC, both SLAMF7 and TREM1 were expressed at significantly higher levels in primary or metastatic tumor tissues than in normal colonic tissue (Supplementary Table 6).
SLAMF7 and TREM1 have functions in immune responses, so their relationships with CD68 and CD73 were assessed by high-throughput imaging. Baseline IHC for these proteins was performed with tissues from the validation cohort of 10 patients with stage IV CRC, and showed TREM1 staining in plasma membrane, extracellular regions and intracellular regions, and SLAMF7 staining in plasma membranes and integral components of membranes (Supplementary Figure 7). CD68 was expressed in stroma (especially membranes), and CD73 was expressed in tumor stroma (Supplementary Figure 8).
CRC cells lines over-expressing SLAMF7 or TREM1 were co-cultured with THP-1 cells, and protein expression in the CRC cells before and after 5 d of co-culturing was compared by high-throughput immunofluorescence imaging. Specifically, expression of CD68 or CD73 was determined in regions expressing SLAMF7 or TREM1. At day 5, co-expression of both CD68 and CD73 with SLAMF7 was significantly lower than at day 0 (both P = 0.029) (Figures 2 and 3). Co-expression of CD68 or CD73 with TREM1 was not significantly different at day 5 from that at day 0 (Supplementary Figure 9).
Our results indicated that TCA19 might be useful for prediction of sensitivity of patients with stage IV CRC to targeted chemotherapy, as demonstrated previously for stage III CRC[8]. Specifically, a high score with the TCA19 classifier suggests that a patient is a possible responder to a targeted regimen. In our cohort of 60 patients with stage IV CRC, a high TCA19 score was associated with a 4-mo survival benefit for targeted regimens compared with the 5-FU regimen. Few efficient biomarkers have been discovered for prediction of responses to targeted regimens (existing marker include the products of KRAS, NRAS, BRAF and PIK3CA genes), although attempts to identify such markers have used laboratory and clinical approaches[11,21-24]. A high TCA19 score might now be considered as a biomarker of the response to targeted chemotherapy in metastatic CRC.
In terms of survival outcomes, our results with multivariate analysis showed that expression levels of SLAM7, TREM1, and CKS2 were independent risk factors of PFS. Expression of these genes was also significantly related to lympho-vascular invasion, perineural invasion, and involvement of circumferential resection margin. In our validation cohort of 10 additional patients, mRNA and protein levels were significantly different from primary tumor and metastatic tumor tissues, compared with normal tissues in SLAMF7 and TREM1. These results might suggest that expression of SLAMF7 and TREM1 was related to progression and metastasis of CRC. A previous report showed that inflammation was a critical component of tumor progression[25] and in this specific view, both SLAMF7 and TREM1 might be associated with the link between inflammation and cancer.
SLAM-family receptors are expressed on hematopoietic cells, and SLAMF7 has an inhibitory role in human monocytes to control pro-inflammatory immune responses[26,27]. A relationship between SLAMF7 and multiple myeloma has previously been demonstrated[28,29], but no report have been published describing correlation between SLAMF7 and CRC. Here, we determined that SLAMF7 was under-expressed in CRC tissue compared with normal tissue, and that SLAMF7 might have an inhibitory role in expression of CD68 and CD73. Immune responses in lymphocytes and mast cells are associated with pathological responses to chemotherapy, and with PFS in metastatic CRC[30]. Although further studies including mechanistic study to investigate the relationships between immune-check point and SLAMF7 or between MHC class and SLAMF7 or clinical studies to be use elotuzumab (SLAMF7-directed immunostimulatory antibody)[29] in CRC are required, SLAMF7 might have potential as an immunotherapeutic target or as a marker for metastatic CRC.
TREM1 enhances degranulation and secretion of pro-inflammatory mediators, and is a potent amplifier of pro-inflammatory innate immune responses[31]. Here, we showed that TREM1 had, overall, higher level of expression in tumor tissue than in normal tissue in RT-qPCR and IHC stain, and TREM1 gave a tendency of enhanced expression of CD68 and CD73. In results derived from experimental colitis and tissue from inflammatory bowel disease, TREM1 inhibition was shown to attenuate inflammation and tumor growth with the colon[32]. High TREM1 expression in tumors is associated with an abundance of neutrophils and high expression of several innate pro-inflammatory genes that might be associated with tumorigenesis in CRC[33]. Our results did not demonstrate a significant difference between day 0 and day 5 in terms of expression of CD68 and CD73 in TREM1-expressing CRC cells, possibly because expression of TREM1 was generally absent in the macrophages of normal colon mucosa, whereas TREM1-expressing macrophages showed significant up-regulation in the diseased colon tissue[34].
To improve the survival rate in metastatic CRC, immunotherapy has been proposed as a treatment option in CRC with microsatellite instability[35], which is associated with immune responses, including up-regulation of immune checkpoint inhibitory molecules such as PD-1, PD-L1 and CTLA4[36,37]. Patients with CRC with microsatellite instability have more mutations and are more responsive to immunotherapy with PD-L1/PD-1 blockade than patients with microsatellite-stable CRC[13,35]. As the number of the patients with CRC with microsatellite instability is limited and therapeutic targeting of microsatellite-stable CRC is difficult, additional immune checkpoints and immunomodulatory molecules still need to be investigated to provide adequate therapeutic coverage for all patients with CRC.
Our study had some limitations, including the small number of patients with the limited use of targeted regimens to assess a clinical implication of the TCA19 risk score and to validate SLAMF7 and TREM1 as suitable candidate. Also, our investigation was limited to assessment of relationship between SLAMF7/TREM1 and CD68/CD73. Further evaluation of other pro-inflammatory mediators with a large study cohort is required and ongoing, to identify any relationship between the expression of SLAMF7/TREM1 and immune-system regulation, and to find possible roles for the targeting of SLAMF7 and TREM1 in new treatment regimens for CRC.
In conclusion, TCA19 may provide prognostic information in patients with metastatic CRC, helping to identify those who will respond to targeted chemotherapy. Expression of SLAMF7 is down-regulated (whereas TREM1 is up-regulated) in primary or metastatic CRC tumors compared with normal colon tissue. SLAMF7 might have an inhibitory role in the immune response in CRC, whereas TREM1, if anything, has a tendency to enhance the immune response. Further mechanistic and functional studies with large cohorts are now required to confirm these relationships.
Our team previously developed a risk score system based a 19 gene-based scoring system (TCA19), worked as a prognostic factor in stage II-III colorectal cancer (CRC). Stage IV CRC is still challenging in the treatment including target-regimen and immunotherapy.
It is needed to identify whether the TCA19 scores predict survival outcomes in patients with stage IV CRC undergoing different chemotherapy regimens including target-regimen and 19 genes are related to immuno-oncology.
The current study aims to determine whether the TCA19 system can be used as a prognostic indicator for stage IV CRC and to identify possible target or marker genes associated with immune functions from 19 genes.
A retrospective review of the medical records of 60 patients with stage IV CRC was conducted, assessing clinico-pathologic variables, and progression-free survival (PFS). TCA19 gene expressions were determined by real-time quantitative polymerase chain reaction (RT-qPCR) in matched normal, primary tumor, and metastatic tumor tissues taken from the 60 study cohort. After selection of genes, related to immuno-oncology, expression of potential target or marker genes were examined by RT-qPCR, immunohistochemistry, western blot, and immunofluorescence staining using tissues from 10 validate set and in CRC cell lines co-cultured with monocytes in vitro.
In the patients with higher TCA19 score, the PFS rates of the patients with target-regimen were significantly higher than the patients with 5-fluorouracil-based regimen. In multivariable analysis, expression of signaling lymphocytic activation molecule family, member 7 (SLAMF7) and triggering receptor expressed on myeloid cells 1 (TREM1) was associated with PFS. From the results of the 10 validate set, down-regulation of SLAMF7 and up-regulation of TREM1 were observed in primary tumor and metastatic tumor tissues compared with normal tissue. In CRC cells expressing SLAMF7 that co-cultured with a monocytic cell line, levels of CD68 and CD73 in IFS imaging were significantly lower at day 5 of co-culture than at day 0. This result suggests that SLAMF7 may have an inhibitory role in the immune response.
TCA19 system may be used as a prognostic indicator for stage IV CRC in terms of use of target-regimen. SLAMF7 and TREM1 may be related to tumorigenesis and progression according to down-regulation of SLAMF7 and up-regulation of TREM1 in tumor tissue.
The current study found an inhibitory role of the SLAMF7 in the immune response. Recently, it is known that the patients with microsatellite-high CRC may respond the immune therapy. In this concept, the direction of the future research is the role of SLAMF7 in the patients with stage IV CRC in terms of the immune therapy.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: South Korea
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Li Y, Maffei F, Wang DR S- Editor: Ma RY L- Editor: A E- Editor: Huang Y
| 1. | Jung KW, Won YJ, Oh CM, Kong HJ, Lee DH, Lee KH. Prediction of Cancer Incidence and Mortality in Korea, 2017. Cancer Res Treat. 2017;49:306-312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 2. | Chang KH, Smith MJ, McAnena OJ, Aprjanto AS, Dowdall JF. Increased use of multidisciplinary treatment modalities adds little to the outcome of rectal cancer treated by optimal total mesorectal excision. Int J Colorectal Dis. 2012;27:1275-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Gennari L, Russo A, Rossetti C. Colorectal cancer: what has changed in diagnosis and treatment over the last 50 years? Tumori. 2007;93:235-241. [PubMed] |
| 4. | Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, Stiller C, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC, Coleman MP; CONCORD Working Group. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1738] [Cited by in RCA: 1735] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 5. | Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2341] [Cited by in RCA: 4397] [Article Influence: 549.6] [Reference Citation Analysis (4)] |
| 6. | Sung SY, Jang HS, Kim SH, Jeong JU, Jeong S, Song JH, Chung MJ, Cho HM, Kim HJ, Kim JG, Lee IK, Lee JH. Oncologic Outcome and Morbidity in the Elderly Rectal Cancer Patients After Preoperative Chemoradiotherapy and Total Mesorectal Excision: A Multi-institutional and Case-matched Control Study. Ann Surg. 2019;269:108-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Benson AB 3rd. Venook AP, Cederquist L, Chan E, Chen YJ, Cooper HS, Deming D, Engstrom PF, Enzinger PC, Fichera A, Grem JL, Grothey A, Hochster HS, Hoffe S, Hunt S, Kamel A, Kirilcuk N, Krishnamurthi S, Messersmith WA, Mulcahy MF, Murphy JD, Nurkin S, Saltz L, Sharma S, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Wu CS, Gregory KM, Freedman-Cass D. Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2017;15:370-398. [PubMed] |
| 8. | Kim SK, Kim SY, Kim JH, Roh SA, Cho DH, Kim YS, Kim JC. A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol Oncol. 2014;8:1653-1666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 9. | Provenzale D, Gupta S, Ahnen DJ, Bray T, Cannon JA, Cooper G, David DS, Early DS, Erwin D, Ford JM, Giardiello FM, Grady W, Halverson AL, Hamilton SR, Hampel H, Ismail MK, Klapman JB, Larson DW, Lazenby AJ, Lynch PM, Mayer RJ, Ness RM, Regenbogen SE, Samadder NJ, Shike M, Steinbach G, Weinberg D, Dwyer M, Darlow S. Genetic/Familial High-Risk Assessment: Colorectal Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2016;14:1010-1030. [PubMed] |
| 10. | Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, Ligon AH, Sholl LM, Kuo FC, MacConaill LE, Lindeman NI, Dong F. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med. 2017;141:751-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 405] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 11. | De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1491] [Cited by in RCA: 1656] [Article Influence: 110.4] [Reference Citation Analysis (1)] |
| 12. | Espenschied CR, LaDuca H, Li S, McFarland R, Gau CL, Hampel H. Multigene Panel Testing Provides a New Perspective on Lynch Syndrome. J Clin Oncol. 2017;35:2568-2575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 13. | Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-2520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6096] [Cited by in RCA: 7233] [Article Influence: 723.3] [Reference Citation Analysis (0)] |
| 14. | Fujiyoshi K, Yamamoto G, Takenoya T, Takahashi A, Arai Y, Yamada M, Kakuta M, Yamaguchi K, Akagi Y, Nishimura Y, Sakamoto H, Akagi K. Metastatic Pattern of Stage IV Colorectal Cancer with High-Frequency Microsatellite Instability as a Prognostic Factor. Anticancer Res. 2017;37:239-247. [PubMed] |
| 15. | Pancione M, Giordano G, Remo A, Febbraro A, Sabatino L, Manfrin E, Ceccarelli M, Colantuoni V. Immune escape mechanisms in colorectal cancer pathogenesis and liver metastasis. J Immunol Res. 2014;2014:686879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 16. | Zhou Q, Peng RQ, Wu XJ, Xia Q, Hou JH, Ding Y, Zhou QM, Zhang X, Pang ZZ, Wan DS, Zeng YX, Zhang XS. The density of macrophages in the invasive front is inversely correlated to liver metastasis in colon cancer. J Transl Med. 2010;8:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 167] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 17. | Wu XR, He XS, Chen YF, Yuan RX, Zeng Y, Lian L, Zou YF, Lan N, Wu XJ, Lan P. High expression of CD73 as a poor prognostic biomarker in human colorectal cancer. J Surg Oncol. 2012;106:130-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 18. | Kim Y, Roh S, Park JY, Kim Y, Cho DH, Kim JC. Differential expression of the LOX family genes in human colorectal adenocarcinomas. Oncol Rep. 2009;22:799-804. [PubMed] |
| 19. | Kim YS, Kim SH, Kang JG, Ko JH. Expression level and glycan dynamics determine the net effects of TIMP-1 on cancer progression. BMB Rep. 2012;45:623-628. [PubMed] |
| 20. | Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817-2826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4524] [Cited by in RCA: 4435] [Article Influence: 211.2] [Reference Citation Analysis (0)] |
| 21. | Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, Krishnamurthi SS, Burris HA, Poplin EA, Hidalgo M, Baselga J, Clark EA, Mauro DJ. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230-3237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 897] [Cited by in RCA: 893] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 22. | Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6:519-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 349] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 23. | To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J Gastroenterol. 2018;24:2949-2973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 116] [Cited by in RCA: 151] [Article Influence: 21.6] [Reference Citation Analysis (2)] |
| 24. | Berger MD, Stintzing S, Heinemann V, Yang D, Cao S, Sunakawa Y, Ning Y, Matsusaka S, Okazaki S, Miyamoto Y, Suenaga M, Schirripa M, Soni S, Zhang W, Falcone A, Loupakis F, Lenz HJ. Impact of genetic variations in the MAPK signaling pathway on outcome in metastatic colorectal cancer patients treated with first-line FOLFIRI and bevacizumab: data from FIRE-3 and TRIBE trials. Ann Oncol. 2017;28:2780-2785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 25. | Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10123] [Cited by in RCA: 11272] [Article Influence: 490.1] [Reference Citation Analysis (2)] |
| 26. | Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol. 2006;6:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 172] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 27. | Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 390] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 28. | Tai YT, Dillon M, Song W, Leiba M, Li XF, Burger P, Lee AI, Podar K, Hideshima T, Rice AG, van Abbema A, Jesaitis L, Caras I, Law D, Weller E, Xie W, Richardson P, Munshi NC, Mathiot C, Avet-Loiseau H, Afar DE, Anderson KC. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood. 2008;112:1329-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 398] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 29. | Lonial S, Kaufman J, Laubach J, Richardson P. Elotuzumab: a novel anti-CS1 monoclonal antibody for the treatment of multiple myeloma. Expert Opin Biol Ther. 2013;13:1731-1740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Tanis E, Julié C, Emile JF, Mauer M, Nordlinger B, Aust D, Roth A, Lutz MP, Gruenberger T, Wrba F, Sorbye H, Bechstein W, Schlag P, Fisseler A, Ruers T. Prognostic impact of immune response in resectable colorectal liver metastases treated by surgery alone or surgery with perioperative FOLFOX in the randomised EORTC study 40983. Eur J Cancer. 2015;51:2708-2717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 31. | Tessarz AS, Cerwenka A. The TREM-1/DAP12 pathway. Immunol Lett. 2008;116:111-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 157] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 32. | Zhou J, Chai F, Lu G, Hang G, Chen C, Chen X, Shi J. TREM-1 inhibition attenuates inflammation and tumor within the colon. Int Immunopharmacol. 2013;17:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 33. | Saurer L, Zysset D, Rihs S, Mager L, Gusberti M, Simillion C, Lugli A, Zlobec I, Krebs P, Mueller C. TREM-1 promotes intestinal tumorigenesis. Sci Rep. 2017;7:14870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 34. | Schenk M, Bouchon A, Seibold F, Mueller C. TREM-1--expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest. 2007;117:3097-3106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 294] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 35. | Boland PM, Ma WW. Immunotherapy for Colorectal Cancer. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 36. | Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, Benson AB, Hamilton SR. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 648] [Cited by in RCA: 639] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 37. | Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, Zhang M, Papadopoulos N, Kinzler KW, Vogelstein B, Sears CL, Anders RA, Pardoll DM, Housseau F. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 1161] [Article Influence: 105.5] [Reference Citation Analysis (0)] |