Published online Mar 7, 2018. doi: 10.3748/wjg.v24.i9.982
Peer-review started: December 9, 2017
First decision: December 20, 2017
Revised: December 26, 2017
Accepted: January 23, 2018
Article in press: January 23, 2018
Published online: March 7, 2018
Processing time: 86 Days and 8.4 Hours
To investigate the potential role of poly(ADP-ribose) polymerase 1 (PARP1) in the development of Barrett’s esophagus (BE).
A BE mouse model was established to examine the esophageal morphological changes and molecular changes. Microarray analysis was performed to compare the gene expression profiles between BE patients and healthy controls. qPCR was used to examine the PARP1 expression in cell lines after treatment with H2O2 and bile acids (pH 4). Immunofluorescence staining, comet assay, and annexin V staining were used to evaluate the impact of PARP1 activity on cell survival and DNA damage response after oxidative stress.
The gene expression profile in normal and BE esophageal epithelial cells showed that PARP1, the major poly(ADP-ribose) polymerase, was overexpressed in BE. In the mouse model of BE, positive staining for NF-κB, γH2AX, and poly(ADP-ribose) (PAR) was observed. H2O2 and bile acids (pH 4) increased the PARP1 mRNA expression level in normal esophageal epithelial cells. Using shRNA-PARP1 to suppress PARP1 activity decreased the cell viability after treatment with H2O2 and bile acids (pH 4), and increased the oxidative damage as demonstrated by an increase in the levels of H2O2, intracellular reactive oxygen species (ROS), oxidative DNA damage, double-strand breaks, and apoptosis (P < 0.01).
The dysfunction of PARP1 in esophageal epithelial cells increases the levels of ROS and oxidative DNA damage, which could be common risk factors for BE and esophageal adenocarcinoma.
Core tip: In this study, we compared the gene expression profile in normal esophagus and Barrett’s esophagus (BE), and found that poly(ADP-ribose) polymerase 1 (PARP1) was overexpressed in BE. In a BE model, positive staining for NF-κB, γH2AX, and PAR was observed. H2O2 and bile acids (pH 4) increased the PARP1 mRNA expression level in normal esophageal epithelial cells. PARP1 inhibition could decrease the cell viability after bile acids treatment, and increased the oxidative damage, double-strand breaks, and apoptosis. Thus, our study demonstrates a novel molecular mechanism for the role of PARP1 in the process of Barrett’s metaplasia, which sheds a potential therapeutic role for PARP1 inhibitor in BE.
- Citation: Zhang C, Ma T, Luo T, Li A, Gao X, Wang ZG, Li F. Dysregulation of PARP1 is involved in development of Barrett’s esophagus. World J Gastroenterol 2018; 24(9): 982-991
- URL: https://www.wjgnet.com/1007-9327/full/v24/i9/982.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i9.982
Gastro-esophageal reflux disease (GERD) is known as reflux of gastric acid, usually mixed with bile acids, into the lower esophagus[1-3]. GERD-associated chronic mucosal injury and inflammation are major risk factors for the development of Barrett’s esophagus (BE), a premalignant condition closely associated with the development of esophageal adenocarcinoma. BE is present in 10%-20% of patients with GERD[4,5]. Although the leading cause of BE is formerly considered a chronic inflammation, increasing evidence suggests that oxidative stress-induced DNA damage induces the apoptosis of esophageal epithelial cells during the pathogenesis of BE. However, it is unclear why esophageal epithelial cells are sensitive to oxidative damage.
Oxidative DNA damage is usually induced by reactive oxygen species (ROS) primarily generated from normal intracellular metabolism in mitochondria and peroxisomes[6,7]. A number of external hazards such as ionizing radiation, chemicals, and solar ultraviolet light can also trigger ROS production. These active free radicals attack double-strand DNA, inducing various types of DNA lesions, including DNA single-strand breaks (SSBs) and DNA double-strand breaks (DSBs), which may lead to genomic instability[6,8]. To cope with these threats, cells have evolved DNA damage response systems to detect and repair DNA lesions. As one of the earliest alarm systems and regulators in DNA damage response, poly(ADP-ribose) (PAR) participates in the repair of numerous types of DNA damage including SSBs and DSBs[9]. Thus, the cellular metabolism of PAR is critical for DNA damage response and genomic stability. The reaction of poly(ADP-ribosyl)ation (PARylation) is catalyzed by a group of PAR polymerases (PARPs). Using NAD+ as the substrate, PARPs covalently adds ADP-ribose to the side chains of arginine, aspartic acid, and glutamic acid residues in target proteins. After catalyzing the first ADP-ribose onto the proteins, other ADP-riboses can be covalently linked and the continuous reactions produce both linear and branched polymers known as PAR[10]. The structure of PAR has been well characterized: the ADP-ribose unit in the polymer is linked by glycosidic ribose-ribose 1’-2’ bonds. The chain length is heterogeneous and can reach around 200 units with 20-50 units in each branch[11].
Although PARylation has been examined both in vivo and in vitro, the function of PARP1 in esophageal epithelial cells remains elusive. In this study, we hypothesized that PARP1 may be involved in the oxidative damage in the development of BE. We aimed to investigate the potential role of PARP1 and PARylation-related DNA damage in the process of BE in GERD through using a BE mouse model and BE cell lines. Elucidating the role of PARP1 in BE will provide a novel therapeutic strategy for this condition.
Experiments in 20 male C57Bl/6 mice with a mean weight of 29 g (27-31 g) were performed. All mice adapted to the environment 7 d to 10 d after purchase, and then water was banned for 24 h before the operation. Chloral hydrate (10%; 3 mL/kg) was administered intraperitoneally for anesthesia. The lower end of the esophagus was separated and a longitudinal cut (about 1-1.5 cm) was performed at the gastroesophageal junction without vascular region. The anti-reflux barrier of the gastroesophageal junction was damaged by the operation.
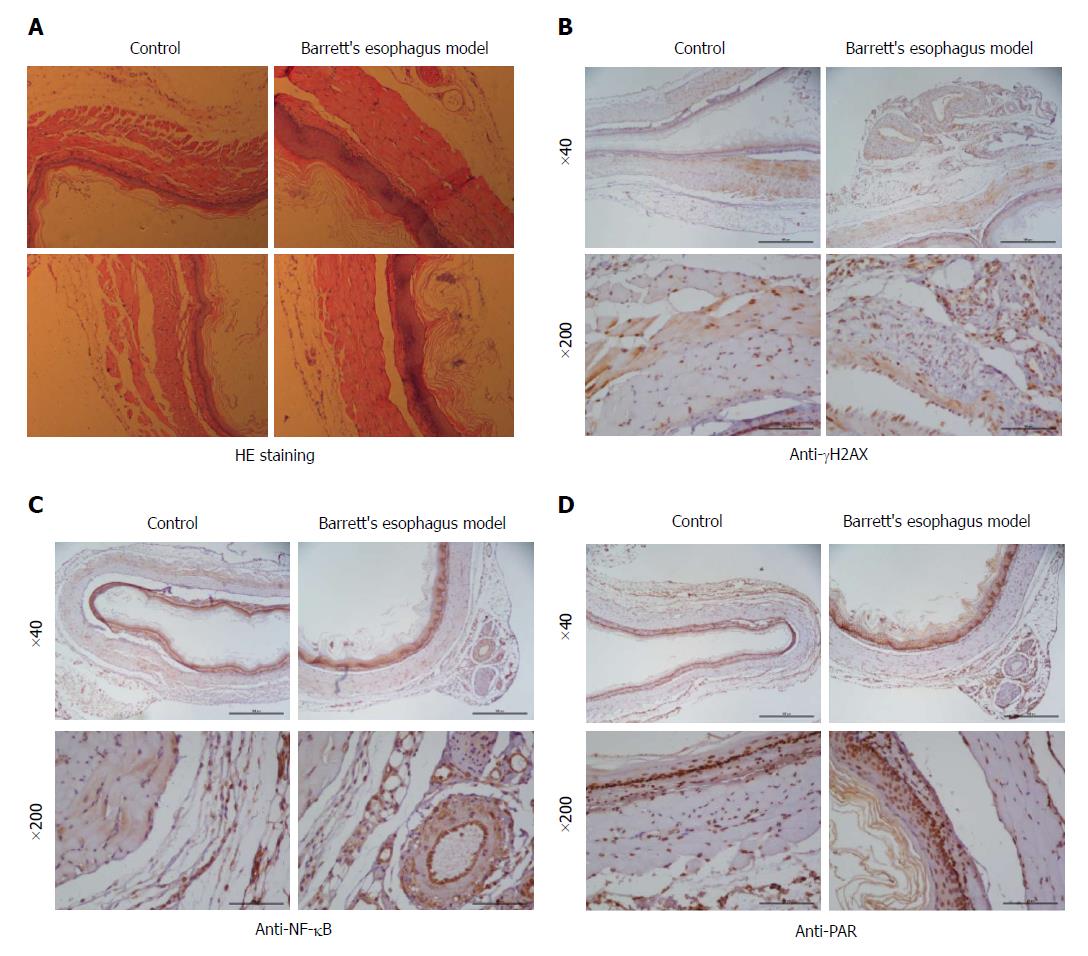
Animals were sacrificed 3 mo after surgery. Samples were taken in the operating room of the animal facility immediately after the animal sacrifice. The entire stomach and esophagus were removed and immediately fixed in formalin for at least 24 h. Subsequently, samples were processed for paraffin embedding and 7 μmol/L sections were obtained through a microtome and put on positive charged slides. Sections of the lower esophagus were then stained with HE following a standardized protocol and observed under a light microscope. The same samples were subjected for immunohistochemistry. For immunohistochemical staining, sections were placed on polylysine-coated slides. After deparaffinization in xylene and rehydration through graded alcohol, each section was treated with 1% hydrogen peroxide for 20 min, and then blocking serum was applied for 20 min. The slides were incubated overnight with the primary antibody (γH2AX and NF-κB at 1:300 and PAR at 1:500) at 4 °C in a closed chamber. Immunohistochemical staining was performed by the streptoavidin-biotin-peroxidase complex technique (Histofine SAB-PO(M) Kit; Nichirei Corp., Tokyo, Japan). Staining was done with 3,3′-diaminobenzidine followed by light counterstaining with methyl green and dehydration.
Immortalised BE cell lines BAR-T and CP-A (American Type Culture Collection, ATCC, Manassas, VA, United States) were cultured with epithelial cell medium 2 (ScienCell, Carlsbad, CA, United States), supplemented with 5% fetal bovine serum and antibiotics on primaria plates and flasks (BD Biosciences, Bedford, MA, United States). The immortalised human esophageal epithelia cell lines HET1A and EPC2 were obtained from ATCC. HET1A cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (Invitrogen). EPC2 cells were grown in keratinocyte SFM medium supplemented with 40 mg/mL bovine pituitary extract and 1.0 ng/mL epidermal growth factor (Invitrogen, Carlsbad, California, United States). All cell lines were grown at 37 °C in 5% carbon dioxide.
Tissue samples of the lower esophagus from 39 individuals, consisting of esophageal squamous epithelia from 19 healthy subjects and 20 specimens from patients with BE, were obtained from the pathological center of the Xuanwu Hospital or the Second Artillery General Hospital of Chinese People’s Liberation Army in China. The baseline characteristics of patients are shown in Table 1. Gene expression was examined by whole-human-genome microarrays (Affymetrix, Santa Clara, CA, United States). Normal healthy esophageal biopsies were collected from patients with esophageal pain but diagnosed with normal squamous epithelia without pathological changes.
| Patient characteristic | n = 20 |
| Age (mean ± SD), yr | 56.0 ± 12.5 |
| Gender, male, n (%) | 16 (80) |
| Body mass index (mean ± SD), kg/m2 | 26.5 ± 3.1 |
| Barrett’s length (median, IQR), cm | 4.7 (3.8-5.5) |
| Circumferential extent (C) | 100% |
All samples were snap-frozen in liquid nitrogen and stored at -80 °C. We obtained tissue specimens from all subjects with informed written consent (approved by the local ethics committees of the Xuanwu Hospital and the Second Artillery General Hospital of Chinese People’s Liberation Army. Each single specimen included in this study was histopathologically approved according to grading and staging results by an experienced pathologist.
Preparation of labeled cRNA and hybridization were done using the gene chip hybridization, wash, and stain kit (Affymetrix, Santa Clara, CA, United States). Two cycle labeling was applied to all samples. In total, 28 chip data were collected using Gene Chip Operation Software (GCOS, Affymetrix, United States). The 39 specimens analyzed consisted of 20 BE and 19 normal esophageal samples. To obtain the relative gene expression measurements, probe set-level data extraction was performed with the GCRMA (Robust Multiarray Average) normalization algorithm implemented in GeneSpring GX10.2 (Agilent). All data were log2 transformed.
RNA extraction was carried out using the RNeasy Mini Kit (Qiagen, Valencia, CA, United States). Total RNA quality and yield were assessed using a bioanalyzer system (Agilent 2100 Bioanalyzer; Agilent Technologies, CA, United States) and a spectrophotometer (NanoDrop ND-1000; NanoDrop Technologies, DE, United States). Only RNA with an RNA integrity number > 9.0 was used for microarray analysis.
For quantification of mRNA expression, qRT-PCR was performed for three genes plus one control, using pre-designed gene-specific TaqMan probes and primer sets purchased from Applied Biosystems.
Single-cell gel electrophoretic comet assays were performed under alkaline conditions. Briefly, cells were treated with 100 μmol/L H2O2 at 37 °C. For cellular lysis, the slides were immersed in the alkaline lysis solution overnight in the dark. Next, the slides were subjected to electrophoresis at 20 V (0.6 V/cm) for 25 min and stained in 2.5 mg/mL propidium iodide for 20 min. All images were taken with a fluorescence microscope and analyzed with Comet Assay IV software.
After treatment with H2O2, cells were fixed in 3% paraformaldehyde and permeabilized in 0.5% Triton X-100 for 30 min at room temperature. Samples were blocked with 5% goat serum and then incubated in primary antibody for 1 h. Samples were then washed with PBS three times and incubated with secondary antibody for 30 min. After PBS washing, the nuclei were stained with Hoechst 33258. The signals were visualized by fluorescence microscopy. We calculated the number of γH2AX foci per nucleus, and for quantitative analysis, 15 nuclear foci were selected to count in each group. For the immunofluorescence assay of anti-8-oxoguanine (8-OXOG), cells were exposed to bile acids cocktail (0.1 mmol/L; pH 4) for 60 min. Cells were then incubated with primary antibody against 8-OXOG (mouse, 1:200; Millipore) overnight at 48 °C, followed by incubation with secondary goat anti-mouse antibody conjugated with TRITC (1:1000) at room temperature for 45 min. The slides were mounted using Vectashield with DAPI (Vector Laboratories, Burlingame, CA, United States) and viewed under a fluorescence microscope.
Cells were harvested and lysed with 100 μL of NETN 300 lysis buffer unless otherwise specified. Soluble fractions were subjected to Western blot analysis.
All experiments were performed in triplicate unless indicated otherwise. Student’s t-test was used for statistical analyses, and P < 0.05 was considered statistically significant.
During the surgical operation, the mortality rate of mice was 10%, the perioperative mortality rate was 15%, and 1 wk later, the mortality rate was 10%. The lower esophagus from the BE mouse model was processed for staining for γH2AX, NF-κB, and PAR. In the mouse BE model, esophagitis with esophageal erosions and squamous epithelial thickening with basal cell layer hyperplasia were seen. Esophagitis was accompanied by the development of papillary hyperplasia of the squamous epithelia (Figure 1A). Columnar intestinal-type metaplasia resembling that of human BE was seen with erosive esophagitis. No adenocarcinoma or squamous cell carcinoma was seen. In the immunohistochemistry staining, more positive staining for γH2AX, NF-κB, and PAR was observed, suggesting the DNA damage response and inflammation in the BE epithelium.
To investigate the gene expression changes during the development of BE, samples from BE patients and normal esophagus controls were subjected to microarray analysis. 20 BE lesions for microarray analysis were resected from 20 patients with a mean age of 56 ± 12 years between February 2013 and January 2016. Main patient characteristics are presented in Table 1.
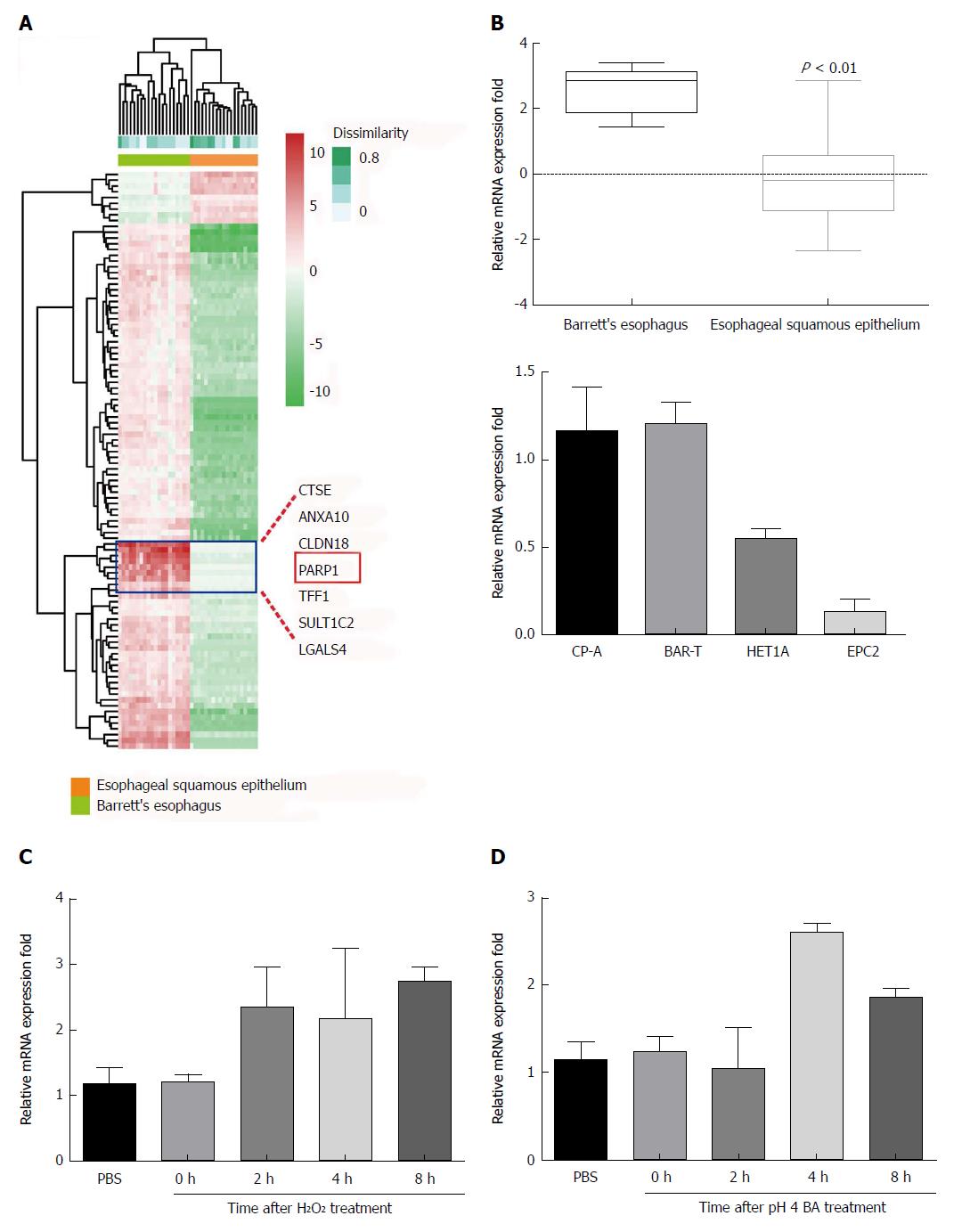
As shown in Figure 2A, PARP1 was one of the upregulated genes. Representative expression profile of PARP1 between the BE patients and normal controls was also showed in Figure 2B. Besides, PARP1 expression in normal esophageal epithelial cell lines (HET1A and EPC2) and BE epithelial cell lines (CP-A and BAR-T) was also examined. PAPR1 was increased in CP-A and BAR-T cell lines.
To investigate whether PARP1 expression levels can be upregulated by exposure to H2O2 and acidic bile acids, we exposed HET1A cells to H2O2 (100 μmol/L) and bile acids (0.1 mmol/L; pH 4) for 30 min. Following this exposure, cells were washed in PBS and cultured in regular medium for 30 min, 1 h, 2 h, 4 h, or 8 h, and mRNA levels were measured by RT-PCR. In response to this treatment, significant upregulation of the mRNA levels of PARP1 was observed at 2 h for H2O2 and at 4 h and 8 h for both H2O2 and bile acids (Figure 2C).
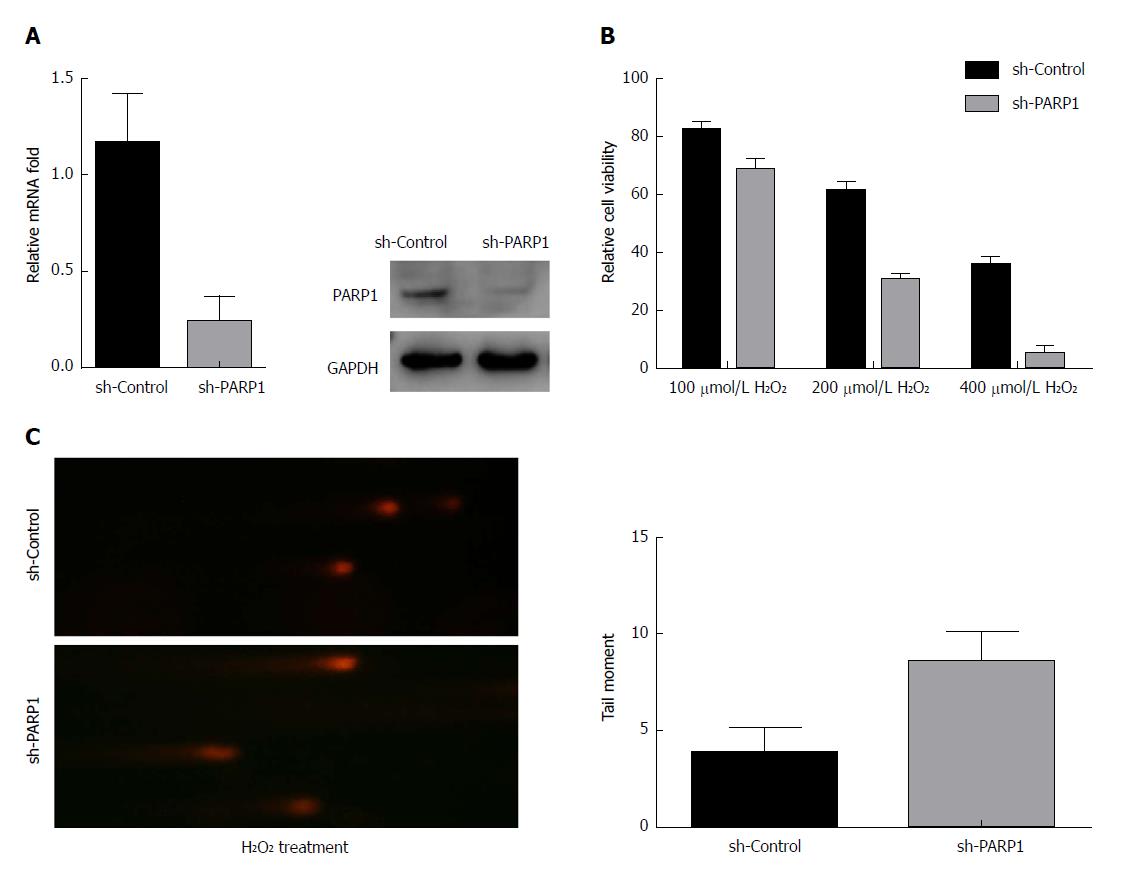
We next tested the protective effect of PARP1 against H2O2-induced oxidative stress. The knockdown of PARP1 in BAR-T cells (Figure 3A) led to a significantly lower cell viability following exposure to H2O2, compared with controls (P < 0.01 for 100 μmol/L, 200 μmol/L, and 400 μmol/L H2O2; Figure 3B), suggesting that the downregulation of endogenous PARP1 sensitized the BAR-T cells to H2O2-induced oxidative stress. To further examine the oxidative DNA damage effect after H2O2 treatment, we performed comet assay analysis. As shown in Figure 3C, BAR-T cells with PARP1 knockdown displayed a significantly severe DNA damage signal upon exposure to 200 μmol/L H2O2 for 2 h (4.3 ± 0.8 vs 8.6 ± 1.4, P < 0.01).
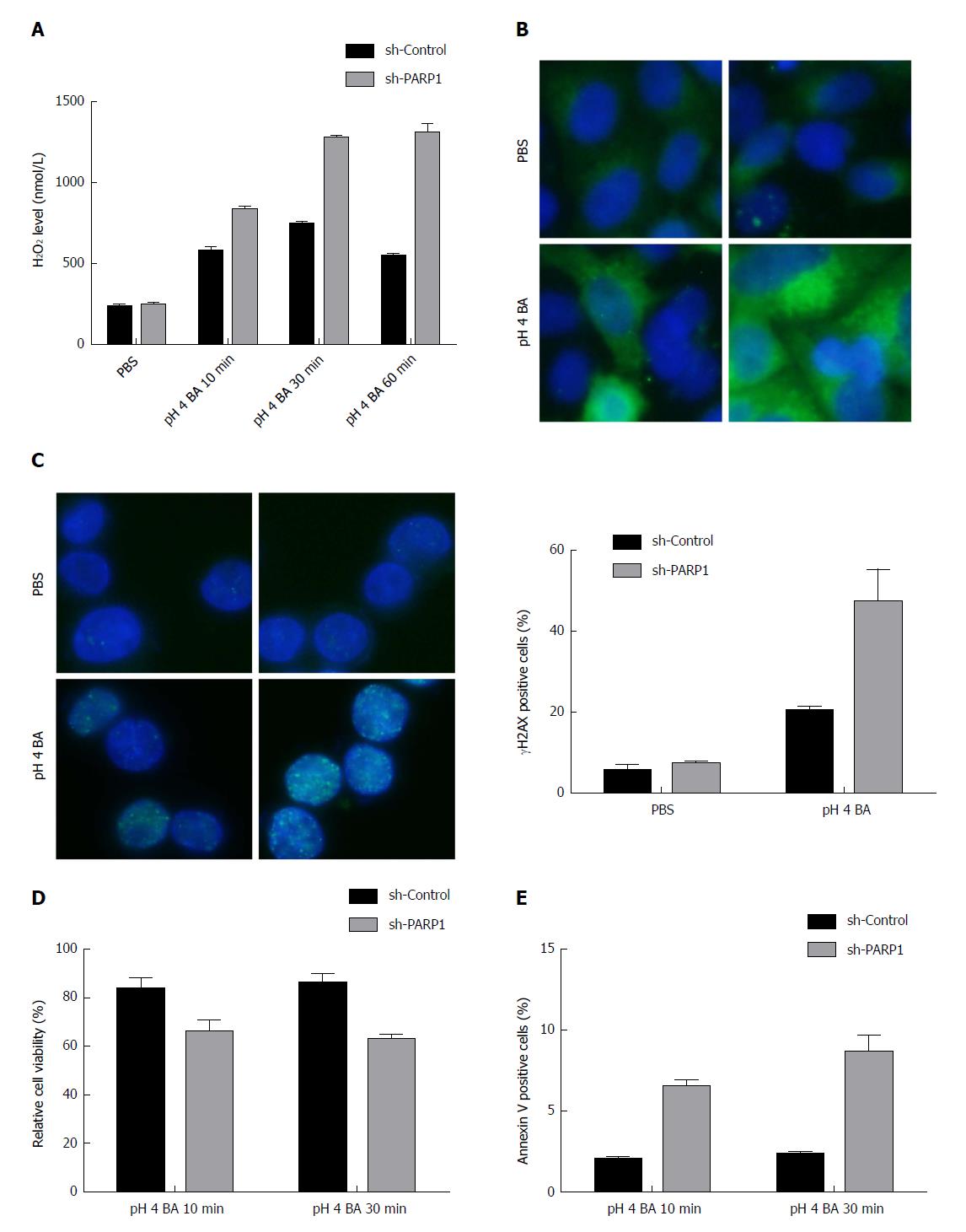
We exposed BE epithelial cells to a bile acids cocktail adjusted to pH 4 and performed the Amplex UltraRed H2O2 assay to measure H2O2 levels. BAR-T cells with knockdown of endogenous PARP1 displayed higher H2O2 levels than control cells transfected with scrambled shRNA (Figure 4A).
We used immunofluorescence staining for 8-OXOG, one of the most identified oxidative DNA damage sites. On exposure of BAR-T cells with PARP1 knockdown to bile acids (pH 4), more oxidative DNA damage was observed than control cells exposed to acidic bile acids (Figure 4B).
Gastric acid, particularly in combination with bile acids, has been shown to induce DSBs in esophageal epithelial cells. The immunocytochemical staining for γH2AX, a reliable marker for DSBs, demonstrated significantly high levels of positive γH2AX sites following knockdown of PARP1 in BAR-T cells (19.8% ± 0.3% vs 42.6% ± 7.2%, P < 0.01, Figure 4C) than in control cells.
Because PARP1 expression protected cells from acidic bile acid-induced DNA damage, it may protect the epithelial cells of BE from acidic bile acid-induced apoptosis. BAR-T cells were treated with bile acids (pH 4) for 10 min or 30 min, followed by replacement with a regular culture medium for 24 h. The ATP-Glo cell viability assay was then used to determine cell viability at this 24 h time point. As shown in Figure 4D, the short-term exposure (10 min) did not change the cell viability at 24 h. However, the exposure for 30 min demonstrated a significant reduction in cell viability in BAR-T cells lacking PARP1 (82.5% ± 2.2% vs 63.0% ± 1.6%, P < 0.05). Consistent with this finding, the cells with PARP1 knockdown displayed significantly more apoptosis after acidic bile acid exposure as indicated by annexin V assay (P < 0.05, 10 min: 2.1 ± 0.2 vs 6.3 ± 0.8; 30 min: 2.3 ± 0.1 vs 8.5 ± 1.2, Figure 4E).
During pathological events such as excessive and unrepairable oxidant DNA damage, highly activated PARPs synthesize large amounts of PAR in a few seconds. Our results showed that excessive PARP1 expression may probably a resistance factor of BE epithelial cells to H2O2 or bile acid-induced oxidative damage and cell death. In recent years, PARP1 inhibitors have shown promising therapeutic effects on ovarian cancer, breast cancer[12], and neurological diseases[13], and this effect is mainly associated with the induction of DSBs. Interestingly, pharmacological inhibition of PARP1 provides significant benefits in animal models of cardiovascular disorders[14]. It is also reported that PARP1 was identified as a direct target gene of miR-223, which is overexpressed in BE-associated esophageal adenocarcinoma[15]. Here, we demonstrated that PARP1 contributes to genomic stability and oxidative stress injury, and positively regulates the viability of esophageal epithelial cells, which reveals a potential therapeutic strategy for BE.
GERD is a chronic inflammation of the esophagus stimulated by repeated acid/bile acids. Esophagitis, of course, is clinically the major pathological manifestation as the result from GERD, which is also characterized with BE[16]. It is known that metaplasia is a pathological condition that commonly occurs in the presence of chronic inflammation[17], including typical Barrett’s metaplasia[18]. In our mouse BE model, esophagitis and papillary hyperplasia of the squamous epithelia was observed, suggesting that the manifestation of GERD occurred. Furthermore, chronic inflammation is likely to carry an increased risk of cancer via oxidative damage pathways[19]. Thus, there is a strong association between GERD-induced ROS accumulation, chronic inflammatory infiltration, and BE formation.
Moreover, the BE mouse model also showed increased positivity for NF-κB, which is a transcription factor that has been established to play an active role in inflammation[20], and there is evidence that it serves as a key determinant of mucosal inflammation and protection[21]. In particular, NF-κB expression has been found in 40%-60% of biopsy specimens of Barrett metaplasia and in 61%-80% of Barrett’s adenocarcinomas, but in only 13% of reflux-injured squamous epithelia[22,23]. The level of NF-κB activation is correlated with the process from Barrett’s mucosal metaplasia to carcinogenesis, and indirectly proves that an inflammatory response might contribute to carcinogenesis[23]. This suggests the possibility that inflammation-mediated activation of the NF-κB pathway in a portion of cells, which may regulate the expression of NF-κB downstream proteins to replace the normal squamous cells for metaplasia and carcinogenesis.
Both the immunohistochemistry staining results and cell line results demonstrated that elevated expression of PARP1 is involved in the pathogenesis of BE. More importantly, PARP1 is known to be a co-activator of NF-κB and play a key role in pro-inflammation to contribute to inflammatory processes through the regulation of transcription factors[24,25]: NF-κB is one of the first mediators of inflammation to be identified as a target for PARP activity, which was proved to be a binding partner and also to be PARylated by PARP1. Further, NF-κB activity was greatly abrogated independent of upstream activation of NF-κB in PARP-/- mice and cell lines[26], and PARP1 inhibition reduced the extent of inflammation by modulating oxidative stress and impairing the activation of NF-κB and activator protein-1 in an inflammation model[27]. Both PAR and NF-κB staining increased in the BE mouse model, indicating the possible correlation between PARP1 activity and NF-κB. Further molecular mechanistic study in cells could clarify whether PARP1 regulates NF-κB activity in the oxidative damage-induced BE.
In conclusion, our study suggests that a high level of endogenous PARP1 in esophageal epithelial cells impairs oxidative DNA damage repair, inducing the death of esophageal epithelial cells during the process of BE formation. PARP inhibition treatment suppresses PARP activity and facilitates DNA damage repair in esophageal epithelial cells by prolonging PARylation. Thus, PARP1 acts as a mediator of esophageal disease, and PARylation may play a pivotal protective role as an antioxidant defense mechanism by maintaining esophageal epithelial cell function under pathological conditions of oxidative stress. Hence, PARP inhibitors may provide a novel clinical treatment for suppressing BE caused by oxidative damage.
Barrett’s esophagus is a major complication of gastro-esophageal reflux disease (GERD) and an important precursor lesion for the development of esophageal adenocarcinoma. However, the cellular and molecular mechanisms of Barrett’s metaplasia remain unclear. It has been demonstrated that poly(ADP-ribose) polymerases (PARPs)-associated ADP-ribosylation plays an important role in DNA damage and inflammatory response. Although PARP1-associated ADP-ribosylation has been examined both in vivo and in vitro, the function of PARP1 in esophageal epithelial cells and Barrett’s esophagus has not been illustrated.
In this study, the potential role of PARP1 and PARP1-related oxidative damage in Barrett’s esophagus was investigated.
The study investigated the potential role of PARP1 in oxidative damage in Barrett’s esophagus, which is urgent and essential for developing therapeutic targets.
Expression of PARP1 gene was analyzed using microarray analysis in patient esophageal tissue samples. A Barrett’s esophagus mouse model was established to examine the esophageal morphological changes and molecular changes. qPCR was used to examine the PARP1 expression in cell lines after treatment with H2O2 and bile acids (pH 4). To evaluate the impact of PARP1 activity on cell survival and DNA damage response after oxidative stress, immunofluorescence staining, comet assay, and annexin V staining were used.
High expression of PARP1 was associated with Barrett’s esophagus. Positive staining for NF-κB, γH2AX, and poly(ADP-ribose) was observed in the mouse model of Barrett’s esophagus. Knockdown of PARP1 decreased the cell viability following treatment with H2O2 and bile acids (pH 4). We further demonstrated that PARP1 inhibition could increase the oxidative damage as demonstrated by an increase in the levels of H2O2, intracellular reactive oxygen species, oxidative DNA damage, double-strand breaks, and apoptosis.
The dysfunction of PARP1 in esophageal epithelial cells increases the levels of reactive oxygen species and oxidative DNA damage, and downregulation of PARP1 or PARP1 inhibitor may be a potential therapeutic strategy for Barrett’s esophagus.
This study will provide an example for investigating the relationship between the oxidative DNA damage and Barrett’s metaplasia, and the underlying role of the PARP1 in Barrett’s esophagus. The direction of the future research is to provide more evidence for developing novel strategies by targeting PARP1 in Barrett’s esophagus. In our future research, the PARP1-related downstream signaling pathway (inflammation or DNA damage) will be tested in Barrett’s esophagus epithelial cells or animal models to observe the inhibitory effect of PARP1.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Zhao JB S- Editor: Wang JL L- Editor: Wang TQ E- Editor: Huang Y
| 1. | Nehra D, Howell P, Williams CP, Pye JK, Beynon J. Toxic bile acids in gastro-oesophageal reflux disease: influence of gastric acidity. Gut. 1999;44:598-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 297] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 2. | Herregods TV, Bredenoord AJ, Smout AJ. Pathophysiology of gastroesophageal reflux disease: new understanding in a new era. Neurogastroenterol Motil. 2015;27:1202-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 124] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 3. | Tack J, Pandolfino JE. Pathophysiology of Gastroesophageal Reflux Disease. Gastroenterology. 2018;154:277-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 218] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 4. | Eluri S, Shaheen NJ. Barrett’s esophagus: diagnosis and management. Gastrointest Endosc. 2017;85:889-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | Shaheen NJ. New Directions in Barrett’s Esophagus. Gastrointest Endosc Clin N Am. 2017;27:xv-xvi. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 6. | Cadet J, Wagner JR. Oxidatively generated base damage to cellular DNA by hydroxyl radical and one-electron oxidants: similarities and differences. Arch Biochem Biophys. 2014;557:47-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 7. | Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6531] [Cited by in RCA: 6485] [Article Influence: 259.4] [Reference Citation Analysis (0)] |
| 8. | Olinski R, Gackowski D, Foksinski M, Rozalski R, Roszkowski K, Jaruga P. Oxidative DNA damage: assessment of the role in carcinogenesis, atherosclerosis, and acquired immunodeficiency syndrome. Free Radic Biol Med. 2002;33:192-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 201] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 9. | Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 250] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 10. | Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1509] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 11. | Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951-1967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 600] [Cited by in RCA: 665] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 12. | Lesueur P, Chevalier F, Austry JB, Waissi W, Burckel H, Noël G, Habrand JL, Saintigny Y, Joly F. Poly-(ADP-ribose)-polymerase inhibitors as radiosensitizers: a systematic review of pre-clinical and clinical human studies. Oncotarget. 2017;8:69105-69124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 107] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 13. | Sriram CS, Jangra A, Kasala ER, Bodduluru LN, Bezbaruah BK. Targeting poly(ADP-ribose)polymerase1 in neurological diseases: A promising trove for new pharmacological interventions to enter clinical translation. Neurochem Int. 2014;76:70-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Xu S, Bai P, Little PJ, Liu P. Poly(ADP-ribose) polymerase 1 (PARP1) in atherosclerosis: from molecular mechanisms to therapeutic implications. Med Res Rev. 2014;34:644-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 15. | Streppel MM, Pai S, Campbell NR, Hu C, Yabuuchi S, Canto MI, Wang JS, Montgomery EA, Maitra A. MicroRNA 223 is upregulated in the multistep progression of Barrett’s esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res. 2013;19:4067-4078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 16. | Vaezi MF, Richter JE. Role of acid and duodenogastroesophageal reflux in gastroesophageal reflux disease. Gastroenterology. 1996;111:1192-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 340] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 17. | Kumar V, Abbas AK, Fausto N, Mitchell R. Robbins Basic Pathology. 8th ed: Publisher: Saunders/Elsevier 2007; . |
| 18. | Colleypriest BJ, Palmer RM, Ward SG, Tosh D. Cdx genes, inflammation and the pathogenesis of Barrett’s metaplasia. Trends Mol Med. 2009;15:313-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Farinati F, Piciocchi M, Lavezzo E, Bortolami M, Cardin R. Oxidative stress and inducible nitric oxide synthase induction in carcinogenesis. Dig Dis. 2010;28:579-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Baeuerle PA. IkappaB-NF-kappaB structures: at the interface of inflammation control. Cell. 1998;95:729-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 372] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 21. | Jobin C, Sartor RB. The I kappa B/NF-kappa B system: a key determinant of mucosalinflammation and protection. Am J Physiol Cell Physiol. 2000;278:C451-C462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 321] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 22. | Abdel-Latif MM, O’Riordan J, Windle HJ, Carton E, Ravi N, Kelleher D, Reynolds JV. NF-kappaB activation in esophageal adenocarcinoma: relationship to Barrett’s metaplasia, survival, and response to neoadjuvant chemoradiotherapy. Ann Surg. 2004;239:491-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 168] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 23. | O’Riordan JM, Abdel-latif MM, Ravi N, McNamara D, Byrne PJ, McDonald GS, Keeling PW, Kelleher D, Reynolds JV. Proinflammatory cytokine and nuclear factor kappa-B expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Gastroenterol. 2005;100:1257-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 24. | Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol Chem. 1999;380:953-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 237] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 25. | Liu L, Ke Y, Jiang X, He F, Pan L, Xu L, Zeng X, Ba X. Lipopolysaccharide activates ERK-PARP-1-RelA pathway and promotes nuclear factor-κB transcription in murine macrophages. Hum Immunol. 2012;73:439-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Oliver FJ, Ménissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446-4454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 452] [Cited by in RCA: 476] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 27. | Brunyánszki A, Hegedus C, Szántó M, Erdélyi K, Kovács K, Schreiber V, Gergely S, Kiss B, Szabó E, Virág L. Genetic ablation of PARP-1 protects against oxazolone-induced contact hypersensitivity by modulating oxidative stress. J Invest Dermatol. 2010;130:2629-2637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |