Published online Mar 14, 2019. doi: 10.3748/wjg.v25.i10.1224
Peer-review started: December 11, 2018
First decision: December 28, 2018
Revised: January 27, 2019
Accepted: January 28, 2019
Article in press: January 28, 2019
Published online: March 14, 2019
Processing time: 93 Days and 17.6 Hours
In the present study, we investigated a suppressive role of microRNA-596 (miR-596) in gastric cancer (GC). Moreover, the downregulation of miR-596 in GC cell lines was associated with an increase of miR-596 promoter methylation. We also established that miR-596 controls the expression of peroxiredoxin 1 (PRDX1), which has never been reported before, suggesting that this interaction could play an important role in GC progression.
To study the potential role and possible regulatory mechanism of miR-596 in GC.
The expression levels of miR-596 and PRDX1 in gastric cancer tissues and cell lines were detected by quantitative real-time PCR (qRT-PCR). Western blot and luciferase reporter assay were used to detect the effect of miR-596 on PRDX1 expression. Then, the proliferation, metastasis, and invasion of GC cell lines transfected with miR-596 mimics were analyzed, respectively, by Cell Counting Kit-8 proliferation assay, wound healing assay, and transwell invasion assay. Meanwhile, the methylation status of the promoter CpG islands of miR-596 in GC cell lines was detected by methylation-specific PCR (MSP).
Expression of miR-596 was decreased and PRDX1 was upregulated in GC tissues and cell lines. Overexpression of miR-596 decreased the expression of PRDX1 and luciferase reporter assays detected the direct binding of miR-596 to the 3'-untranslated region (UTR) of PRDX1 transcripts. Furthermore, we found that overexpression of miR-596 remarkably suppressed cell proliferation, migration, and invasion in GC cells. We further analyzed miR-596 promoter methylation by MSP and qRT-PCR, and found the downregulation of miR-596 was associated with promoter methylation status in GC cell lines. Moreover, DNA demethylation and reactivation of miR-596 after treatment with 5-Aza-2’-deoxycytidine inhibited the proliferative ability of GC cells.
MiR-596 has a tumor suppressive role in GC and is downregulated partly due to promoter hypermethylation. Furthermore, PRDX1 is one of the putative target genes of miR-596.
Core tip: In the present study, we investigated a suppressive role of microRNA-596 (miR-596) in gastric cancer (GC). MiR-596 was downregulated in human specimens of GC and GC cell lines, and overexpression of miR-596 significantly reduced GC cell proliferation. Downregulation of miR-596 in GC cell lines was associated with an increase of miR-596 promoter methylation. We also established that miR-596 controls the expression of peroxiredoxin 1, which has never been reported before, suggesting that this interaction could play an important role in GC progression. Overall, this study has an impact in understanding the role of miRNAs in cancer progression.
- Citation: Zhang Z, Dai DQ. MicroRNA-596 acts as a tumor suppressor in gastric cancer and is upregulated by promotor demethylation. World J Gastroenterol 2019; 25(10): 1224-1237
- URL: https://www.wjgnet.com/1007-9327/full/v25/i10/1224.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i10.1224
Gastric cancer (GC) is one of the most common digestive malignancies and remains the second most common cause of cancer-related mortality worldwide[1]. Currently, most patients are diagnosed at an advanced stage, leading to a low cure rate and a low 5-year survival rate. Therefore, it is necessary to further study the molecular mechanism of GC and identify more effective biomarkers for the diagnosis and treatment of GC. A greater understanding of these factors will play an important role in early diagnosis and treatment of GC and improvement of prognosis.
MicroRNAs (miRNAs) are a class of endogenous non-coding RNA with a length of approximately 23 nucleotides, which can bind to complementary sequences in the 3'-untranslated regions (UTRs) of specific mRNAs, resulting in degradation of target mRNAs or inhibition of their translation into functional proteins[2]. Accumulating studies have indicated that miRNAs have been associated with almost all known physiological and pathological processes, including cancer[3,4]. In addition, it is suggested in recent studies that epigenetic modification, especially DNA methylation, is one of many mechanisms of miRNA suppression in human cancer[5,6]. MicroRNA-596 (miR-596) is located on human chromosome 8p23.3 and is an intergenic miRNA gene. In recent years, studies have found that miR-596 is downregulated in a variety of cancers, such as oral cancer[7], endometrial cancer[8], melanoma[9], and bladder cancer[10,11]. Additionally, miR-596 was previously found to be silenced by promoter CpG island hypermethylation in hepatocellular carcinoma[12,13], endometrial cancer[14], and oral cancer[7]. In GC, Song et al[15] reported that miR-596 had low expression and could target CLDN4 to inhibit the invasion of GC cells. However, whether the expression of miR-596 is associated with promoter methylation alteration in GC remains unclear and whether other targets of miR-596 exist in GC remains to be studied.
In this study, we confirmed that the expression of miR-596 was markedly downregulated in GC tissues and cell lines. Upregulation of miR-596 suppressed the proliferation, migration, and invasion of GC cells. The decreased expression of miR-596 was associated with promoter DNA methylation in GC. Moreover, it was indicated that peroxiredoxin 1 (PRDX1) is one of the putative targets of miR-596 in GC. All these results suggest that there is a critical role for miR-596 in the pathogenesis of GC and it may serve as a potential therapeutic target for patients with this disease.
A total of 55 paired surgically resected GC tissues and adjacent normal gastric tissues, which were diagnosed by an independent pathologist, were collected from the Fourth Affiliated Hospital of China Medical University, between 2014 and 2015. The specimens were promptly collected following surgery and all were pathologically confirmed. None of the patients enrolled in this study had previously received preoperative chemotherapy or radiotherapy. Informed consent was obtained from all patients. The Medical Association Ethics Committee of the Fourth Affiliated Hospital of China Medical University approved all aspects of the present study in accordance with the Helsinki Declaration.
One immortalized normal gastric cell line (GES-1) and four human GC cell lines (AGS, SGC-7901, MKN-45, and MGC-803) were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). These cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, United States) containing 10% fetal bovine serum (Invitrogen) in a humidified atmosphere of 5% CO2 at 37 °C.
Total RNA was extracted from tissues or cultured cells using Trizol reagent (Invitrogen). According to the protocol of the Poly(A) Tailing Kit (Tiangen, Beijing, China), poly(A) tails were added to the miRNA. The PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Dalian, China) and gene-specific primers or random primers were used to generate cDNA. Real-time PCR was performed in a Light Cycler 480 II Real-Time PCR system (Roche Diagnostics, Basel, Switzerland) using SYBR® Green (Takara). Glyceraldehyde phosphate dehydrogenase (GAPDH) and U6 snRNA were employed as endogenous controls for mRNA and miRNA, respectively. The comparative Ct method was used to calculate the relative expression of RNA. Primer sequences are displayed in Table 1.
| Name | Sequence (5'-3') |
| Primers used for mRNA detection | |
| miR-596 (forward) | AAGCCTGCCCGGCTCCT |
| miR-596 (reverse) | GCTGTCAACGATACGCTACGT |
| miR-596 (RT) | (GCTGTCAACGATACGCTACGTAACGGCATGACA; GTGTTTTTTTTTTTTTTTTTTTTTTTTC) |
| U6 (forward) | CGCTTCGGCAGCACATATAC |
| U6 (reverse) | TTCACGAATTTGCGTG TCAT |
| U6 (RT) | (GCTGTCAACGATACGCTACGTAACGGCATGACAG; TGTTTTTTTTTTTTTTTTTTTTTTTTG) |
| PRDX1 (forward) | AAGAAACTCAACTGCCAAGTG |
| PRDX1 (reverse) | CAGCCTTTAAGACCCCATAAT |
| GAPDH (forward) | CGGATTTGGTCGTATTGGG |
| GAPDH (reverse) | CTGGAAGATGGTGATGGGATT |
Transfected cells were lysed with lysis buffer (Beyotime, Shanghai, China) containing 1 mmol/L PMSF. The protein concentrations were measured using the bicinchoninic-acid (BCA) protein assay kit (Beyotime). Equal amounts (30 μg) of protein were separated by 15% SDS-PAGE and electro-transferred to PVDF membranes (Millipore), which were then incubated with primary antibodies against PRDX1 (1:500 dilution; Proteintech, Wuhan, China) and β-actin (1:1000 dilution; Proteintech) overnight at 4 °C. After incubation with peroxidase-conjugated affinipure goat anti-rabbit IgG or peroxidase-conjugated affinipure goat anti-mouse IgG (Beyotime), the bands were visualized using an electrochemiluminescence (ECL) detection kit (ThermoBiotech Inc, Rockford, IL, United States). Protein bands were scanned and quantified using densitometric software (Bio-Rad, California, United States).
Transfections were performed using the Lipofectamine3000 Reagent (Invitrogen) according to the manufacturer’s protocol. Final concentrations of 50 nmol/L of miR-596 mimics (5'-AAGCCUGCCCGGCUCCUCGGG-3’)/ miR-NC (5'-UUGUACUA CACAAAAGUACUG-3’) and 0.75 μg/mL plasmids were used for each transfection in a six-well plate with 2 mL culture medium. Total RNA and protein were collected 48 h after transfection.
MiR-596 mimics or miR-NC, reporter construct, or control vector was cotransfected into MGC-803 cells for 48 h. Dual luciferase Reporter GeneMAssay Kit (Tiangen, Beijing, China) was used to detect luciferase activity. Each test was repeated three times.
Cell proliferation was determined with the Cell Counting Kit-8 (CCK-8) kit (Dojindo, Japan); 3000 cells were seeded into 96-well plates for 24 h and then treated with NC, miR-NC, or miR-596 mimics. After 0, 24, 48, and 72 h of treatment, 100 μL cultural supernatant was collected to another 96-well plate and then 10 μL CCK-8 solution was added for incubation at 37 °C for 4 h. The absorbance was measured at 450 nm wavelength using spectrophotometry (BioTek, United States).
Cells were seeded in six-well plates and treated with NC, miR-NC, or miR-596 mimics. Linear scratch wounds were created on cell monolayers with a sterile 200 μL pipette tip, and the scratched areas were photographed at × 100 magnification using a Leica DMI3000B computer-assisted microscope (Leica, Buffalo Grove, United States). Images were captured at 0, 24, 48, and 72 h after the scratch was made. Images were analyzed using Image-Pro Plus v6.0 image analysis software (Media Cybernetics, Rockville, MD, United States).
Transwell plates (BD, Biosciences, United States) were used for GC cell invasion assay. The bottom chamber contained complete medium and the upper chamber has serum-free medium. Matrigel (BD) was added to the RPMI 1640 medium for detecting invading cells. After transfection with NC, miR-NC, or miR-596 mimics, GC cells were appropriately seeded into the cell culture. After incubation at 37 °C for 24 h, the invaded cells were fixed and stained using Giemsa. The images of cells were photographed with a microscope (Leica) at × 100 magnification and the cell number counted in three random fields of view. The results are presented as a column graph with statistics.
Genomic DNA was extracted from the cultured cells by SDS/proteinase K treatment, followed by phenol–chloroform extraction and ethanol precipitation. The bisulfite treatment was performed using the kit EZ DNA Methylation-Gold Kit (Zymo Research, CA, United States) according to the manufacturer’s protocol. The methylated primers were 5’-GAG GTT CGG GAT GTA TCG TT-3’ (forward) and 5’-TAA CTT CCG CAA TAA CCG TAT-3’ (reverse), which result in a 193 bp band; the unmethylated primers were 5’-GTG GAG GTT TGG GAT GTA TTG-3’(forward) and 5’-CTC TTA ACT TCC ACA ATA ACC ATA-3’ (reverse), which result in a 200 bp band. The PCR reaction conditions were as follows: 94 °C for 5 min, 40 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 45 s, and 72 °C for 10 min. Agarose gel electrophoresis and ethidium bromide staining were then performed. Data from the gels were collected with a laser density scanner (Pharmacia LKB Ultroscan) and subsequently analyzed. All experiments were repeated three times, and the mean value was used for statistical analysis.
GC cells were seeded at 5 × 105 cells per well in six-well culture plates and cultured in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum in a humidified atmosphere of 5% CO2 at 37 °C for 24 h. The cells were incubated in culture medium with 0, 2.5, 5, or 10 μmol/L of 5-Aza-dC (Sigma, Shanghai, China) for 3 d, with daily medium replacement. After that, total RNA and DNA were isolated from these treated cells for qRT-PCR and MSP analysis, respectively.
GC cells were seeded on 96-well plates and treated with different concentrations of 5-Aza-dC in triplicate. After 24, 48, 72, or 144 h of incubation, cells were washed using PBS and cultured in 100 mL RPMI 1640, including 10 mL CCK-8 solution for another 4 h. The absorbance at 450 nm was measured using spectrophotometry (BioTek).
All statistical analyses were performed with SPSS 17.0 software package (SPSS, Chicago, IL, United States). Data are expressed as the mean ± SD. χ2 test, Student’s t test, and one-way ANOVA analysis were used for comparisons. P-values < 0.05 were considered statistically significant.
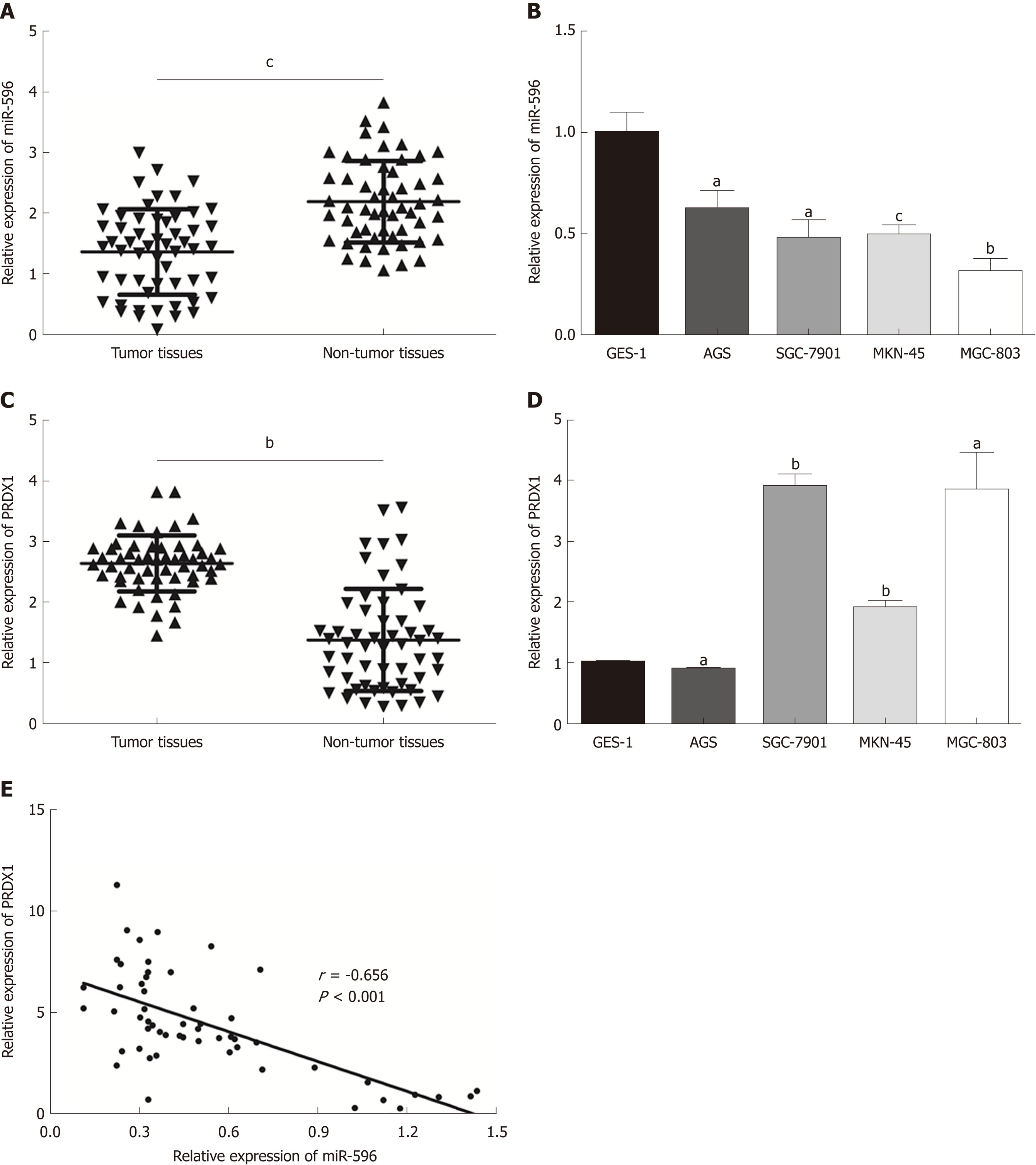
In the present study, the levels of miR-596 expression in 55 pairs of GC and normal control tissues and cell lines were initially assessed using qRT-PCR. The results demonstrated that miR-596 was significantly downregulated in GC tissues compared with paired control tissues (Figure 1A). Furthermore, endogenous expression of miR-596 was investigated in different gastric cell lines, including AGS, SGC-7901, MKN-45, MGC-803, and GES-1. It was shown that the GC cell lines (AGS, SGC-7901, MKN-45, and MGC-803) exhibited relatively low miR-596 expression levels compared to the normal gastric cell line GES-1 (Figure 1B). In addition, PRDX1 expression was analyzed in 55 paired GC and control tissues and cell lines by qRT-PCR. The results indicated that PRDX1 expression was significantly upregulated in GC tissues and cell lines when compared to corresponding non-tumorous tissue and the normal gastric cell line GES-1 (Figure 1C and D). Moreover, Pearson's correlation analysis revealed that the expression of miR-596 was inversely correlated with PRDX1 in GC tissues (Figure 1E). Then, we further examined the relationship between miR-596 expression and clinicopathological factors in 55 GC tissues by Pearson's χ2 test. MiR-596 expression was significantly related to tumor differentiation grade and TNM stage, but not with age, sex, tumor size, tumor site, Borrmann type, or lymph node metastasis (Table 2).
| Variable | No. of Patients | Low expression | High expression | P-value |
| Age (yr) | 0.667 | |||
| < 60 | 18 | 14 | 4 | |
| ≥ 60 | 37 | 32 | 5 | |
| Gender | 1.000 | |||
| Male | 36 | 30 | 6 | |
| Female | 19 | 16 | 3 | |
| Size (cm) | 0.227 | |||
| < 5 | 18 | 13 | 5 | |
| ≥ 5 | 37 | 33 | 4 | |
| Tumor differentiation | 0.043a | |||
| Well/moderate | 23 | 16 | 7 | |
| Poor | 32 | 30 | 2 | |
| Tumor location | 0.827 | |||
| Upper | 5 | 4 | 1 | |
| Middle | 12 | 11 | 1 | |
| Lower | 38 | 31 | 7 | |
| Borrmann type | 0.545 | |||
| I + II | 6 | 4 | 2 | |
| III + V | 49 | 42 | 7 | |
| TNM stage | 0.031a | |||
| I + II | 22 | 15 | 7 | |
| III + IV | 33 | 31 | 2 | |
| Lymph node metastasis | 0.239 | |||
| No | 13 | 9 | 4 | |
| Yes | 42 | 37 | 5 | |
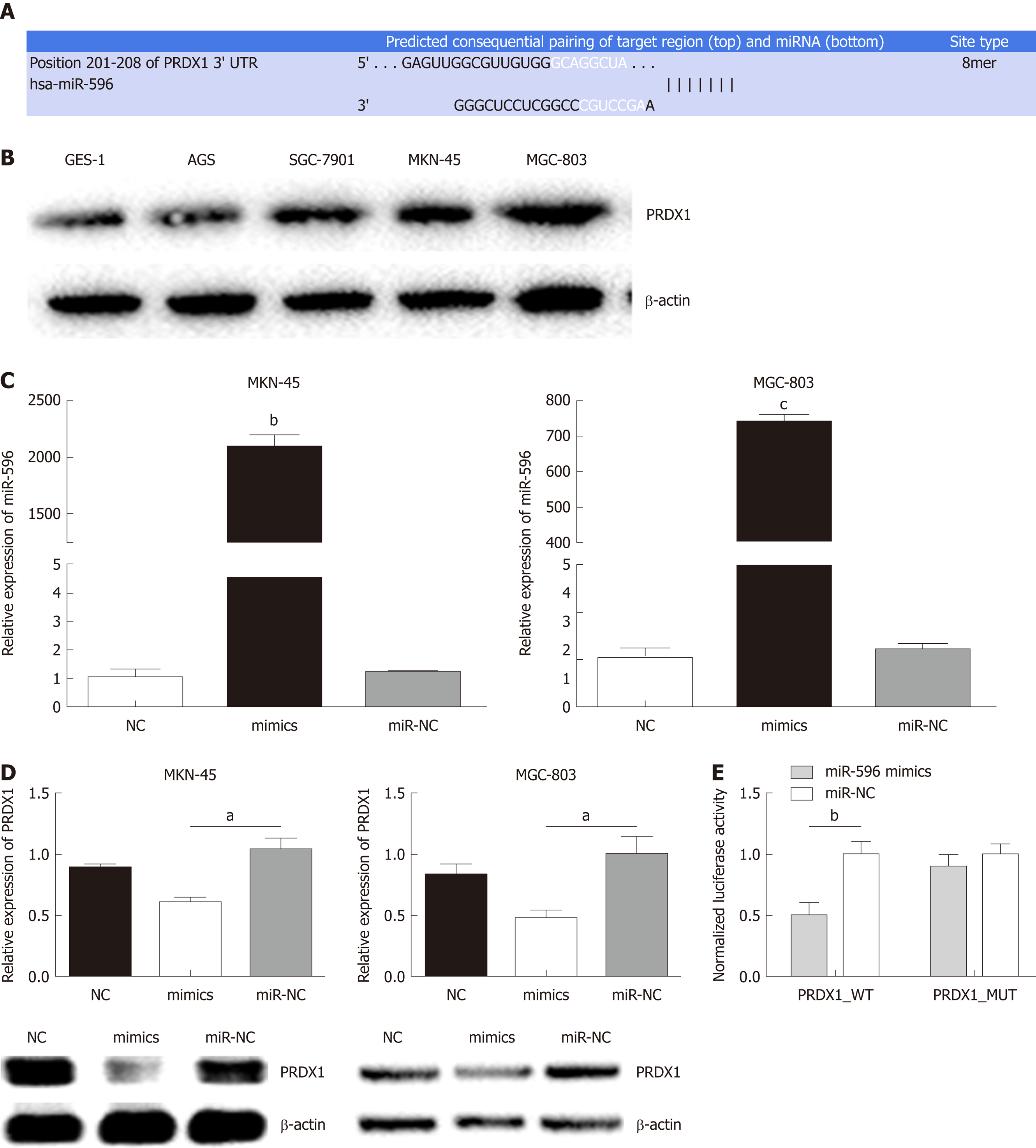
Using three bioinformatic databases (TargetScan, miRWalk, and miRanda), PRDX1 was selected as a predicted target gene of miR-596 (Figure 2A). PRDX1 expression was analyzed in GC cell lines and the GES-1 cell line by Western blot. The results indicated that PRDX1 expression was significantly upregulated in GC cell lines when compared to the normal gastric cell lines GES-1 (Figure 2B). To further validate this prediction, miR-NC or miR-596 mimics was co-transfected into MKN-45 and MGC-803 cell lines and cultured for 48 h. qRT-PCR and Western blot analysis approved that overexpression of miR-596 (Figure 2C) significantly inhibited PRDX1 expression at the mRNA and protein levels in MKN-45 and MGC-803 cell lines (Figure 2D). Furthermore, the luciferase activity of the PRDX1_WT vector was significantly suppressed by the miR-596 mimics, but miR-596 mimics could not affect the luciferase activity of PRDX1_MUT vector or the miR-NC (Figure 2E). These results suggested that miR-596 may bind directly to PRDX1 and inhibit its expression.
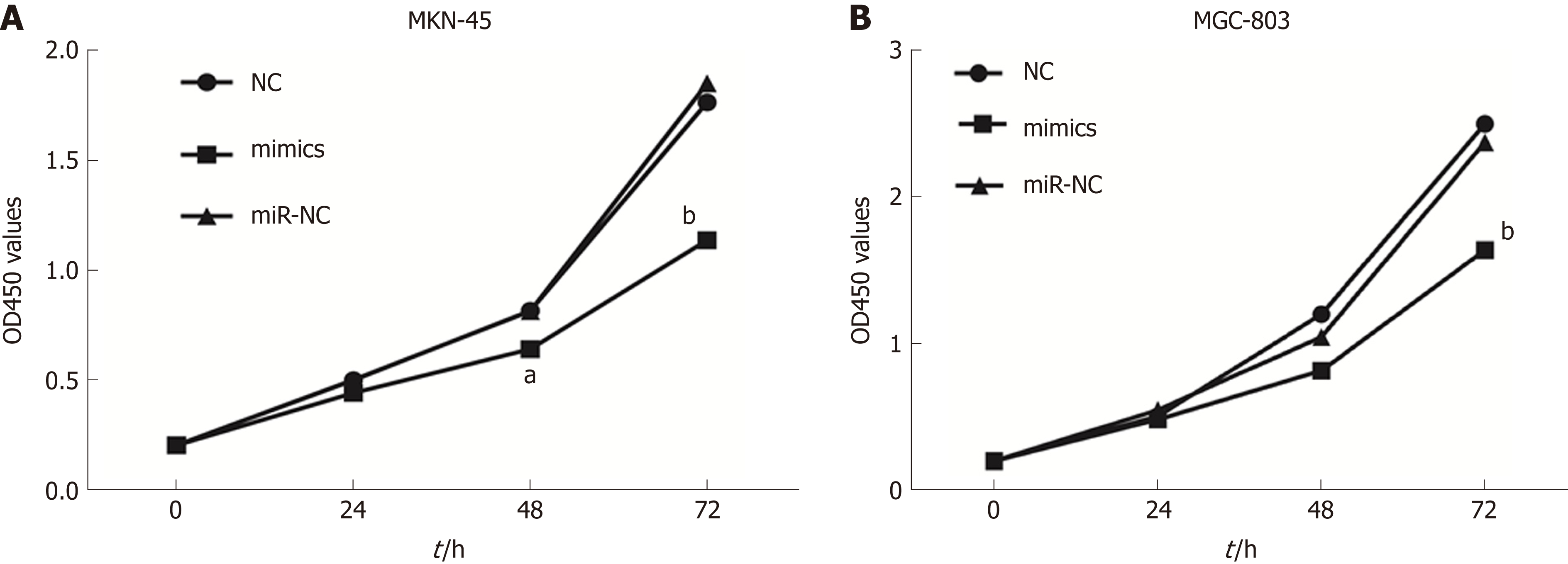
To investigate the role of miR-596 in GC cell proliferation, we utilized ectopic expression of miR-596 and measured cell growth of two GC cell lines MKN-45 and MGC-803 using the CCK-8 method. The results revealed that cell proliferation was clearly suppressed in MKN-45 (Figure 3A) and MGC-803 (Figure 3B) cells after manipulation of miR-596 mimics at 48 and 72 h, but no significant difference was observed at 24 h.
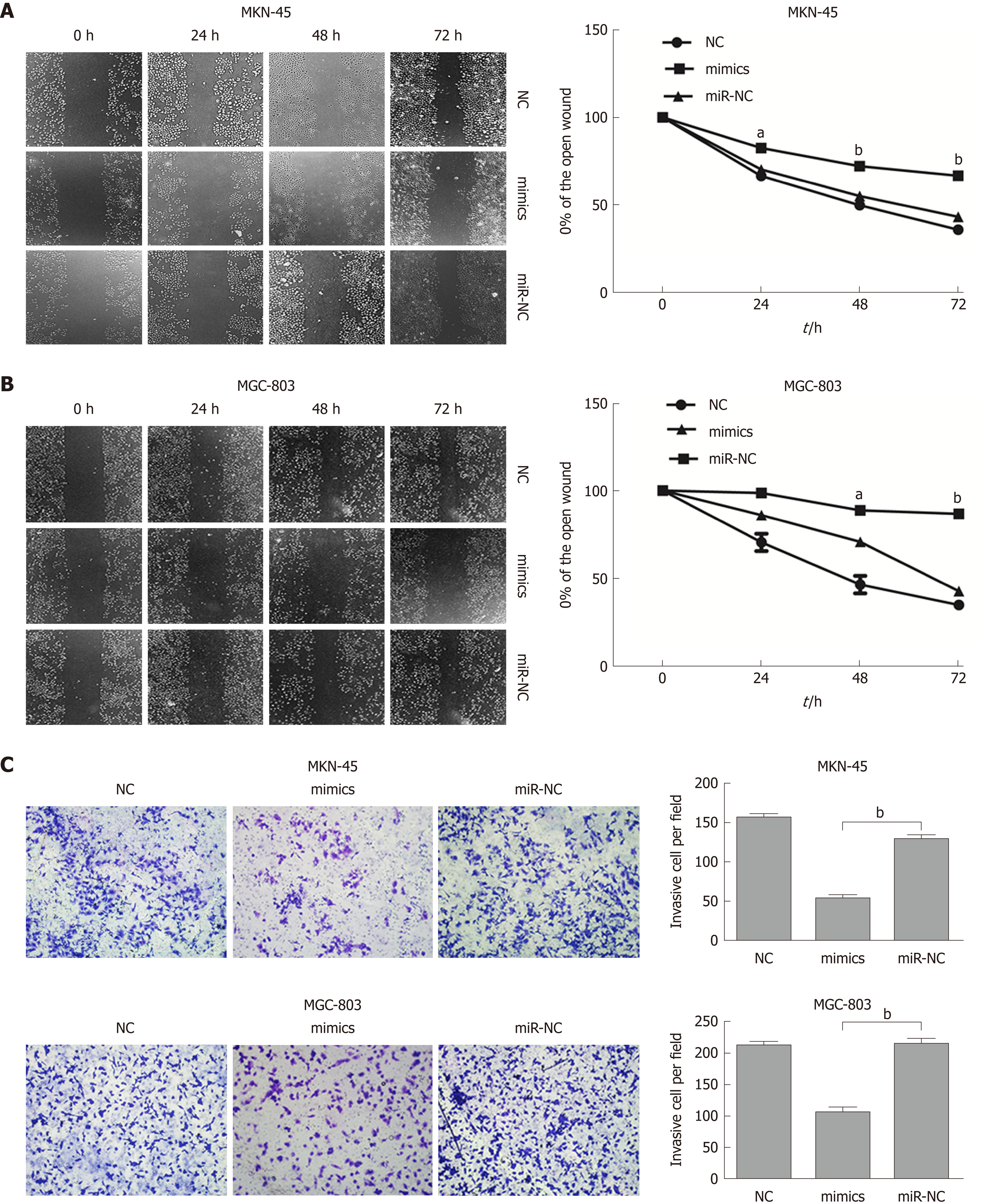
To further evaluate the role of miR-596 in the progression and metastasis of GC, we performed wound healing and Transwell invasion assays in MKN-45 and MGC-803 cells transfected with miR-NC or miR-596 mimics. Transfection with miR-596 mimics significantly suppressed cell migration (Figure 4A and B) and invasion (Figure 4C) in the MKN-45 and MGC-803 cells compared to the NC and miR-NC groups.
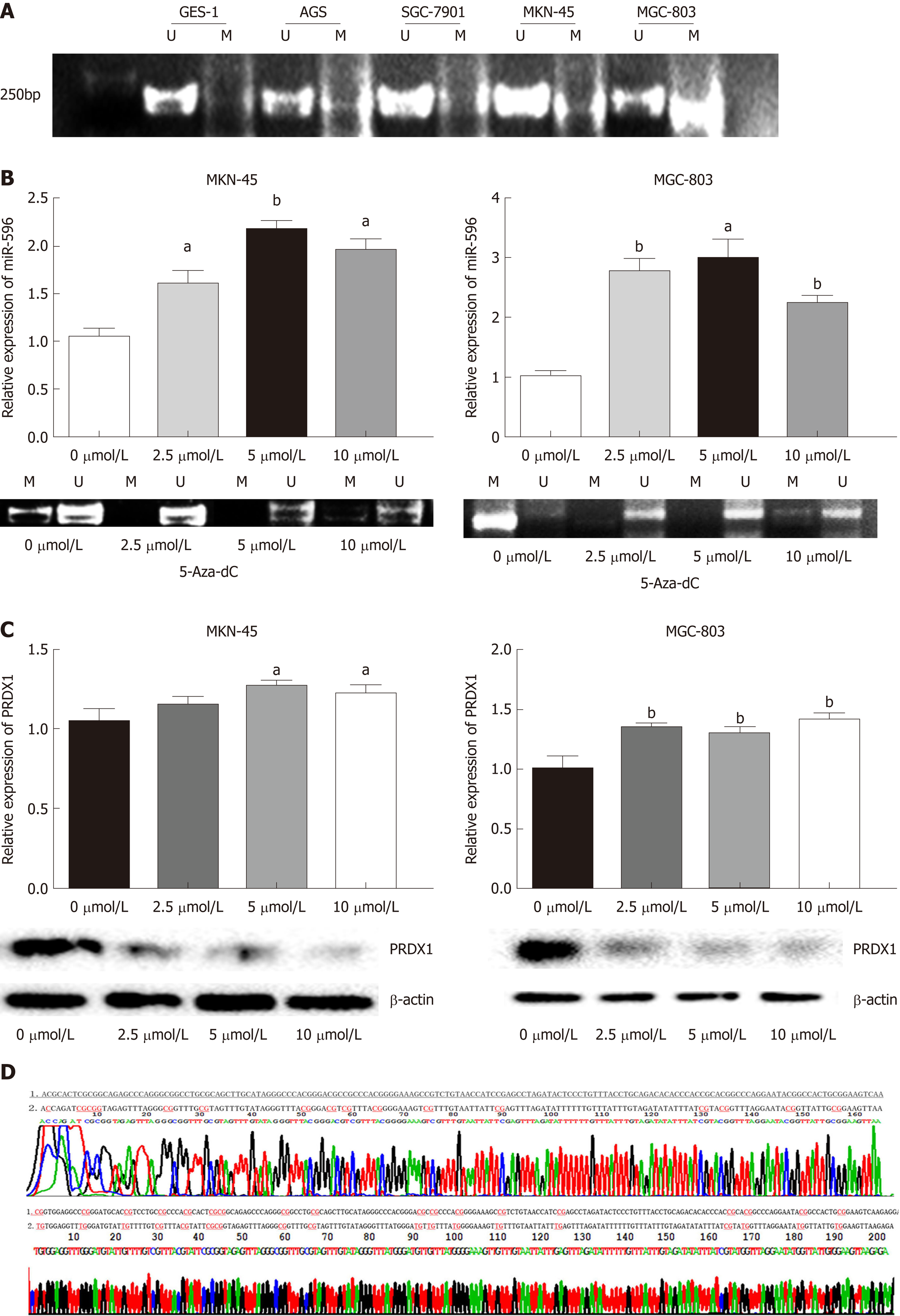
First, to detect whether low expression of miR-596 is related to DNA methylation, we detected the DNA methylation status of miR-596 promoter region in GC cell lines. Data from our MSP analysis displayed that there was a significant negative correlation between miR-596 promoter methylation and expression levels in GC cell lines (Figure 5A). Meanwhile, we treated the miR-596-silenced cell lines, MKN-45 and MGC-803, with the demethylating agent 5-Aza-dC, to confirm whether miR-596 expression could be restarted. We observed that the methylation status of miR-596 promoter region was significantly decreased in MKN-45 and MGC-803 cells after treatment with 5-Aza-dC, the expression of miR-596 was obviously increased, and the highest expression occurred at a concentration of 5 μmol/L (Figure 5B). Moreover, it was found that the expression of PRDX1 mRNA was slightly upregulated in MKN-45 and MGC-803 cells after treatment with 5-Aza-dC, but the expression of PRDX1 protein was significantly downregulated (Figure 5C). MSP products were sequenced, which confirmed that sodium bisulfite modification was sufficient for DNA (Figure 5D).
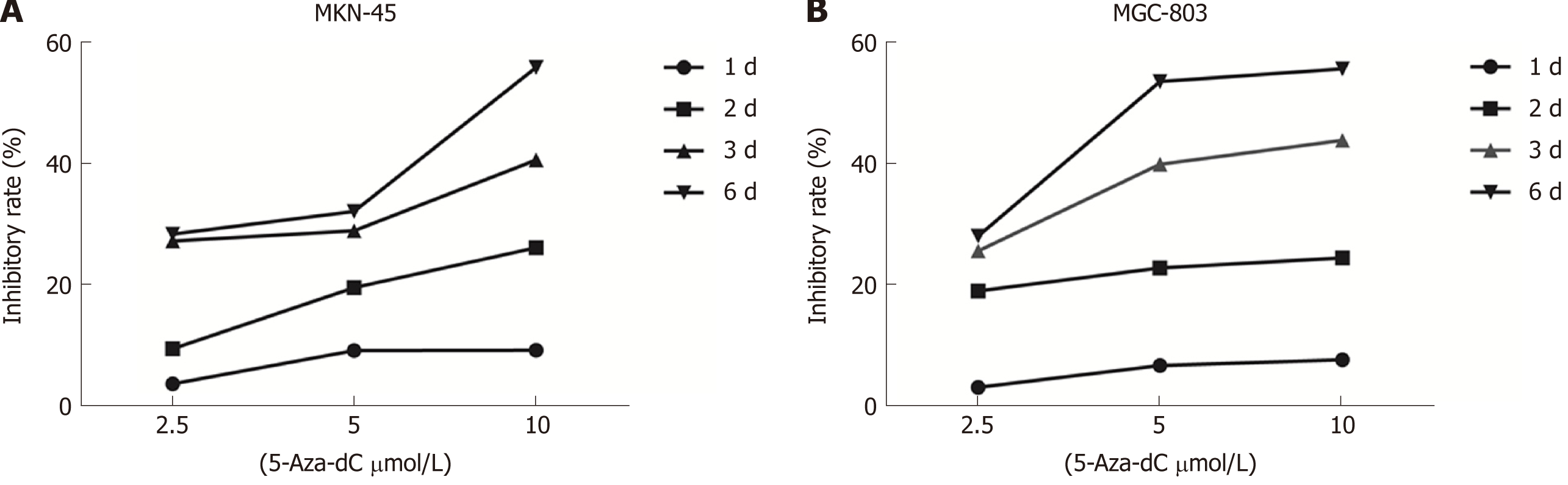
To evaluate whether 5-Aza-dC treatment affects GC cell growth, we analyzed the proliferation of MKN-45 (Figure 6A) and MGC-803 (Figure 6B) cells treated with 5-Aza-dC. The results of the CCK-8 assay showed that the cell inhibition rate was significantly increased as the 5-Aza-dC concentration or the culture time was increased as compared with the untreated control group.
Accumulating evidence has demonstrated that miRNAs may play an important role in GC initiation and development[16,17]. In the present study, we found that miR-596 expression was significantly decreased in GC tissue and cell lines. Moreover, artificial ectopic expression of miR-596 potentially suppressed GC cell proliferation, migration, and invasion. This indicates that miR-596 may function as a tumor suppressor in GC and that miR-596 expression may contribute to the development of GC.
MiRNAs are dysregulated in nearly all human tumours and can function as either tumour suppressors or oncogenes, depending on their target transcripts[18,19]. They work by binding to complementary sequences in the 3'-UTRs of specific mRNAs, leading to translation inhibition[20]. Moreover, due to a non-strict hybridization of the seed match region, one miRNA can bind to multiple mRNA targets, allowing simultaneous downregulation of multiple target mRNAs. Similarly, multiple miRNAs can bind to the same mRNA target and enhance translational repression[21,22]. In our research, bioinformatic analysis show that PRDX1 is a potential target of miR-596. PRDX1 belongs to the peroxiredoxin family and is composed of thiol-specific antioxidant enzymes, which can reduce H2O2 and alkyl hydroperoxide[23], and is related to the reduction of oxidative damage[24]. PRDX1 overexpression was associated with tumor growth and poor prognosis in numerous cancers including breast cancer[25], lung cancer[26], and esophageal squamous cell cancer[27]. However, the expression profile and potential role of PRDX1 in GC remain to be investigated. In this study, we found that the expression of PRDX1 was substantially upregulated in cancer tissues and cell lines by qRT-PCR and was inversely correlated with miR-596 in GC tissues. Additionally, the results showed that overexpression of miR-596 decreased the expression of PRDX1 and luciferase reporter assays detected the direct binding of miR-596 to the 3'-UTR of PRDX1 transcripts. Our results provided a theoretical basis for further study of the mechanism of miR-596 and PRDX1 in GC.
Previous studies showed that transcription of miRNAs can also be epigenetically regulated by methylation in CpG islands[28,29]. In addition, DNA methylation is a reversible signal, similar to other physiological biochemical modifications[30]. The silencing of tumor suppressor genes is closely related to DNA hypermethylation and can be effectively reversed by DNA methyltransferase inhibitors, thereby inhibiting tumor growth[31]. In the present research, DNA methylation analysis by MSP indicated that there was a markedly negative correlation between miR-596 promoter methylation and expression levels in GC cell lines. Furthermore, treatment with the demethylating agent 5-Aza-dC raised the expression of miR-596 in GC cell lines. In addition, the expression of its target gene PRDX1 protein was significantly downregulated. These results showed that the downregulation of miR-596 in GC is attributed, at least in part, to the hypermethylation of CpG sequences in its promoter.
5-Aza-dC as a demethylating agent has been recently used for treatment of myelodysplastic syndromes and acute myelomonocytic leukemia[32]. The main mechanism of 5-Aza-dC is to reduce its activity by binding to DNA methyltransferase I (DNMT1), selectively induce DNMT degradation, and inhibit tumor growth by inducing apoptosis, affecting cell cycle and cytotoxicity[33]. Moreover, 5-Aza-dC is cytotoxic at high concentrations and demethylated at low concentrations[34]. Therefore, our study suggested that the inhibitory effect of 5-Aza-dC on the proliferative ability of GC cells may be due in part to the demethylation and reactivation of miR-596.
In conclusion, our current study demonstrated that the expression of miR-596 was downregulated in GC cells and GC tissues. The in vitro data further suggested that miR-596 expression was able to inhibit GC cell growth, migration, and invasion. Furthermore, miR-596 expression was regulated by epigenetic mechanisms. The downregulation of miR-596 was associated with promoter methylation status in GC cell lines. DNA demethylation and reactivation of miR-596 after treatment with 5-Aza-dC inhibited the proliferative ability of GC cells. Moreover, we identified that PRDX1 is one of the putative target genes of miR-596. Our results further emphasize that miR-596 functions as a crucial tumor suppressor that is regulated by epigenetic mechanisms in GC and may offer a promising novel therapeutic approach for GC. Nonetheless, more studies are required to determine the precise mechanism of miR-596 in the progression of GC.
Gastric cancer (GC) is one of the most common digestive malignancies and remains the second most common cause of cancer-related mortality worldwide. Currently, most patients are diagnosed at an advanced stage, leading to a low cure rate and a low 5-year survival rate. Therefore, it is necessary to further study the molecular mechanism of GC and identify more effective biomarkers for the diagnosis and treatment of GC.
Accumulating studies have indicated that microRNAs (miRNAs) have been associated with almost all known physiological and pathological processes, including cancer. In addition, it is suggested in recent studies that epigenetic modification, especially DNA methylation, is one of many mechanisms of miRNA suppression in human cancer. A greater understanding of these factors will play an important role in early diagnosis and treatment of GC and improvement of prognosis.
To study the potential role and possible regulatory mechanism of microRNA-596 (miR-596) in GC, and whether other targets of miR-596 exist in GC.
In the present study, the levels of miR-596 expression in 55 pairs of GC and normal control tissues and cell lines were initially assessed using quantitative real-time (qRT-PCR). We further examined the relationship between miR-596 expression and clinicopathological factors in 55 GC tissues by Pearson's χ2 test. In addition, peroxiredoxin 1 (PRDX1) expression was analyzed in 55 paired GC and control tissues and cell lines by qRT-PCR. Western blot and luciferase reporter assay were used to detect the effect of miR-596 on PRDX1 expression. Then, the proliferation, metastasis, and invasion of GC cells transfected with miR-596 mimics were analyzed, respectively, by Cell Counting Kit-8 proliferation assay, wound healing assay, and transwell invasion assay. The methylation status of the promoter CpG islands of miR-596 in GC cell lines was detected by methylation-specific PCR. Meanwhile, we treated GC cells with the demethylating agent 5-Aza-2’-deoxycytidine (5-Aza-dC) to confirm whether miR-596 expression could be restarted. Finally, in order to evaluate whether 5-Aza-dC treatment affects GC cell growth, we analyzed the proliferation of GC cells treated with 5-Aza-dC.
Our current study demonstrated that the expression of miR-596 was downregulated in GC cells and GC tissues. The in vitro data further suggested that miR-596 expression was able to inhibit GC cell growth, migration, and invasion. Furthermore, miR-596 expression was regulated by epigenetic mechanisms. Moreover, we identified that PRDX1 is one of the putative target genes of miR-596.
In the present study, we have investigated a suppressive role of microRNA-596 in GC. We reported that miR-596 was downregulated in human specimens of GC and GC cell lines, and that overexpression of miR-596 significantly reduced GC cell proliferation. Moreover, the downregulation of miR-596 in GC cell lines was associated with an increase of miR-596 promoter methylation. We also established that miR-596 controls the expression of PRDX1, which has never been reported before, suggesting that this interaction could play an important role in GC progression.
Our results further emphasize that miR-596 functions as a crucial tumor suppressor that is regulated by epigenetic mechanisms in GC and may offer a promising novel therapeutic approach for GC. Nonetheless, more studies are required to determine the precise mechanism of miR-596 in the progression of GC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Dalay N, Kopljar M S- Editor: Ma RY L- Editor: Wang TQ E- Editor: Yin SY
| 1. | Global Burden of Disease Cancer Collaboration; Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, Hamadeh RR, Moore A, Werdecker A, Te Ao B, Gessner BD, McMahon B, Karimkhani C, Yu C, Cooke GS, Schwebel DC, Carpenter DO, Pereira DM, Nash D, De Leo D, Woodbrook R, Plass D, Ukwaja KN, Thurston GD, Yun Jin K, Simard EP, Mills E, Park EK, Catalá-López F, deVeber G, Gotay C, Khan G, Hosgood HD 3rd, Santos IS, Leasher JL, Singh J, Leigh J, Jonas JB, Sanabria J, Beardsley J, Jacobsen KH, Takahashi K, Franklin RC, Ronfani L, Montico M, Naldi L, Tonelli M, Geleijnse J, Petzold M, Shrime MG, Younis M, Yonemoto N, Breitborde N, Yip P, Pourmalek F, Lotufo PA, Esteghamati A, Hankey GJ, Ali R, Lunevicius R, Malekzadeh R, Dellavalle R, Weintraub R, Lucas R, Hay R, Rojas-Rueda D, Westerman R, Sepanlou SG, Nolte S, Patten S, Weichenthal S, Abera SF, Fereshtehnejad SM, Shiue I, Driscoll T, Vasankari T, Alsharif U, Rahimi-Movaghar V, Vlassov VV, Marcenes WS, Mekonnen W, Melaku YA, Yano Y, Artaman A, Campos I, MacLachlan J, Mueller U, Kim D, Trillini M, Eshrati B, Williams HC, Shibuya K, Dandona R, Murthy K, Cowie B, Amare AT, Antonio CA, Castañeda-Orjuela C, van Gool CH, Violante F, Oh IH, Deribe K, Soreide K, Knibbs L, Kereselidze M, Green M, Cardenas R, Roy N, Tillmann T, Li Y, Krueger H, Monasta L, Dey S, Sheikhbahaei S, Hafezi-Nejad N, Kumar GA, Sreeramareddy CT, Dandona L, Wang H, Vollset SE, Mokdad A, Salomon JA, Lozano R, Vos T, Forouzanfar M, Lopez A, Murray C, Naghavi M. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2055] [Article Influence: 205.5] [Reference Citation Analysis (0)] |
| 2. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16089] [Article Influence: 1005.6] [Reference Citation Analysis (2)] |
| 3. | Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 912] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 4. | Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172-1187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1233] [Cited by in RCA: 1371] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 5. | Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609-1622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 273] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 6. | Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455-1459. [PubMed] |
| 7. | Endo H, Muramatsu T, Furuta M, Uzawa N, Pimkhaokham A, Amagasa T, Inazawa J, Kozaki K. Potential of tumor-suppressive miR-596 targeting LGALS3BP as a therapeutic agent in oral cancer. Carcinogenesis. 2013;34:560-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Ma M, Yang J, Wang B, Zhao Z, Xi JJ. High-Throughput Identification of miR-596 Inducing p53-Mediated Apoptosis in HeLa and HCT116 Cells Using Cell Microarray. SLAS Technol. 2017;22:636-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Liu SM, Lin CH, Lu J, Lin IY, Tsai MS, Chen MH, Ma N. miR-596 Modulates Melanoma Growth by Regulating Cell Survival and Death. J Invest Dermatol. 2018;138:911-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Olivieri M, Ferro M, Terreri S, Durso M, Romanelli A, Avitabile C, De Cobelli O, Messere A, Bruzzese D, Vannini I, Marinelli L, Novellino E, Zhang W, Incoronato M, Ilardi G, Staibano S, Marra L, Franco R, Perdonà S, Terracciano D, Czerniak B, Liguori GL, Colonna V, Fabbri M, Febbraio F, Calin GA, Cimmino A. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget. 2016;7:20636-20654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 11. | Terreri S, Durso M, Colonna V, Romanelli A, Terracciano D, Ferro M, Perdonà S, Castaldo L, Febbraio F, de Nigris F, Cimmino A. New Cross-Talk Layer between Ultraconserved Non-Coding RNAs, MicroRNAs and Polycomb Protein YY1 in Bladder Cancer. Genes (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Anwar SL, Albat C, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer. 2013;133:660-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 13. | Potapova A, Albat C, Hasemeier B, Haeussler K, Lamprecht S, Suerbaum S, Kreipe H, Lehmann U. Systematic cross-validation of 454 sequencing and pyrosequencing for the exact quantification of DNA methylation patterns with single CpG resolution. BMC Biotechnol. 2011;11:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Huang YW, Kuo CT, Chen JH, Goodfellow PJ, Huang TH, Rader JS, Uyar DS. Hypermethylation of miR-203 in endometrial carcinomas. Gynecol Oncol. 2014;133:340-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, Chen XW, Gao P, Miao ZF, Wang ZN. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 225] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 16. | Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 676] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 17. | Kiga K, Mimuro H, Suzuki M, Shinozaki-Ushiku A, Kobayashi T, Sanada T, Kim M, Ogawa M, Iwasaki YW, Kayo H, Fukuda-Yuzawa Y, Yashiro M, Fukayama M, Fukao T, Sasakawa C. Epigenetic silencing of miR-210 increases the proliferation of gastric epithelium during chronic Helicobacter pylori infection. Nat Commun. 2014;5:4497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 112] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 18. | Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1357] [Cited by in RCA: 1381] [Article Influence: 115.1] [Reference Citation Analysis (0)] |
| 19. | Kwan JY, Psarianos P, Bruce JP, Yip KW, Liu FF. The complexity of microRNAs in human cancer. J Radiat Res. 2016;57 Suppl 1:i106-i111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 365] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 21. | Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 865] [Cited by in RCA: 918] [Article Influence: 76.5] [Reference Citation Analysis (0)] |
| 22. | Pinweha P, Rattanapornsompong K, Charoensawan V, Jitrapakdee S. MicroRNAs and oncogenic transcriptional regulatory networks controlling metabolic reprogramming in cancers. Comput Struct Biotechnol J. 2016;14:223-233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 23. | Kim JH, Lee JM, Lee HN, Kim EK, Ha B, Ahn SM, Jang HH, Lee SY. RNA-binding properties and RNA chaperone activity of human peroxiredoxin 1. Biochem Biophys Res Commun. 2012;425:730-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Aeby E, Ahmed W, Redon S, Simanis V, Lingner J. Peroxiredoxin 1 Protects Telomeres from Oxidative Damage and Preserves Telomeric DNA for Extension by Telomerase. Cell Rep. 2016;17:3107-3114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 25. | Cha MK, Suh KH, Kim IH. Overexpression of peroxiredoxin I and thioredoxin1 in human breast carcinoma. J Exp Clin Cancer Res. 2009;28:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 26. | Jiang H, Wu L, Mishra M, Chawsheen HA, Wei Q. Expression of peroxiredoxin 1 and 4 promotes human lung cancer malignancy. Am J Cancer Res. 2014;4:445-460. [PubMed] |
| 27. | Gong F, Hou G, Liu H, Zhang M. Peroxiredoxin 1 promotes tumorigenesis through regulating the activity of mTOR/p70S6K pathway in esophageal squamous cell carcinoma. Med Oncol. 2015;32:455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Poddar S, Kesharwani D, Datta M. Interplay between the miRNome and the epigenetic machinery: Implications in health and disease. J Cell Physiol. 2017;232:2938-2945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 29. | Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, Kimura T, Kudo T, Harada E, Sugai T, Takamaru H, Niinuma T, Maruyama R, Yamamoto H, Tokino T, Imai K, Toyota M, Shinomura Y. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31:2066-2073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 157] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 30. | Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci USA. 1999;96:6107-6112. [PubMed] |
| 31. | Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 365] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 32. | Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632-4642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 797] [Cited by in RCA: 875] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 33. | Mai A, Altucci L. Epi-drugs to fight cancer: from chemistry to cancer treatment, the road ahead. Int J Biochem Cell Biol. 2009;41:199-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 146] [Article Influence: 8.6] [Reference Citation Analysis (0)] |