Published online Feb 14, 2018. doi: 10.3748/wjg.v24.i6.657
Peer-review started: December 21, 2017
First decision: December 27, 2017
Revised: January 3, 2018
Accepted: January 18, 2018
Article in press: January 18, 2018
Published online: February 14, 2018
Processing time: 47 Days and 9.9 Hours
To establish the relationship of protein tyrosine phosphatase non-receptor type 2 and 22 (PTPN2/22) polymorphisms and mycobacterial infections in Crohn’s disease (CD).
All 133 subjects’ blood samples were genotyped for nine single nucleotide polymorphisms (SNPs) in PTPN2/22 using TaqMan™ genotyping, while the effect of the SNPs on PTPN2/22 and IFN-γ gene expression was determined using RT-PCR. Detection of Mycobacterium avium subspecies paratuberculosis (MAP) IS900 gene was done by nPCR after DNA extraction from the isolated leukocytes of each subjects’ blood samples. T-cells isolated from the patient samples were tested for response to phytohematoagglutonin (PHA) mitogen or mycobacterial antigens by BrdU proliferation assays for T-cell activity.
Out of the nine SNPs examined, subjects with either heterozygous (TC)/minor (CC) alleles in PTPN2:rs478582 occurred in 83% of CD subjects compared to 61% healthy controls (P-values < 0.05; OR = 3.03). Subjects with either heterozygous (GA)/minor (AA) alleles in PTPN22:rs2476601 occurred in 16% of CD compared to 6% healthy controls (OR = 2.7). Gene expression in PTPN2/22 in CD subjects was significantly decreased by 2 folds compared to healthy controls (P-values < 0.05). IFN-γ expression levels were found to be significantly increased by approxiately 2 folds in subjects when either heterozygous or minor alleles in PTPN2:rs478582 and/or PTPN22:rs2476601 were found (P-values < 0.05). MAP DNA was detected in 61% of CD compared to only 8% of healthy controls (P-values < 0.05, OR = 17.52), where subjects with either heterozygous or minor alleles in PTPN2:rs478582 and/or PTPN22:rs2476601 had more MAPbacteremia presence than subjects without SNPs did. The average T-cell proliferation in CD treated with PHA or mycobacteria antigens was, respectively, 1.3 folds and 1.5 folds higher than healthy controls without any significant SNP.
The data suggests that SNPs in PTPN2/22 affect the negative regulation of the immune response in CD patients, thus leading to an increase in inflammation/apoptosis and susceptibility of mycobacteria.
Core tip: Knowledge of the pathophysiology of Crohn’s disease (CD) is vital in the development of new diagnosis techniques and treatments for the disease. Our study involves the investigation of single nucleotide polymorphisms (SNPs) in protein tyrosine phosphatase non-receptor type 2 and 22 (PTPN2/22) and their effects on susceptibility to mycobacteria species and the elevation of pro-inflammatory cytokines. Our data demonstrates that SNPs in PTPN2/22 lead to less negative regulation in T-cells and increase susceptibility to mycobacteria, thus increasing inflammation and apoptosis in intestinal tissues. Personalized treatment could be accomplished by genetic testing and antibiotic treatment for mycobacteria in CD patients.
- Citation: Sharp RC, Beg SA, Naser SA. Role of PTPN2/22 polymorphisms in pathophysiology of Crohn’s disease. World J Gastroenterol 2018; 24(6): 657-670
- URL: https://www.wjgnet.com/1007-9327/full/v24/i6/657.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i6.657
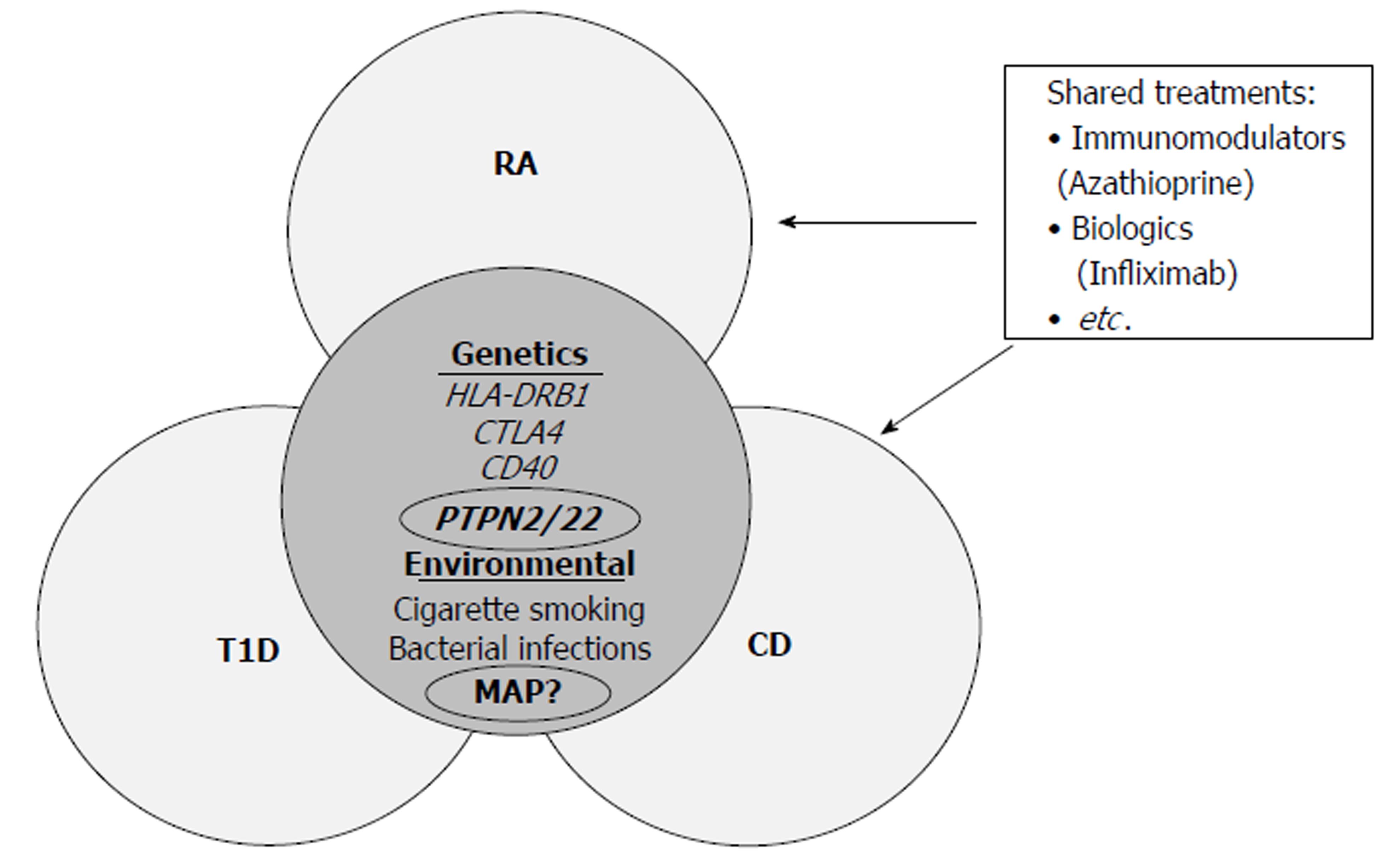
Single nucleotide polymorphisms (SNPs) play a significant role in the pathogenic process of inflammatory autoimmune disorders. These SNPs affect several immunity genes, leading to an overactive immune system. Consequently, self-tolerance mechanisms fail in a variety of immune cells including T-cells, B-cells, and antigen-presenting cells[1,2]. Along with these genetic defects, environmental factors such as bacterial and viral infections have also been associated with inflammatory autoimmune disorders. These factors trigger phenotypical response to occur in the defected immune cells[1-3]. Some of these diseases, such as Rheumatoid Arthritis (RA), Type 1 Diabetes (T1D), and Crohn’s Disease (CD), share some of the same genetic SNPs with each other[1-4]. An example of regulatory immune genes that these diseases share SNPs in are in the protein tyrosine phosphatases non-receptor type 2 (PTPN2) and type 22 (PTPN22) genes[1-5]. PTPN2/22 are genes found more frequently in T-cells, where they encode enzymatic phosphatase proteins (PTPN2/22) that negatively regulate the T-cell receptor (TCR)[4,5]. PTPN2 and its protein product (PTPN2) are also found in a majority of epithelial cell types including synovial joint tissue, β-cells, and intestinal tissues, where they control apoptosis and chemokine production[4,5]. SNPs in PTPN2/22 have been hypothesize to cause a dysregulation of the immune system that is brought upon by overactive T-cells and increased pro-inflammatory cytokine production due to lack of negative regulation[1-5].
With a majority of inflammatory autoimmune disorders sharing the same genetic pre-dispositions, it is possible that the pathogenesis of these disorders could also share some of the same common environmental triggers with each other as well (Figure 1)[6-10]. Recent studies have shown that Mycobacterium avium subspecies paratuberculosis (MAP) infections have been associated with a variety of different inflammatory disorders including CD[4,11-14]. Mycobacterial infections causes problems in these inflammatory autoimmune patients when the patient is genetically predisposed, causing the immune system to become dysregulated[4,11-14]. This dysregulation will lead to high amounts of pro-inflammatory cytokines, production of autoantibodies, and high amounts of apoptosis occurring in a variety of cell types, thus leading to chronic inflammation[4,11-14].
In addition to sharing the same genetic predispositions and environmental triggers, many inflammatory autoimmune disorders share the same medical treatments as well. For instance, anti-TNF-α therapeutics such as adalimumab and infliximab are used for RA and CD[6,7]. However, anti-TNF-α medications along with non-steroid anti-inflammatory drugs (NSAIDs), glucocorticoids, and other disease-modifying drugs cause several side effects[6-10]. These side effects include osteoporosis, hypertension, GI intolerance, autoantibodies against medications, and increased risk of developing opportunistic infections, especially mycobacterial infections[6-10]. With the undesirable side effects of these medications, it is important that inflammatory autoimmune disorders pathogeneses is thoroughly examined in order to develop more accurate detection of disease and to develop more personal treatment with little side effects.
In this study, we focus on the pathogenesis of CD, where we explore the effect of both the genetic predisposition of SNPs in PTPN2/22 and the environmental trigger of MAP infection. We hypothesize that SNPs in PTPN2/22 lead to loss of negative regulation in T-cells and, with a MAP infection, increases production of pro-inflammatory cytokines such as IFN-γ. This leads to an increase inflammation and apoptosis in the intestinal tissues of CD patients.
A total of 133 consented CD subjects and healthy controls donated two to three 4.0 mL K2-EDTA coded blood tubes for us in this study. The study was approved by the University of Central Florida Institutional Review Board #IRB00001138. Each subject completed and signed a written consent form before samples were collected. Healthy control subjects completed a survey that question if said subjects had any medical abnormality (CD, T1D, RA or “other diseases”). No healthy control subjects had any type of medical conditions to the best of their knowledge. The severities of the CD subjects’ symptoms were scored from moderate to severe symptoms. The average age of CD subjects was 39.6 ± 14.3 with a gender ratio of 48.6% male and 51.4% female. The average age of healthy controls was 30.7 ± 13.4 with a gender ratio of 41.9% male and 58.1% female subjects. Table 1 lists age, gender and other demographic information for all CD subjects in this study. From the blood tubes, the following procedures were done to the samples: PTPN2/22 genotyping, gene expression profiling, MAP IS900 nested PCR (nPCR) detection, and T-cell proliferation assays.
| Sample code | Gender | Age | Diagnosis | MAP +/- | PTPN2:rs478582 | PTPN22:rs2476601 |
| RCS1 | M | 50 | CD | - | TC | GA |
| RCS2 | F | 25 | CD | - | TC | GA |
| RCS3 | F | 68 | CD | + | TC | GG |
| RCS4 | M | 26 | CD | + | CC | GG |
| RCS5 | F | 56 | CD | + | CC | GG |
| RCS6 | NA | NA | CD | + | TC | GG |
| RCS7 | M | 60 | CD | + | TC | GG |
| RCS8 | M | 43 | CD | + | TC | GG |
| RCS9 | F | 54 | CD | - | CC | GG |
| RCS10 | F | 31 | CD | NA | TC | GG |
| RCS11 | M | 21 | CD | + | NA | GG |
| RCS12 | M | 25 | CD | + | CC | GG |
| RCS13 | F | 40 | CD | + | TC | GG |
| RCS14 | M | 36 | CD | + | TC | GG |
| RCS15 | NA | NA | CD | - | CC | GA |
| RCS16 | F | 25 | CD | + | TC | GG |
| RCS17 | F | 27 | CD | + | TC | GG |
| RCS18 | M | 20 | CD | - | TT | GG |
| RCS19 | M | 25 | CD | + | CC | GA |
| RCS20 | F | 41 | CD | - | TC | GG |
| RCS21 | M | 20 | CD | - | TT | GG |
| RCS22 | M | 40 | CD | - | TC | GG |
| RCS23 | M | 30 | CD | - | TC | GG |
| RCS24 | F | 60 | CD | + | TC | GG |
| RCS25 | F | 39 | CD | + | TT | GG |
| RCS26 | F | 30 | CD | + | CC | GA |
| RCS27 | F | 43 | CD | + | CC | GG |
| RCS28 | M | 30 | CD | + | TC | GA |
| RCS29 | M | 28 | CD | + | TC | GG |
| RCS30 | M | 66 | CD | + | TT | GG |
| RCS31 | M | 53 | CD | - | TT | GG |
| RCS32 | M | 28 | CD | - | TC | GA |
| RCS33 | F | 38 | CD | + | CC | GG |
| RCS34 | M | 44 | CD | - | CC | GA |
| RCS35 | M | 53 | CD | - | TC | GG |
| RCS36 | M | 24 | CD | + | TC | GG |
| RCS37 | F | 51 | CD | + | TC | GG |
| RCS38 | F | 46 | CD | + | TC | GG |
| RCS39 | M | 24 | CD | - | CC | GG |
| RCS40 | F | 63 | CD | + | TC | GG |
| RCS41 | F | 25 | CD | - | TC | GG |
| RCS42 | F | 66 | CD | - | TC | GG |
| RCS43 | F | 27 | CD | + | TC | GG |
| RCS44 | F | 25 | CD | + | TC | GG |
| RCS45 | F | 38 | CD | + | TC | GG |
| RCS46 | F | 26 | CD | - | CC | AA |
| RCS47 | M | 54 | CD | + | TT | GA |
| RCS48 | F | 31 | CD | + | TC | GG |
| RCS49 | M | 56 | CD | - | CC | GG |
| RCS50 | F | 53 | CD | - | TC | GG |
| RCS51 | F | 51 | CD | - | TT | GA |
| RCS52 | F | 23 | CD | + | TC | GG |
| RCS53 | M | 26 | CD | + | TC | GG |
| RCS54 | M | 38 | CD | - | TT | GG |
| RCS55 | F | 31 | CD | + | TC | GG |
| RCS56 | M | 61 | CD | + | TC | GG |
| RCS57 | F | 24 | CD | + | TC | GG |
| RCS58 | M | 57 | CD | - | CC | GG |
| RCS59 | F | 30 | CD | + | TT | GG |
| RCS60 | M | 51 | CD | - | CC | GG |
| RCS61 | F | 55 | CD | - | CC | GG |
| RCS62 | F | 61 | CD | - | TT | GG |
| RCS63 | F | 31 | CD | + | TC | GG |
| RCS64 | F | 56 | CD | NA | TC | GG |
| RCS65 | M | 25 | CD | + | NA | NA |
| RCS66 | F | 53 | CD | + | NA | NA |
| RCS67 | M | 30 | CD | - | TC | GG |
| RCS68 | F | 49 | CD | - | CC | GG |
| RCS69 | M | 28 | CD | + | TT | GG |
| RCS70 | M | 26 | CD | + | TT | GG |
| RCS71 | M | 26 | CD | + | CC | GG |
| RCS72 | M | 58 | CD | + | CC | GG |
TaqMan™ SNP Genotyping Assays (Applied Biosystems™) were used to genotype nine SNPs in PTPN2/22 from the isolated DNA from subjects’ blood samples. Samples and reagents were sent to the University of Florida Pharmacotherapy and Translational Research Department (Gainesville, FL) to perform genotyping assays. Out of the nine SNPs, four SNPs were specific to PTPN2 that includes rs1893217, rs2542151, rs7234029, rs478582 along with five SNPs that were specific to PTPN22 that includes rs2476601, rs2488457, rs33996649, rs34209542, rs2476599. Briefly, DNA was extracted from whole blood samples using QIAamp® DNA Blood Mini Kit (Qiagen™) following manufacturer’s protocol. TaqMan™ genotyping assays for PTPN2/22 SNPs were performed on DNA samples following manufacturer protocol (Applied Biosystems™). Briefly, DNA samples and the TaqMan™ SNP Genotyping Assays mixtures (primers with Vic and Fam fluorophore attachment) were transferred into a 384-well plate along with 2 × TaqMan™ Master Mix and 20 × Assay Working Stock in each well. Plates were treated to an RT-PCR protocol consisting of 95 °C for 10 min for 1 cycle, 92 °C for 15 s and 58 °C for 1 min for 50 cycles. The plates were then read for VIC (551 nm) and FAM (517 nm) fluorescence, where VIC or FAM alone determined allele 1 or allele 2 in the samples, while VIC and FAM together determined heterozygous for each allele in the samples.
Gene expression of PTPN2/22 and IFN-γ was performed by converting RNA from subjects’ whole blood samples to cDNA and performing RT-PCR. RNA from the subjects’ blood samples were isolated from peripheral leukocytes via TRIzol® Reagent (Invitrogen) per manufacturer’s instruction. Briefly, 1.0 mL of whole blood from subjects’ samples were transferred into a microcentrifuge tubes and centrifuged for 3000 rpm for 15 min until the leukocytes formed a buffy coat layer, which was then transferred to new 2.0 mL RNase free microcentrifuge tubes. Tubes containing the leukocytes from subjects’ samples were then suspended in 1.0 mL of TRIzol®, where the tubes were incubated and gently rocked for 15 min at room temperature. Next, 0.2 mL of chloroform was then mixed in each tube and then incubated at room temperature for 3 min. Tubes were then centrifuged at 11400 rpm for 15 min at 4 °C, where afterwards the upper aqueous phase containing RNA was transferred to new 2.0 mL RNase free microcentrifuge tubes. Next, 0.5 mL of 100% isopropanol was added to the tubes containing subjects’ RNA samples, where they were incubated at room temperature for 10 min. Tubes were then centrifuged at 11400 rpm for 10 min at 4 °C, where afterwards the RNA pellets were washed in 1 mL of 75% ethanol. Washed RNA pellets were then centrifuged for 8700 rpm for 5 min at 4 °C and then air-dried until fully dried. Dried RNA pellets were then suspended in 20 μL of RNase free H2O and boiled to 60 °C for 10 min.
Conversion of RNA to cDNA was done following the iScript™ Reverse Transcription (Bio-Rad®) manufacturer’s instruction. RNA concentration from each subjects’ samples were first quantified via NanoDrop ND-1000 Spectrophotometer (ThermoFisher Scientific®) and then diluted to 600 ng of total RNA. Next, diluted RNA samples were then added to PCR reaction tubes that contained 0.2 mL PCR reaction, 4 μL of iScript™ Reverse Transcription (Bio-Rad®), and up to 20 μL RNase free H2O. The PCR reaction tubes then underwent a PCR protocol consisting of 5 min at 25 °C, 20 min at 46 °C and 1 min at 95 °C, where the final concentration of cDNA for each sample was 30 ng/μL.
For the RT-PCR reaction, 1 μL of cDNA (30 ng) was added to a 96-well microamp plates along with 10 μL of Fast SYBR Green Mastermix (ThermoFisher Scientific®), 1 μL of PrimePCR SYBR Green Assay mix (Bio-Rad®) specific to target gene, and 8 μL of sterile H2O. For the positive control for the RT-PCR reactions, the 18s RNA gene was the target to determine if the reaction work and to obtain baseline CT readings. The oligonucleotide primers for the 18s RNA gene that were used for the RT-PCR reaction was the following: forward primer: 5’-GTA ACC CGT TGA ACC CCA TT-3’ and reverse primer: 5’-CCA TCC AAT CGG TAG TAG CG-3’. RT-PCR reactions were performed using the 7500 Fast Real-Time PCR System (Applied Biosystems®), where relative gene expression levels were calculated using ∆CT (sample gene CT reading-18s RNA gene CT baseline reading) and using the equation (2(∆CT) × 1000).
MAP IS900 DNA was detected via nPCR from cultured peripheral leukocytes that were isolated from the subjects’ blood samples as described previously[15]. Briefly, subjects’ blood sample tubes were centrifuged for 3000 rpm for 10 min at room temperature, where the buffy coat layer containing peripheral leukocytes was present and transferred to new sterile 2.0 mL microcentrifuge tubes. The peripheral leukocytes were then washed twice by adding double the volume of red cell lysis buffer (ammonium chloride solution, G-Biosciences®) to each tube and incubating/gently rocking for 10 min and then centrifuged at 5000 rpm for 5 min at room temperature. The supernatant from each subjects’ samples were then removed and the isolated peripheral leukocyte pellets were re-suspended in Tris-EDTA (TE) buffer. The isolated pellets were then cultured in BD Bactec™ MGIT™ Para-TB medium (Becton, Dickinson and Company©) tubes supplemented with 800 uL of Bactec™ MGIT™ Para-TB Supplement (Becton, Dickinson and Company©) for six months at 37 °C in a BD Bactec™MGIT™ 320 Analyzer (Becton, Dickinson and Company©).
After six months of culturing, subjects’ cultured samples underwent DNA extraction by using a modified DNAzol® (ThermoFisher Scientific®) extraction protocol as follows. A 2.0 mL sampling of culture from each subjects’ tubes were obtained and pipetted into new sterile 2.0 mL microcentrifuge tubes. The tubes were then centrifuged at 13000 rpm for 2.5 min, where afterwards the supernatant was discarded from the tubes and the culture pellets were saved. The subjects’ culture pellet tubes were then mixed with 1.0 mL DNAzol® reagent and then mixed with 400 μL of 100% isopropanol. The tubes were then incubated for 15 min at room temperature followed by centrifugation at 8000 rpm for 6 min, where afterwards the supernatant was discarded, leaving a DNA pellet. DNA pellets from the subjects’ samples were then washed once with 500 μL DNAzol® reagent and centrifuged at 8000 rpm for 5 min. Supernatant was then discarded from the tubes and the DNA pellets were then washed again with 1.0 mL of 75% ethanol, where they were centrifuged at 8000 rpm for 5 min. DNA pellets were then dried after supernatant was removed via speedvac for 5 min. The dried DNA pellets were then dissolved in 50 μL of TE buffer.
MAP IS900 DNA was then detected in each subjects’ samples by the use of our nPCR protocol and nucleotide primers as described previously[15]. Subjects were considered to have MAP presence when a 298 bp band on a 2% agarose gel is shown after nPCR reaction. The positive MAP DNA control that was used originated from our laboratory cultured clinical strain UCF4, which was isolated from a CD patient. The negative controls for each PCR step that was used contained all PCR reagents except for the DNA template used in the reactions.
T-cells were fully isolated from subjects’ whole blood samples by the use of RosetteSep™ Human T-cell Enrichment Cocktail (StemCell™ Technology) as per manufacture’s instruction. For the T-cell isolation and proliferation assays, the entire T-cell populations were examined in this study and were not segregated by subpopulations. Briefly, 50 μL/mL of RosetteSep™ Human T-cell Enrichment Cocktail was added to each subjects’ whole blood samples and was incubated at 20 min at room temperature. Samples were then diluted with equal volumes of PBS with 2% fetal bovine serum (FBS, Sigma-Aldrich®) and mixed gently. The mixtures from each subjects’ samples were then layered on top of a Lymphoprep™ (Axis-Shield®) density medium in a separated tube and centrifuged for 20 min at 2500 rpm at room temperature. Separated T-cells from each subjects’ samples were then found on top of the density medium layer and were collected into new sterile 2.0 mL microcentrifuge tubes and washed twice with PBS with 2% FBS.
Subjects’ isolated T-cells were then plated on a 96-well plate, where T-cell proliferation assays were done using bromodeoxyuridine (BrdU) labeling proliferation ELISA kit (Roche Molecular Biochemicals®) as described previously[16]. To stimulate the subjects’ isolated T-cells, phytohematoagglutunin (PHA) was used as a positive control mitogen. The test mitogen used in the T-cell proliferation assays was purified protein derivative-like (PPD-like) from UCF4 MAP bacterial cultures that were prepared by purification of supernatant from sonicated protein extract. Briefly, 1 × 105 isolated T-cells from each subjects’ samples were transferred in triplicates to 96-well plates and incubated in the following conditions: RPMI-1640 (Sigma-Aldrich®) only, PHA (10 μg/mL, Sigma-Aldrich®) or MAP PPD-like (5 μg/mL) along with respected subjects’ plasma. The plates were then incubated for 72 h at 37 °C and 5% CO2 and then labeled with 20 μL/well of BrdU and incubated again for 24 h at 37 °C and 5% CO2. The T-cell proliferation assay was done through the Roche BrdU proliferation ELISA kit as described previously[16]. Relative T-cell proliferation levels of samples were compared to the control group (isolated T-cells in RPMI only) by examining the fold change in the absorbance reading of each well at 450 nm.
Samples were analyzed for significance using unpaired, two-tailed t-tests; unpaired, two-tailed z-scores; and odds ratio. GraphPad Prism 7 was used for statistical analysis and creation of graphs. P-values < 0.05 were considered significant.
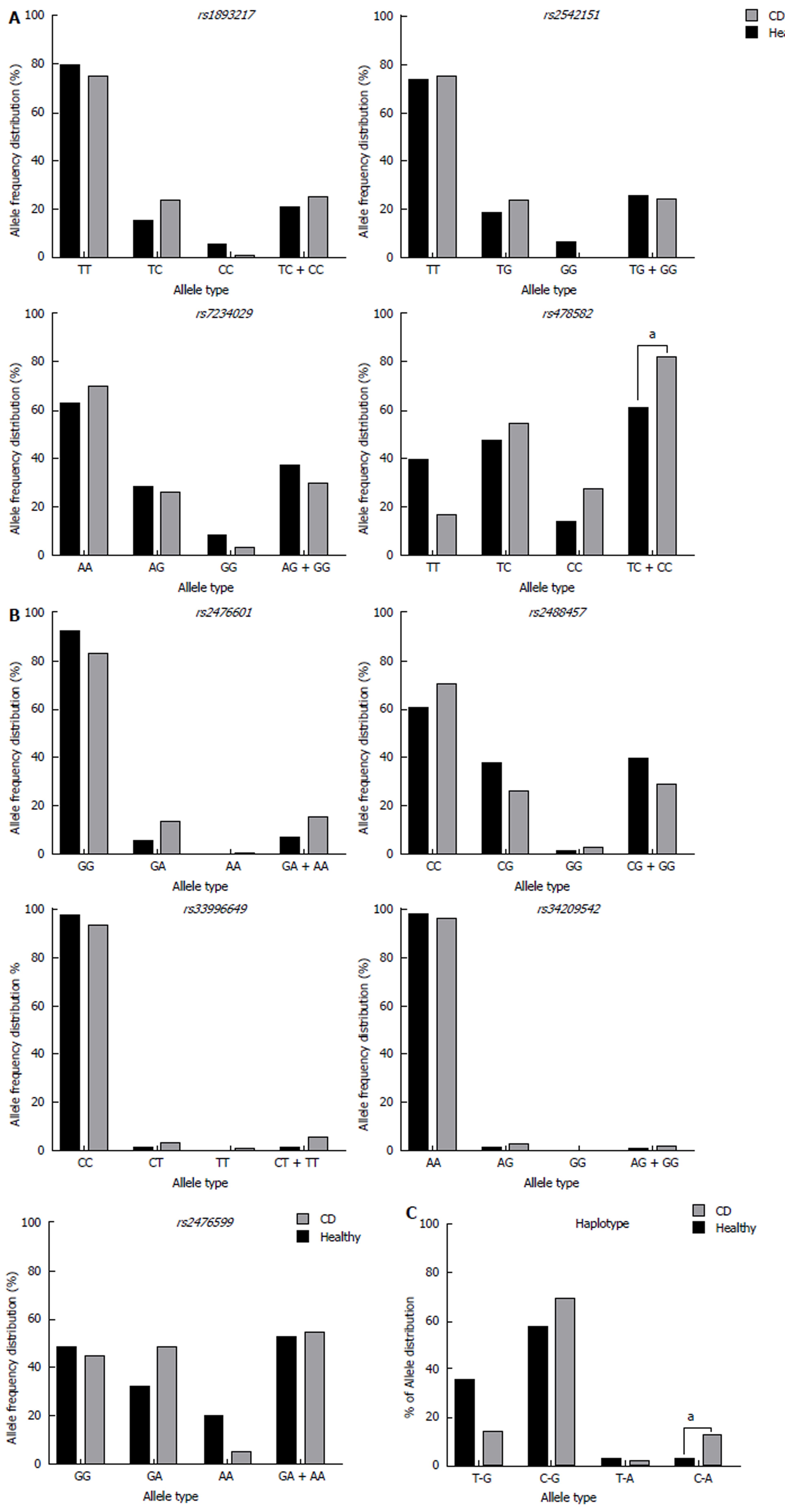
Allele frequency of the nine SNPs examined in PTPN2/22 found in both CD subjects and healthy controls are shown in Figure 2. All genotyped samples were found in Hardy-Weinberg equilibrium. Out of the four SNPs found in PTPN2 (rs1893217, rs2542151, rs7234029, and rs478582), rs478582 was significant in the CD, where heterozygous (TC) or minor (CC) alleles when examined together were detected in 57/69 (82.6%) in CD compared to 36/59 (61.0%) healthy controls (OR = 3.03, 95%CI: 1.35-6.84, P-values < 0.05, Figure 2A). Specifically, the heterozygous (TC) alleles were detected in 38/69 (55.1%) CD compared to the 28/59 (47.5%) of healthy controls, while homozygous (CC) alleles were detected in 19/69 (27.5%) CD compared to 8/59 (13.6%) healthy controls. SNPs rs1893217, rs2542151, and rs7234029 were found to be not significant in CD compared to the healthy controls. Out of the five SNPs specific to PTPN22 (rs2476601, rs2488457, rs33996649, rs34209542, and rs2476599), none of SNPs were considered significant in CD compared to the healthy controls (Figure 2B). However, since PTPN22:rs2476601 is found significantly in various inflammatory autoimmune diseases, we continued to investigate the SNP in more detail along with PTPN2:rs478582[3-5,17-19]. For PTPN22:rs2476601, CD with either heterozygous (GA) or minor (AA) alleles were detected in 11/70 (15.7%) subjects, while 4/62 (6.45%) was detected in healthy controls (OR = 2.7, 95%CI: 0.81-8.98, P-values > 0.05). Specifically, the heterozygous (GA) alleles were detected in 10/70 (14.3%) CD compared to the 4/62 (6.45%) of healthy controls, while homozygous (AA) alleles were rare in all samples.
For confirmation that CD subjects were significant in having SNP alleles for PTPN2:rs478582 and PTPN22:rs2476601, determination of haplotype combinations were done (Figure 2C). Examination of the following haplotype combinations between PTPN2:rs478582 and PTPN22:rs2476601 were examined: T-G, C-G, T-A, and C-A. The T-G haplotype (major/major) was found more significantly in the healthy controls (21/59 = 35.6%) than in CD (10/69 = 14.5%, P-values < 0.05). The C-G haplotype (heterozygous or minor/major) and the C-A (heterozygous or minor/heterozygous or minor) were found more in CD (48/69 = 69.6%; 9/69 = 13.0%, respectively) than in healthy controls (34/59 = 57.6%; 2/59 = 3.39%, respectively). The C-A haplotype was found more significantly in CD than the healthy controls (P-values < 0.05).
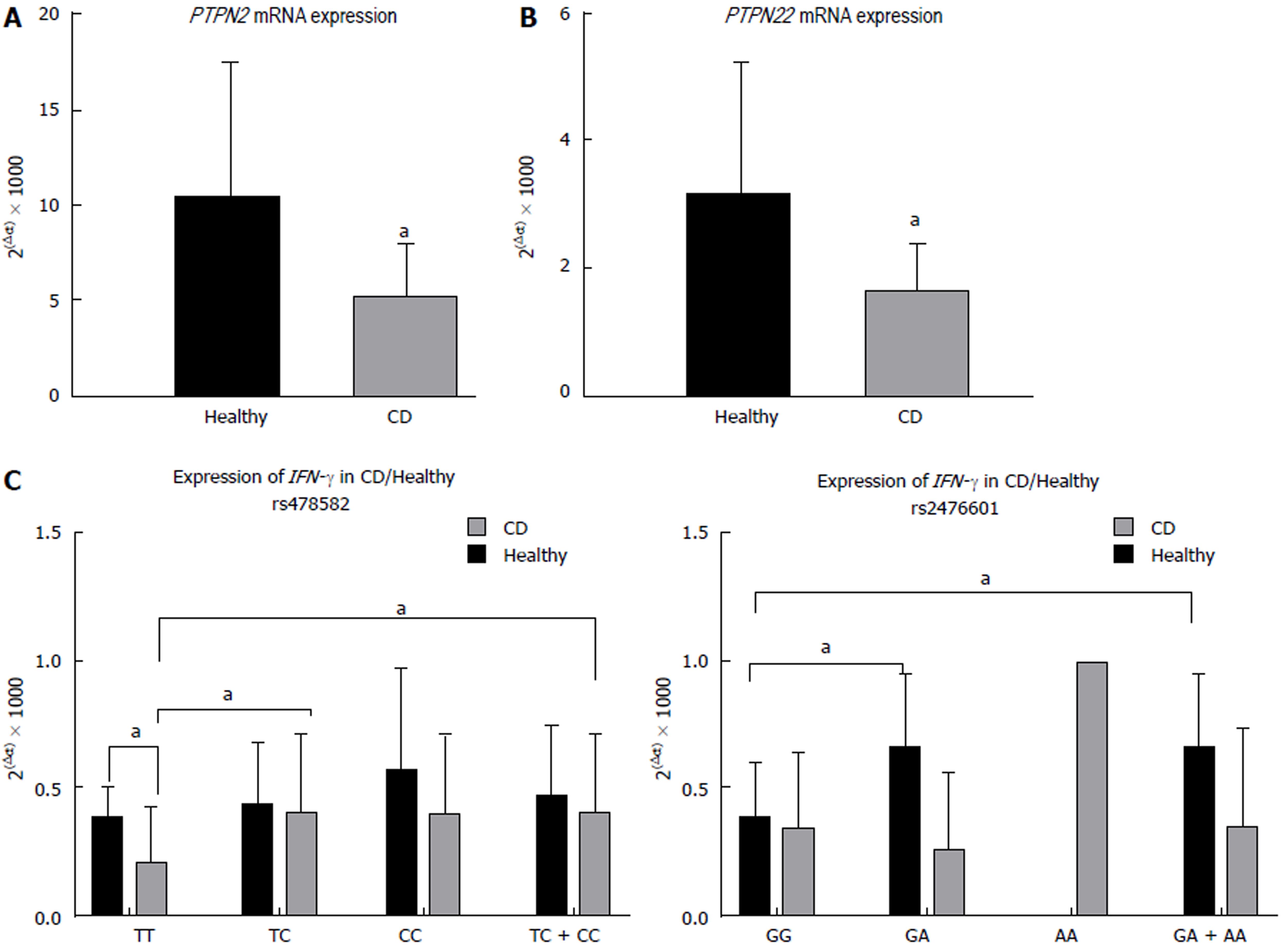
The average relative gene expression (2(-∆CT) × 1000) of PTPN2, regardless of SNPs, in CD was significantly lower (5.27 ± 2.68, n = 38) than in healthy controls (10.5 ± 6.95, n = 30, P-values < 0.05, Figure 3A). Similarly, the average relative gene expression of PTPN22, regardless of SNPs, was also significantly lower in CD (1.76 ± 1.12, n = 38) than in healthy controls (3.24 ± 1.84, n = 30, P-values < 0.05, Figure 3B). The evaluation of the effect of PTPN2:rs478582 and PTPN22:rs2476601 on expression of PTPN2/22 and IFN-γ was determined.
For subjects with either heterozygous (TC) or minor (CC) alleles in PTPN2:rs478582, regardless of disease, expression of PTPN2 did not change when compared to the normal (TT) subjects. However, when examining the CD and healthy control subjects in each allele group, CD overall had a lower average relative gene expression of PTPN2. The average relative gene expression in CD with heterozygous (TC) or minor (CC) alleles in PTPN2:rs478582 was significantly lower (5.34 ± 2.77, n = 31) compared to 10.2 ± 7.15 (n = 21) in healthy controls with similar SNPs (P-values < 0.05). Specifically, when examining subjects with heterozygous (TC) alleles in PTPN2:rs478582, CD average relative gene expression was 5.22 ± 2.57 (n = 22), which was significantly lower than the healthy controls with heterozygous (TC) alleles (10.5 ± 7.15, n = 17, P-values < 0.05). When examining subjects with homozygous (CC) alleles in PTPN2:rs478582, CD average relative gene expression was 5.64 ± 3.37 (n = 9), which was lower than the healthy controls with homozygous (CC) alleles (8.89 ± 8.03, n = 4).
For subjects with either heterozygous (GA) or minor (AA) alleles in PTPN22:rs2476601, regardless of disease, expression of PTPN22 did not change when compared to the normal (GG) subjects. However, when examining the CD and healthy control subjects in each allele group, CD overall had a lower average relative gene expression of PTPN22. The average relative gene expression in CD with heterozygous (GA) or minor (AA) alleles in PTPN22:rs2476601 was significantly lower (1.58 ± 0.93, n = 6) compared to 3.40 ± 1.19 (n = 4) in healthy controls with similar SNPs (P-values < 0.05). Specifically, when examining subjects with heterozygous (GA) alleles in PTPN22:rs2476601, CD average relative gene expression was 1.48 ± 1.00 (n = 5), which was significantly lower than the healthy controls with heterozygous (GA) alleles (3.40 ± 1.19, n = 4, P < 0.05). Minor (AA) alleles in PTPN22:rs2476601 was rare in all subjects.
Correlation analyses were performed to determine if expression of relative gene expression of IFN-γ changed in subjects with PTPN2:rs478582 or PTPN22:rs2476601 (Figure 3C and 3D, respectively). The average relative gene expression of IFN-γ in CD subjects with the PTPN2:rs478582 heterozygous (TC) or minor (CC) allele was 0.41 ± 0.31 (n = 38), which was significantly higher compared to the CD subjects with normal (TT) alleles (0.21 ± 0.22, n = 12, P < 0.05). Specifically, CD subjects with the heterozygous (TC) allele had significantly higher (0.41 ± 0.31, n = 24, P < 0.05) IFN-γ relative gene expression than CD subjects with normal (TT) alleles, while CD subjects with the minor (CC) alleles had higher gene expression as well (0.40 ± 0.31, n = 14). There was no significant change in IFN-γ relative gene expression in the CD subjects with the PTPN22:rs2476601 heterozygous (GA) or minor (AA) alleles. However, in healthy controls, subjects with the heterozygous (GA) or minor (AA) alleles had a significantly higher gene expression (0.67 ± 0.28, n = 4, P < 0.05) than healthy controls with normal (GG) alleles (0.40 ± 0.21, n = 20).
Overall detection of MAP IS900 DNA was found in CD and healthy control subjects and were correlated with PTPN2:rs478582 and PTPN22:rs2476601 (Table 2). Out of 70 CD subjects, 43 (61.4%) were positive for MAPbacteremia compared to only 4/48 (9.33%) of healthy controls (P < 0.05, OR = 17.5, 95%CI: 5.65-54.3).
| MAP presence | |||
| Healthy | CD | OR (95%CI) | |
| Overall | 4/48 (9.33%) | 43/70 (61.4%)a | 17.5 (5.65–54.3)a |
| rs478582 | |||
| TT | 2/17 (11.8%) | 6/12 (50%)a | 7.5 (1.17-48.2)a |
| TC | 0/22 (0%) | 25/37 (67.6%) | 91.8 (5.14-1604.3)a |
| CC | 2/8 (25%) | 9/19 (47.4%) | 2.7 (0.43-16.9) |
| TC + CC | 2/30 (6.67%) | 34/56 (60.7%)a | 21.6 (4.68-100.1)a |
| rs2476601 | |||
| GG | 4/59 (6.78%) | 33/59 (55.9%)a | 17.6 (5.59-54.4)a |
| GA | 0/4 (0%) | 3/10 (30.0%) | 4.2 (0.17-101.5) |
| AA | 0/1 (0%) | 0/1 (0%) | 1.00 (0.02-92.4) |
| GA + AA | 0/5 (0%) | 3/11 (27.3%) | 4.53 (0.19-105.8) |
| Haplotypes | |||
| T-G | 2/15 (13.3%) | 5/10 (50.0%)a | 6.5 (0.94-45.1)a |
| C-G | 2/29 (6.90%) | 31/46 (67.4%)a | 30.0 (6.3-142.6)a |
| T-A | 0/2 (0%) | 1/2 (50.0%) | 5.00 (0.11-220.6) |
| C-A | 0/2 (0%) | 3/9 (33.3%) | 2.69 (0.1-73.2) |
Correlation analyses with PTPN2:rs478582 and PTPN22:rs2476601 along with MAP infection was done on CD and healthy controls to see if these SNPs increase MAP susceptibility (Table 2). For CD subjects with heterozygous (TC) or minor (CC) alleles in PTPN2:rs478582, 34/56 (60.7%) had MAPbacteremia presence compared to only 2/30 (6.67%) in healthy controls with similar SNPs (P < 0.05, OR = 21.6, 95%CI: 4.68-100.1). Specifically, CD subjects with heterozygous (TC) alleles in PTPN2:rs478582 was 25/37 (67.6%) compared to 0/22 (0.00%) in healthy controls with heterozygous (TC) alleles (P < 0.05, OR = 91.8, 95%CI: 5.14-1640.3). The CD subjects with heterozygous (TC) or minor (CC) alleles group (34/56 = 60.7%) and CD subjects with heterozygous (TC) allele group (25/37 = 67.6%) in PTPN2:rs478582 had higher MAPbacteremia compared to CD subjects with normal (TT) alleles (6/12 = 50%).
For CD subjects with heterozygous (GA) alleles in PTPN22:rs2476601, 3/10 (30.0%) had MAPbacteremia compared to 0/4 (0.00%) in healthy controls with heterozygous (GA) alleles (OR = 4.2, 95%CI: 0.17-101.5). Presence of MAPbacteremia was rare in all subjects with the minor (AA) allele.
Correlation of haplotype combinations of PTPN2:rs478582 and PTPN22:rs2476601 alleles on susceptibility to MAPbacteremia was analyzed, where CD subjects with the C-G haplotype (heterozygous or minor/major) had 31/46 (67.4%) with MAPbacteremia presence compared to 2/29 (6.90%) of healthy controls with the C-G haplotype (P-values < 0.05, OR = 30.0, 95%CI: 6.3-142.6). The T-A haplotype (major/heterozygous or minor) and the C-A haplotype (heterozygous or minor/heterozygous or minor) was rare in all samples. However, CD subjects with the T-A haplotype had 1/2 (50.0%) with MAPbacteremia presence compared to the 0/2 (0.00%) in healthy controls with the T-A haplotype, while CD subjects with the C-A haplotype had 3/9 (33.3%) with MAPbacteremia presence compared to the 0/2 (0.00%) in healthy controls with the C-A haplotype.
When examining CD and healthy control subjects with or without MAPbacteremia presence alone, there was no change in PTPN2/22 and IFN-γ relative gene expression when examining correlation data. However, PTPN2 was significantly lower in CD subjects than in the health control subjects regardless of MAPbacteremia presence or not. CD subjects who had MAPbacteremia presence had an average relative gene expression of 5.25 ± 2.58 (n = 21) in PTPN2 compared to the healthy controls with MAPbacteremia presence (11.9 ± 10.5, n =3, P < 0.05). CD subjects who had an absence of MAPbacteremia presence had an average relative gene expression of 5.28 ± 2.87 (n = 17) in PTPN2 compared to the healthy controls without MAPbacteremia presence (10.3 ± 6.71, n = 27, P < 0.05). For PTPN22 average relative gene expression, CD subjects with MAPbacteremia presence had 1.73 ± 0.97 (n = 21) compared to healthy controls with MAPbacteremia presence (2.83 ± 1.94, n = 3). CD subjects without MAPbacteremia presence had an average relative gene expression of 1.81 ± 1.31 (n = 17) in PTPN22 compared to the healthy controls without MAPbacteremia presence (3.29 ± 1.86, n = 27, P-values < 0.05).
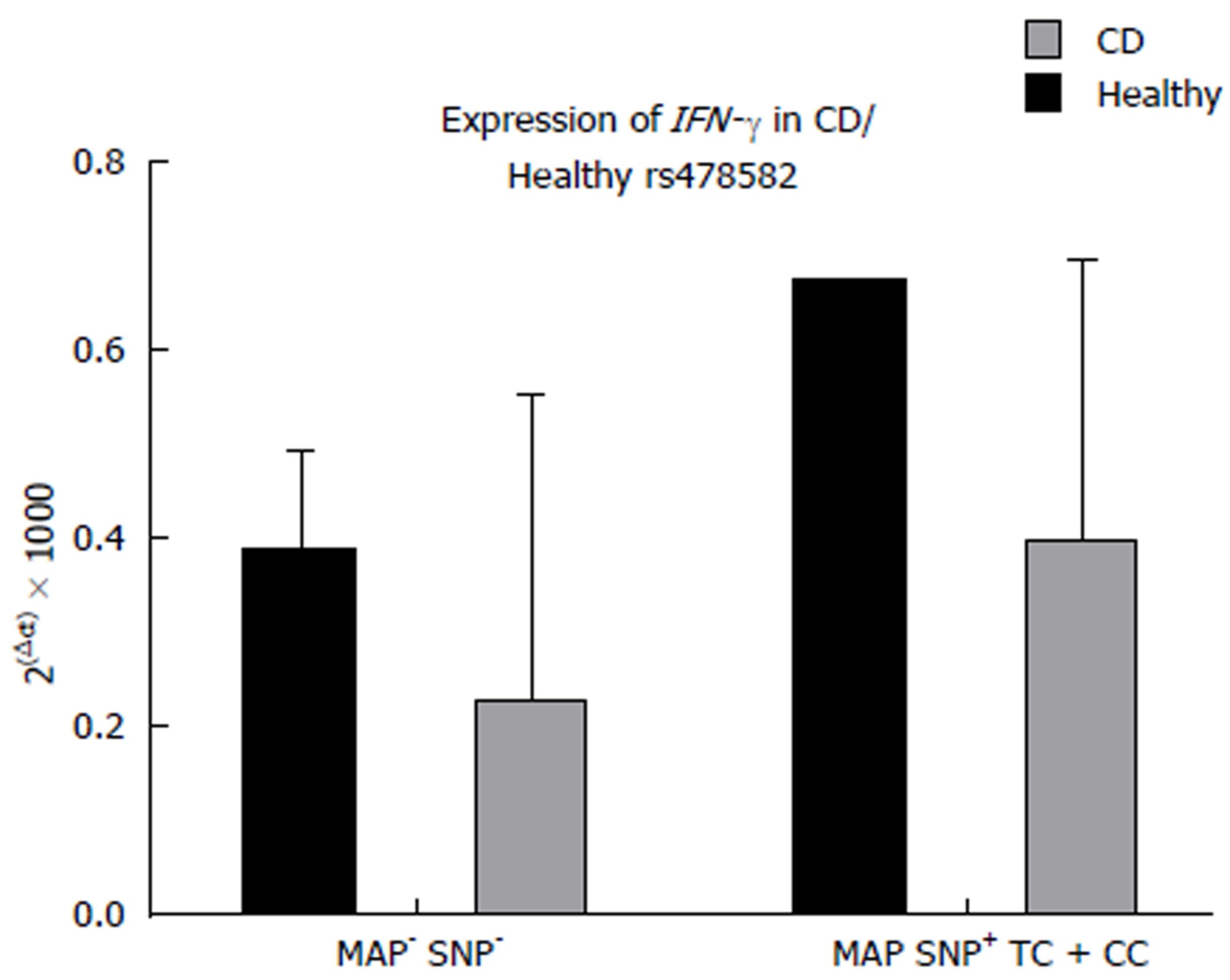
The effect of combined MAPbacteremia presence and either PTPN2:rs478582 or PTPN22:rs2476601 did not significantly change PTPN2/22 expression in all CD and healthy control samples. However, when examining the combined effects of MAPbacteremia presence and either PTPN2:rs478582 or PTPN22:rs2476601, the average relative gene expression of IFN-γ does increase in subjects compared to subjects without MAPbacteremia presence and no SNPs. For CD subjects with both MAPbacteremia and heterozygous (TC) or minor (CC) alleles in PTPN2:rs478582, the average relative gene expression of IFN-γ was higher (0.40 ± 0.29 n = 22) compared to the CD subjects without MAPbacteremia and PTPN2:rs478582 (0.23 ± 0.31, n = 5, Figure 4). For CD subjects with both MAPbacteremia and heterozygous (GA) or minor (CC) alleles in PTPN22:rs2476601, the average relative gene expression of IFN-γ was higher (0.42 ± 0.32, n = 4) compared to the CD subjects without MAPbacteremia and PTPN22:rs2476601 (0.37 ± 0.31, n = 18).
T-cell functionality when SNPs and MAPbacteremia was presented in subjects was determined in five CD and five healthy control subjects. All five CD subjects that had their T-cell response tested had SNPs in either PTPN2:rs478582 and/or PTPN22:rs2476601, while the five healthy control subjects had no observed SNPs present. Overall, when the subjects’ T-cells were treated with PHA, the average overall fold change in the CD subjects was 2.22 ± 1.36 (n = 5) fold increase compared to the healthy controls (1.67 ± 0.51 fold increase, n = 5). Similarly, when the same T-cells were treated with MAP PPD-like, the average overall fold change in CD subjects was 2.01 ± 0.79 (n = 5) compared to the healthy controls (1.39 ± 0.24 fold increase, n = 5).
Out of the five CD subjects, 3/5 were tested for having MAPbacteremia presence. When examining T-cells treated with PHA from CD subjects tested positive for MAPbacteremia presence, the average overall fold change was 2.7 ± 1.65 (n = 3) compared to the CD subjects’ T-cells that were absence of MAPbacteremia presence and treated with PHA (1.51 ± 0.51 fold increase, n = 2). Similarly, when the same T-cells were treated with MAP PPD-like, the average overall fold change in CD subjects with MAPbacteremia was 2.5 ± 0.59 (n = 3) compared to the CD subjects’ T-cells without MAPbacteremia presence (1.27 ± 0.12 fold increase, n = 2).
The pathogenesis of CD, as with other inflammatory autoimmune disorders, involves both genetic pre-disposition leading to higher immune responses and an environmental trigger that exacerbates the immune response. However, with current diagnosis and treatment, it has been difficult to treat CD symptoms due to loss of treatment response and many side effects[6-10]. Thus, understanding the key elements of CD pathogenesis (genetic SNPs and environmental triggers), it is possible to find new treatment targets for the disease and new diagnosis techniques as well. CD pathogenesis is very dependent on the overproduction of pro-inflammatory cytokines such as TNF-α and IFN-γ, which promote chronic inflammation, increased granuloma formation, and increased apoptosis of intestinal tissues[8,9,20,21]. Since the majority of CD medications are blocking pro-inflammatory cytokines such as TNF-α and IFN-γ, other types of targets has been ignored[6-10,20,21]. This study is focused on finding new targets for both diagnosis and treatment of CD, where we looked into the SNPs of negative regulatory genes PTPN2/22 and their impact on: increased production of pro-inflammatory cytokines, apoptosis, mycobacterial susceptibility, and inflammation. To our knowledge, this is the first study to look into SNPs in both PTPN2/22 together along with correlation with gene expression and MAP susceptibility in CD.
The effect of SNPs in PTPN2/22 in CD pathogenesis has been highly debated in the literature, thus we selected nine SNPs that not only was found associated with CD, but with other diseases as well[4,5,17-19,22-24]. Out of the nine SNPs examined in this study, PTPN2:rs478582 was found to be significant in CD (P-values < 0.05, OR = 3.03) compared to the healthy controls (Figure 2A). Although PTPN22:rs2476601 was found to not be significant to CD (P > 0.05, OR = 2.7) compared to the healthy controls, we continued to study the effects of the SNP along with PTPN2:rs478582 due to PTPN22:rs2476601 being associated with inflammatory autoimmune diseases in general (Figure 2B)[3-5,17-19,22-24]. Since a diverse population (no restriction on race, place of origin, age, or gender) was used in this study, alterations of allele distribution in the SNPs could possibly happen due to SNPs overall fluctuating between different population groups[3-5,17-19,22-24]. Further isolated population studies on PTPN2/22 SNPs in CD subjects need to be investigated more. Knowledge of which SNP is more associated with CD could possibly be used as a diagnosis tool for clinicians when examining patients with CD like symptoms.
Gene expression of PTPN2/22 correlated with the SNPs PTPN2:rs478582 and PTPN22:rs2476601 was also done to determine if the SNPs did change PTPN2/22 levels. Although overall PTPN2/22 expression was significantly decreased in CD subjects (P < 0.05, Figure 3A and 3B), the SNPs PTPN2:rs478582 and PTPN22:rs2476601 did not change gene expression between normal, heterozygous, or minor alleles. However, IFN-γ gene expression was found significantly higher in both CD and healthy controls (P < 0.05) along with an overall increased T-cell activity in subjects that had heterozygous/minor alleles in either PTPN2:rs478582 and/or PTPN22:rs2476601 (Figure 3C and 3D). These correlation analyzes shows that the SNPs PTPN2:rs478582 and PTPN22:rs2476601 may not necessarily change the regulation of the PTPN2/22 gene, but could possible disrupt the protein activity of PTPN2/22. For the PTPN2:rs478582 SNP, a base change (T > C) in intron 3 occurs, where it is theorized that splicing problems could occur during the RNA splicing[25-28]. This could lead to loss of activity in the protein once fully translated[25-28]. The PTPN22:rs2476601 SNP is a base change (G > A) that occurs in exon 14, which physically changes the amino acid arginine (R) to a tryptophan (W) on the 620 amino acid residue on the catalytic portion of the PTPN22 protein[19,26-28]. It has been highly debated what the R620W does to the PTPN22 protein, but it is suspected to cause the protein to be less active[19,26-28]. Overall, the SNPs PTPN2:rs478582 and PTPN22:rs2476601 seem to cause a loss of function in PTPN2/22, thus leading to less negative regulated T-cells. This will lead to a high production of pro-inflammatory cytokines, which will lead to increased inflammation/apoptosis in intestinal tissues in CD subjects. Other SNPs in PTPN2/22 will need to be studied further to see if those SNPs will alter gene expression of PTPN2/22 instead of PTPN2:rs478582 and PTPN22:rs2476601 just altering protein activity. Although we only examined the effect of PTPN2/22 on the expression of IFN-γ, other factors do control IFN-γ expression and production. These include cytokines, such as TNF-α and IL-12, which stimulate T-cell production of IFN-γ and cytokines, such as IL-6 and IL-10, which decrease T-cell production of IFN-γ[29]. However, since CD and other inflammatory autoimmune disorders are T-cell mediated, we focused only on PTPN2/22 regulation on IFN-γ expression. This is due to PTPN2/22 ultimately acting as negative regulators of T-cell activity and thus controlling IFN-γ production from T-cells. Further investigation of the effect of these other regulatory IFN-γ production cytokines in subjects with SNPs in PTPN2/22 is needed.
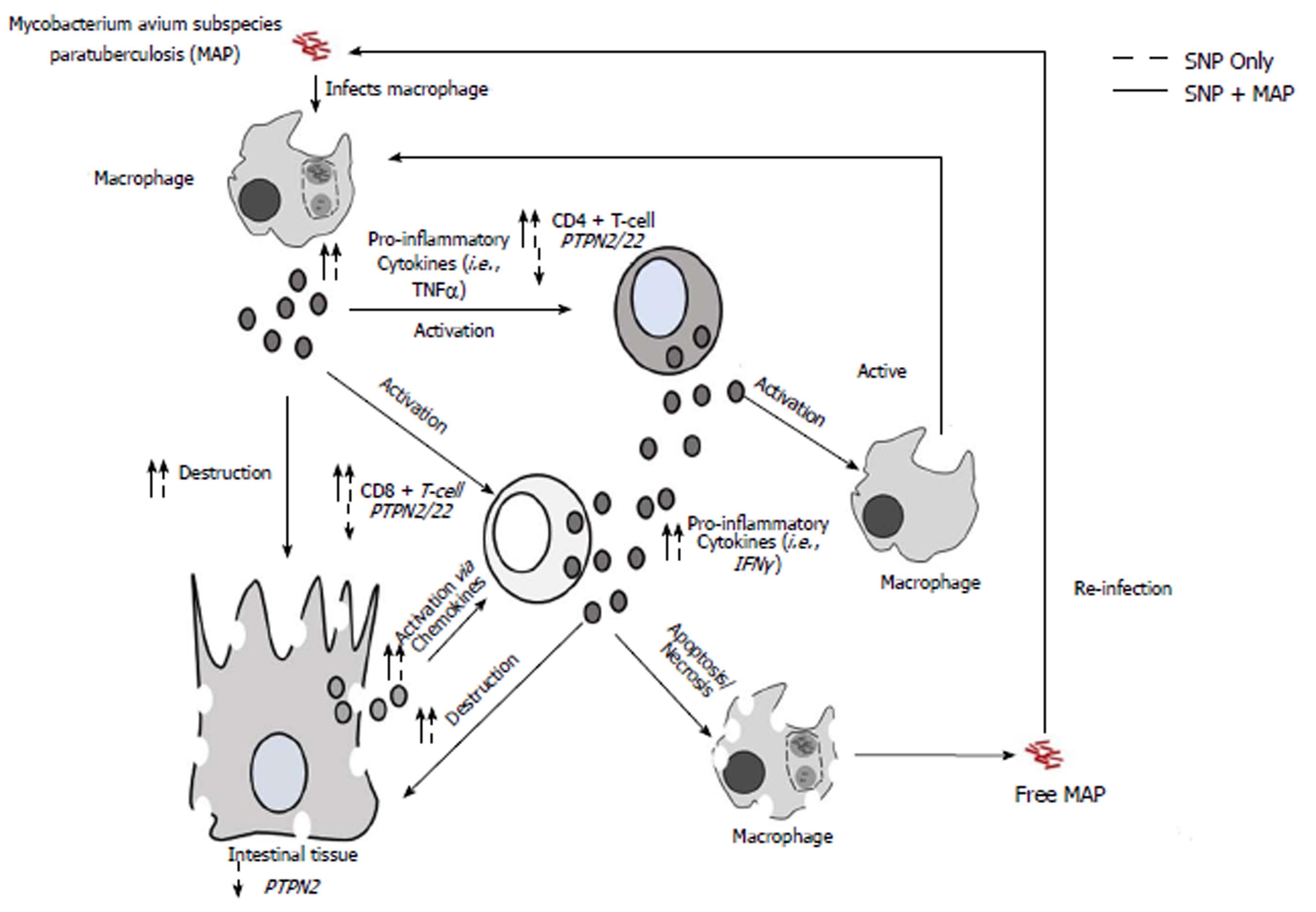
Although the role MAP has been studied in CD pathogenesis extensively, correlation studies with SNPs in PTPN2/22 and MAP susceptibility have not been done before until this study[4,9,11-16,20]. Overall, the correlation analyzes of SNPs in PTPN2/22 and MAPbacteremia presence showed that the SNPs might have increased susceptibility in CD subjects (Table 2). Specifically, 60.7% (OR = 21.6, P < 0.05) of CD subjects with PTPN2:rs478582 SNP (heterozygous or minor group) had MAPbacteremia presence, while 27.3% (OR = 4.53) subjects with the PTPN22:rs2476601 SNP (heterozygous or minor group) had MAPbacteremia. Limitations however in the detection of MAP IS900 DNA from the blood of subjects’ samples do not provided the information that the MAP bacteria is alive or dead, thus does not show active infection or previous infection. Further culturing of the blood from the subjects is necessary to determine live MAP infection in the subjects examined. The findings found in this study suggest that SNPs in PTPN2/22 increases susceptibility to MAPbacteremia, which is possible due to the lack of negative regulation in the T-cells. Since T-cells control macrophage activity and mycobacterial species such as MAP can survive in infected macrophages, it is important that the T-cells are regulated correctly in order to prevent MAP infection[30-34]. If problems involving the PTPN2/22 gene regulation or function the PTPN2/22 protein occurs, T-cells will be overactive and in turn will make macrophages overactive as well (Figure 5)[30-34]. This increased activity of macrophages will not only lead to increased pro-inflammatory cytokines like TNF-α, but could allow MAP and other intracellular pathogens to survive and grow faster due to the increased activation of newer macrophages[30-34]. This is why SNPs in PTPN2/22 and the hyperactivity of T-cells should increase susceptibility to intracellular pathogens such as MAP.
To further test if T-cells from the CD subjects with the PTPN2:rs478582 and the PTPN22:rs2476601 were overactive, we induced isolated T-cells from CD subjects with either PHA or MAP PPD-like. Although we did not isolate out total T-cell populations from mucosal intestinal tissues and instead from peripheral blood draws, we believe that T-cell proliferation will be the same regardless of the source of origin. This is possible due to PTPN2/22 being found in every T-cell population, regardless of the site of isolation, thus SNPs in PTPN2/22 should affect all T-cells in the body in the same way. Overall, CD subjects with the SNPs proliferated more than healthy controls without the SNPs. In addition, CD subjects who had MAPbacteremia presence and SNPs in PTPN2/22 proliferated more than CD subjects who did not have MAPbacteremia presence. These analyzes showed that for T-cells to become overactive, both SNPs in PTPN2/22 and the presence of MAPbacteremia is required to induce the pathogenesis process of CD. This is further evidence that for the pathogenesis of any inflammatory autoimmune disorder, both genetic predisposition and an environmental trigger are needed to cause disease. Further investigation in gene expression of pro-inflammatory cytokines produced (IFN-γ for example) by T-cells after being induced with antigens need to be examined. Along with this, further investigation of subpopulations of T-cell activity is needed to determine which T-cell population is more active in subjects with SNPs in PTPN2/22.
Overall, SNPs in PTPN2/22 lead to overactive T-cell activity and increased susceptibility to intracellular pathogens such as MAP. With genetic testing for SNPs and detection/treatment for mycobacterial infections such as MAP, it is possible for personalized treatment of CD to be an option. Further studies in SNPs in PTPN2/22 and other immunity specific genes need to be researched and correlated with bacterial infections to improve CD diagnosis and treatment.
Single nucleotide polymorphism (SNPs) and environmental triggers have been associated with a variety inflammatory autoimmune disorders including Crohn’s disease (CD). Specifically, SNPs in the negative regulatory immune genes Protein Tyrosine Phosphatase Non-receptor type 2 and 22 (PTPN2/22) have been associated with CD along with mycobacterial infections. Although both elements have been examined separately, correlation analysis have not been done to determine if SNPs in PTPN2/22 along with a Mycobacterium avium subspecies paratuberculosis (MAP) infections do cause a dysregulation in the immune system that could lead to CD symptoms.
Due to the flaws of current diagnosis and treatments of CD, new and better methods need to be determined. Investigating the pathogenesis of CD via SNPs analysis and MAP presence could lead to the possibility of individualized diagnosis/treatment for CD patients via genetic testing and antibiotic treatments. Our research could potentially propose newer routes of CD treatment for clinicians in the near future.
In this study, we examined the allele distribution in nine SNPs found in PTPN2/22 along with MAP presence in CD and healthy control subjects. Along with this, we determined gene expression of PTPN2/22 and correlated with both SNPs and MAP presence. Lastly, we examined T-cell proliferation of the subjects and correlated that with both SNPs and MAP presence as well. This study overall examined the effects of both SNPs in PTPN2/22 and MAP presence in CD subjects.
We obtained K2-EDTA coded blood tubes from both CD and healthy control subjects. Each subjects’ blood was examined for PTPN2/22 genotyping by TaqMan™ SNP genotyping, PTPN2/22 and IFN-γ gene expression by real-time PCR (RT-PCR), MAP presence by MAP IS900 nested PCR (nPCR), and T-cell proliferation by BrdU treatment.
We found in this study that the PTPN2:rs478582 SNP and the haplotype combination of PTPN2:rs478582 and PTPN22:rs2476601 SNPs were found significant in CD subjects compared to healthy control subjects. Gene expression of PTPN2/22 was also found to be decreased significantly in CD subjects as well. IFN-γ gene expression was found to be significantly higher in subjects with either PTPN2:rs478582 or PTPN22:rs2476601. MAP presence was found significantly in CD subjects compared to the healthy control subjects, were CD subjects with either PTPN2:rs478582 or PTPN22:rs2476601 had higher MAP presence than subjects without SNPs. Overall T-cell proliferation was higher in CD subjects with either SNPs and induced with MAP antigens than subjects who didn’t. These findings should provide more background to the pathogenesis of CD. Further studies into the gene expression of pro-inflammatory cytokines produced by T-cells with SNPs in PTPN2/22 after proliferation needs to be investigated.
This study was done in order to provide answers on the pathogenesis of CD. We have demonstrated that SNPs found in PTPN2/22 are found significantly in CD subjects and the SNPs have the following effects on the immune system: increases T-cell proliferation due to loss of negative regulation, increases pro-inflammatory cytokines such as IFN-γ, and increases susceptibility to mycobacterial infections. This is further evidence that both a genetic predisposition and an environmental trigger are needed to cause disease in inflammatory autoimmune disorders such as CD.
This study has provided us with new, possible targets that could be used in diagnosis methods and treatment for CD. With the data found in this study, the possibility of personalized treatment for CD could be possible with genetic testing for SNPs and antibiotic treatments for MAP. Further testing for other immune gene SNPs are needed in order to fully understand the genetic profile of CD patients. Additional research in MAP’s relationship with CD pathogenesis is also needed to fully understand the effect of MAP in CD patients.
Our thanks are due to Latifa Abdelli, PhD, Ahmad Qasem, and all members of Dr. Naser’s lab. Special thanks to the staff of Dr. Shazia Beg’s office and all of those who participated in this study.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Aytac E, Gassler N S- Editor: Ma YJ L- Editor: A E- Editor: Ma YJ
| 1. | Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat Rev Genet. 2016;17:160-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 166] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 2. | Richard-Miceli C, Criswell LA. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med. 2012;4:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 3. | Hewagama A, Richardson B. The genetics and epigenetics of autoimmune diseases. J Autoimmun. 2009;33:3-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 216] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 4. | Sharp RC, Abdulrahim M, Naser ES, Naser SA. Genetic Variations of PTPN2 and PTPN22: Role in the Pathogenesis of Type 1 Diabetes and Crohn’s Disease. Front Cell Infect Microbiol. 2015;5:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 5. | Gurzov EN, Stanley WJ, Brodnicki TC, Thomas HE. Protein tyrosine phosphatases: molecular switches in metabolism and diabetes. Trends Endocrinol Metab. 2015;26:30-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 6. | Cheifetz AS, Feuerstein JD. Treatment of Inflammatory Bowel Disease with Biologics. Springer International Publishing. 2017;. |
| 7. | Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388:2023-2038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2186] [Cited by in RCA: 3099] [Article Influence: 344.3] [Reference Citation Analysis (0)] |
| 8. | Kuek A, Hazleman BL, Ostör AJ. Immune-mediated inflammatory diseases (IMIDs) and biologic therapy: a medical revolution. Postgrad Med J. 2007;83:251-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 242] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 9. | Qasem A, Naser AE, Naser SA. The alternate effects of anti-TNFα therapeutics and their role in mycobacterial granulomatous infection in Crohn’s disease. Expert Rev Anti Infect Ther. 2017;15:637-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Allen PB, Olivera P, Emery P, Moulin D, Jouzeau JY, Netter P, Danese S, Feagan B, Sandborn WJ, Peyrin-Biroulet L. Review article: moving towards common therapeutic goals in Crohn’s disease and rheumatoid arthritis. Aliment Pharmacol Ther. 2017;45:1058-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 11. | Naser SA, Thanigachalam S, Dow CT, Collins MT. Exploring the role of Mycobacterium avium subspecies paratuberculosis in the pathogenesis of type 1 diabetes mellitus: a pilot study. Gut Pathog. 2013;5:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Naser SA, Collins MT, Crawford JT, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis (MAP) from the Blood of Patients with Crohn’s disease: A Follow-Up Blind Multi Center Investigation. Open Inflamm J. 2010;3:22-23. [RCA] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Masala S, Paccagnini D, Cossu D, Brezar V, Pacifico A, Ahmed N, Mallone R, Sechi LA. Antibodies recognizing Mycobacterium avium paratuberculosis epitopes cross-react with the beta-cell antigen ZnT8 in Sardinian type 1 diabetic patients. PLoS One. 2011;6:e26931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 14. | Sechi LA, Dow CT. Mycobacterium avium ss. paratuberculosis Zoonosis - The Hundred Year War - Beyond Crohn’s Disease. Front Immunol. 2015;6:96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 103] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 15. | Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn’s disease. Lancet. 2004;364:1039-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 429] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 16. | Naser SA, Romero C, Elwasila S, Ghonaim M, Naser N, Valentine JF. Functional Dysregulation of PBMC and PMN in Crohn’s Disease. Open Inflamm J. 2009;2:24-33. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 17. | Serrano A, Márquez A, Mackie SL, Carmona FD, Solans R, Miranda-Filloy JA, Hernández-Rodríguez J, Cid MC, Castañeda S, Morado IC. Identification of the PTPN22 functional variant R620W as susceptibility genetic factor for giant cell arteritis. Ann Rheum Dis. 2013;72:1882-1886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 18. | Spalinger MR, Lang S, Weber A, Frei P, Fried M, Rogler G, Scharl M. Loss of protein tyrosine phosphatase nonreceptor type 22 regulates interferon-γ-induced signaling in human monocytes. Gastroenterology. 2013;144:978-988.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Qu H, Tessier MC, Hudson TJ, Polychronakos C. Confirmation of the association of the R620W polymorphism in the protein tyrosine phosphatase PTPN22 with type 1 diabetes in a family based study. J Med Genet. 2005;42:266-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Naser SA, Sagramsingh SR, Naser AS, Thanigachalam S. Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients. World J Gastroenterol. 2014;20:7403-7415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 94] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 21. | Manuc TE, Manuc MM, Diculescu MM. Recent insights into the molecular pathogenesis of Crohn’s disease: a review of emerging therapeutic targets. Clin Exp Gastroenterol. 2016;9:59-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Glas J, Wagner J, Seiderer J, Olszak T, Wetzke M, Beigel F, Tillack C, Stallhofer J, Friedrich M, Steib C. PTPN2 gene variants are associated with susceptibility to both Crohn’s disease and ulcerative colitis supporting a common genetic disease background. PLoS One. 2012;7:e33682. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 23. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2114] [Cited by in RCA: 2043] [Article Influence: 120.2] [Reference Citation Analysis (0)] |
| 24. | Waterman M, Xu W, Stempak JM, Milgrom R, Bernstein CN, Griffiths AM, Greenberg GR, Steinhart AH, Silverberg MS. Distinct and overlapping genetic loci in Crohn’s disease and ulcerative colitis: correlations with pathogenesis. Inflamm Bowel Dis. 2011;17:1936-1942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Espino-Paisan L, de la Calle H, Fernández-Arquero M, Figueredo MA, de la Concha EG, Urcelay E, Santiago JL. A polymorphism in PTPN2 gene is associated with an earlier onset of type 1 diabetes. Immunogenetics. 2011;63:255-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Hendriks WJ, Pulido R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim Biophys Acta. 2013;1832:1673-1696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 27. | Vang T, Miletic AV, Bottini N, Mustelin T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity. 2007;40:453-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 123] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Zikherman J, Weiss A. Unraveling the functional implications of GWAS: how T cell protein tyrosine phosphatase drives autoimmune disease. J Clin Invest. 2011;121:4618-4621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (1)] |
| 29. | Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1083] [Cited by in RCA: 1203] [Article Influence: 70.8] [Reference Citation Analysis (0)] |
| 30. | Janeway Jr CA, Travers P, Walport M, Shlomchik MJ. Macrophage activation by armed CD4 TH1 cells. Immunobiology: The Immune System in Health and Disease, 5th edition 2001; . |
| 31. | Prezzemolo T, Guggino G, La Manna MP, Di Liberto D, Dieli F, Caccamo N. Functional Signatures of Human CD4 and CD8 T Cell Responses to Mycobacterium tuberculosis. Front Immunol. 2014;5:180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 200] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 32. | Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Helper T Cells and lymphocyte activation. Molecular Biology of the Cell, 4th edition. 2002;. |
| 33. | Bermudez LE, Danelishvili L, Early J. Mycobacteria and macrophage apoptosis: complex struggle for survival. Microbe Wash DC. 2006;1:372-375. |
| 34. | Early J, Fischer K, Bermudez LE. Mycobacterium avium uses apoptotic macrophages as tools for spreading. Microb Pathog. 2011;50:132-139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |