Published online Dec 14, 2018. doi: 10.3748/wjg.v24.i46.5234
Peer-review started: September 4, 2018
First decision: October 4, 2018
Revised: October 31, 2018
Accepted: November 7, 2018
Article in press: November 8, 2018
Published online: December 14, 2018
Processing time: 100 Days and 17 Hours
To determine whether cell division cycle (Cdc)42 is regulated by microRNA (miR)-15a in the development of pediatric inflammatory bowel disease (IBD).
We cultured 293T cells, used plasmids and performed dual-luciferase assay to determine whether Cdc42 is a miR-15a target gene. We cultured Caco-2 cells, and stimulated them with tumor necrosis factor (TNF)-α. We then employed lentiviruses to alter the expression of miR-15a and Cdc42. We performed quantitative reverse transcription polymerase chain reaction (qRT-PCR) and immunofluorescence to determine whether Cdc42 is regulated by miR-15a in Caco-2 cells. Finally, we collected ileocecal tissue by endoscopy from patients and performed qRT-PCR to examine the expression of miR-15a and Cdc42 in pediatric IBD patients.
Target Scan and dual-luciferase assay revealed that Cdc42 was a miR-15a target gene. MiR-15a expression increased (P = 0.0038) and Cdc42 expression decreased (P = 0.0013) in cells stimulated with TNF-α, and the expression of the epithelial junction proteins zona occludens (ZO)-1 (P < 0.05) and E-cadherin (P < 0.001) decreased. Cdc42 levels decreased in miR-15a-mimic cells (P < 0.001) and increased in miR-15a inhibitor cells (P < 0.05). ZO-1 and E-cadherin decreased in miR-15a-mimic cells (P < 0.001) but not in the miR-15a inhibitor + TNF-α cells. In Lv-Cdc42 + TNF-α cells, ZO-1 and E-cadherin expression increased compared to the Lv-Cdc42-NC + TNF-α (P < 0.05) or miR-15a-mimic cells (P < 0.05). Fifty-four pediatric IBD patients were included in this study, 21 in the control group, 19 in the Crohn’s disease (CD) active (AC) group, seven in the CD remission (RE) group, and seven in the ulcerative colitis (UC) group. MiR-15a increased and Cdc42 decreased in the CD AC group compared to the control group (P < 0.05). miR-15a decreased and Cdc42 increased in the CD RE group compared to the CD AC group (P < 0.05). miR-15a was positively correlated with the Pediatric Crohn’s disease Activity Index (PCDAI) (P = 0.006), while Cdc42 was negatively correlated with PCDAI (P = 0.0008). Finally, miR-15a expression negatively correlated with Cdc42 in pediatric IBD patients (P = 0.0045).
MiR-15a negatively regulates epithelial junctions through Cdc42 in Caco-2 cells and pediatric IBD patients.
Core tip: This study aimed to determine whether cell division cycle (Cdc)42 was regulated by microRNAs in pediatric inflammatory bowel disease (IBD) development. We studied microRNA (miR)-15a - Cdc42 signaling in pediatric IBD patients and found that miR-15a negatively regulated Cdc42 in pediatric IBD patients, and both were strongly correlated with the severity of IBD. Additionally, this study provides evidence that Cdc42 is a target gene of miR-15a, and this will provide a critical clue for understanding IBD development.
- Citation: Tang WJ, Peng KY, Tang ZF, Wang YH, Xue AJ, Huang Y. MicroRNA-15a - cell division cycle 42 signaling pathway in pathogenesis of pediatric inflammatory bowel disease. World J Gastroenterol 2018; 24(46): 5234-5245
- URL: https://www.wjgnet.com/1007-9327/full/v24/i46/5234.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i46.5234
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder with an increasing prevalence. It includes ulcerative colitis (UC) and Crohn’s disease (CD). Epidemiological studies have demonstrated a rising incidence of IBD in Chinese children. In 2001, it was reported that the incidence of pediatric IBD was 0.5/million, while in 2011, the incidence increased to 6.1/million[1]. Pediatric IBD has severe phenotypes, extensive intestinal involvement, rapid progression, and has several side issues, including growth failure, poor bone density, and delayed puberty. Thus, it is important to investigate the pathogenesis of pediatric IBD.
The etiology of IBD remains unknown, and multiple factors may be involved in its development, including infection, the immune system, genetics, and the environment[2,3]. High expression of tumor necrosis factor (TNF)-α has been found in most pediatric IBD patients. TNF-α plays an important role in microRNA expression.
MicroRNAs are families of small (approximately 20 nucleotides long), noncoding RNAs. They play a definitive regulatory role in innate and adaptive immune processes[4,5]. microRNA expression has been shown to be altered in adult patients suffering from CD and UC. Wu et al[6] reported that the levels of 11 microRNAs were altered in UC patient colons and that microRNA (miR)-192 negatively regulated TNF-α in IBD patients. Iborra et al[7] showed that the levels of several microRNAs were increased in CD and UC patients. Takagi et al[8] reported that miR-21 and miR-15 were upregulated in UC patients. However, most studies have been conducted in adults, and there are only a limited number of studies examining microRNAs in children with IBD. Béres et al[9] showed that the level of miR-155 was increased in pediatric IBD patients and that its levels increased after TNF-α stimulation in vitro.
Cell division cycle (Cdc)42 is a small GTPase of the Rho family that acts as a molecular switch by cycling between an active GTP-bound form and an inactive GDP-bound form. It controls various cellular functions, including adhesion, migration, transcription and growth[10,11]. It has a special role in hematopoietic stem cell regulation[12,13] and is critical for intestinal stem cell division, survival and differentiation in mice[14]. It has recently been reported that Cdc42 is critical for intestinal epithelial stem cell differentiation and formation of a functional intestinal barrier[15,16].
The epithelial barrier plays a critical role in maintaining intestinal homeostasis and is formed by multiple intercellular junctions, including the apical junctional complex (AJC), tight junction (TJ) and adherens junction (AJ), which are the major protein complexes of the AJC[17]. TJs are composed of occludin, claudins, junctional adhesion molecules (JAMs), and the zona occludens (ZO) family of proteins. AJs are formed by E-cadherin[18]. A number of studies in IBD patients have demonstrated that intestinal barrier function is disrupted in both active and quiescent disease states[16,19,20].
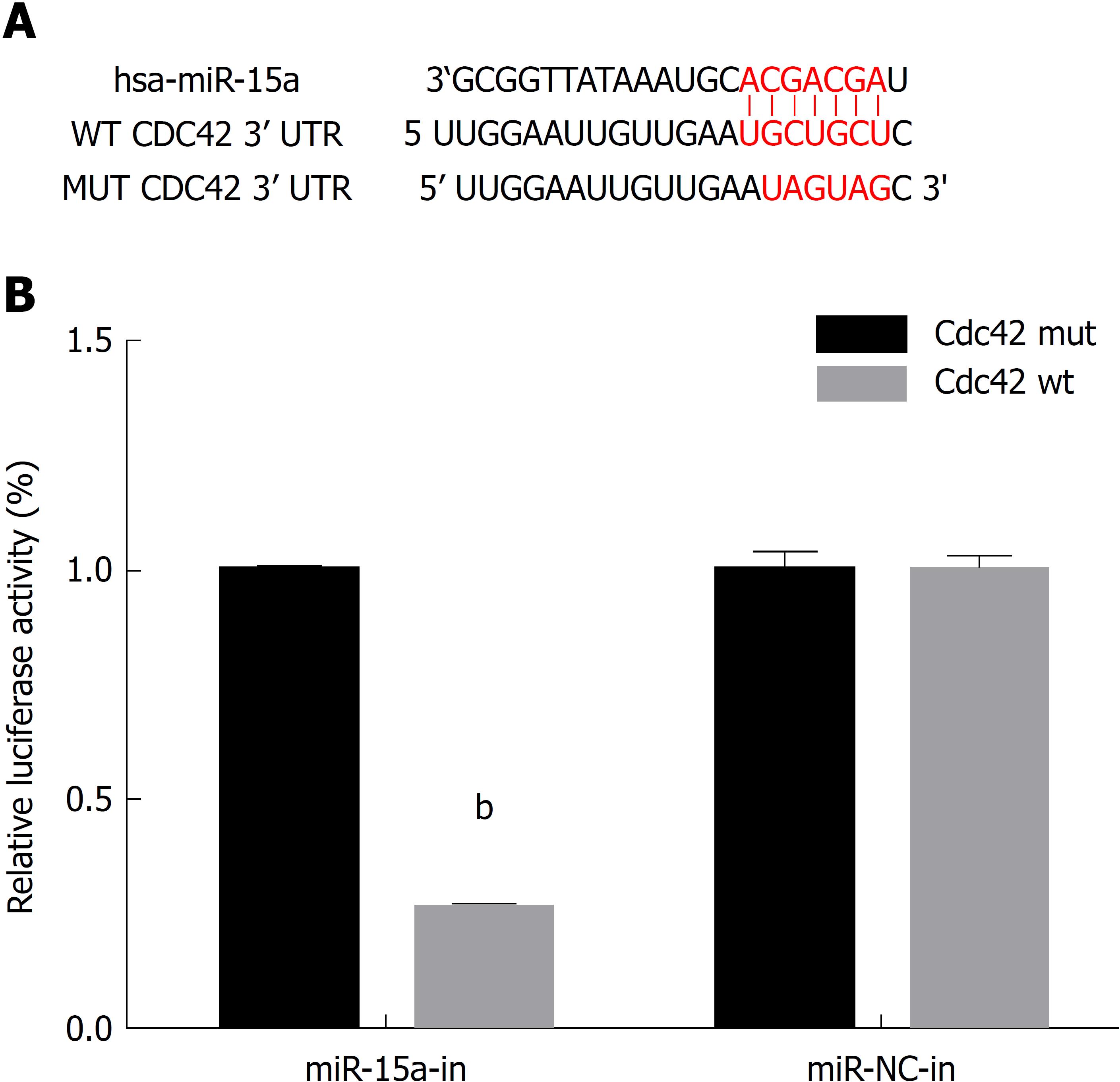
Several studies have shown that microRNAs participate in the development of IBD, and miR-15 has an altered expression level in IBD patients, but the underlying mechanisms are not fully elucidated. Recently, microRNA-Cdc42 signaling has been studied by some researchers; they have shown that miR-137 negatively regulates H2O2-induced cardiomyocyte apoptosis through Cdc42[21]. MiR-17 is involved in Japanese flounder development by targeting Cdc42 mRNA[22], and miR-106b negatively regulates Cdc42 in human colorectal cancer cells[23]. We used Target Scan and found the putative seed region of miR-15a in the wild-type 3’-UTR of Cdc42 (Figure 1A). As Cdc42 is critical for intestinal barrier function, we hypothesized that miR-15a regulates Cdc42 to disrupt the intestinal barrier in IBD development.
Chinese Han pediatric IBD patients were included in this study. The diagnosis of CD and UC was based on clinical symptoms, endoscopic findings, and histopathology according to the Porto criteria[24]. Disease activity score was calculated by Pediatric Crohn’s Disease Activity Index (PCDAI) and Pediatric Ulcerative Colitis Activity Index (PUCAI). Age- and sex-matched Chinese Han juvenile polyp patients were enrolled as a control group. Colonoscopy was used for diagnosis, and the ileocecal mucosa showed normal macroscopic appearance with normal histology in the biopsy specimens.
Ileocecal tissues of all patients were collected by endoscopy, frozen in liquid nitrogen, and stored at -80 °C until further analysis. All tissues had been histologically confirmed.
293T cells were a generous gift from Dr. Hua-Qing Zhong (Institute of Pediatrics, Children’s Hospital of Fudan University), and Caco-2 cells were purchased from Genechem (Shanghai, China). They were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% nonessential amino acids, 1% penicillin-streptomycin antibiotic solution (Gibco) and 1 mmol/L L-glutamine at 37 °C in 5% CO2. The cells were passaged at 80% confluence with a 0.25% trypsin-EDTA solution for 3-5 min.
Plasmid transfection. The following plasmids were used: hsa-mir-15a and its negative control, Cdc42 [NM-001791-3UTR (miR-15a-5p)] and its negative control, Cdc42 [NM-001791-3UTR (miR-15a-5p)]-mut. All of the plasmids were purchased from Genechem. Transfection was performed using X-tremegene HP (Roche). At 48 h after transfection, cells were harvested for further experiments.
Lentiviral transfection. The following lentiviruses were used: miR-15a mimic and its negative control (Ubi-MCS-SV40-EGFP-IRES-puromycin LVcon220), miR-115a inhibitor and its negative control (hU6-MCS-ubiquitin-EGFP-IRES-puromycin LVcon137), and Cdc42 and its negative control (Ubi-EGFP-MCS-IRES-puromycin LVcon129). All of the lentiviruses were purchased from Genechem. At 12 h after transfection, cells were cultured in regular media, and after 48 h, cells were harvested for further experiments.
TNF-α-stimulated cells
Caco-2 cells (1 × 105) were passaged in 35-mm dishes, or 5 × 104 cells were grown in 35-mm confocal dishes for imaging. After 24 h, medium containing 35 ng/mL TNF-α was used to culture the cells for 60 h, and then the cells were harvested.
Total RNA was extracted using TRIzol reagent (Thermo Scientific). For analysis of microRNA expression, the MiScript II RT Kit (Qiagen) was used to convert RNA into cDNA. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using the SYBR Premix Ex Taq II kit (Takara). The PCR conditions were set as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. U6 was used as a reference gene for microRNA-15a, GAPDH was used as a reference gene for all other genes. Relative expression was analyzed by the 2−ΔΔCt method. The primers used in this study are listed in Table 1.
| Primer | Primer sequences (5' → 3') |
| miR-15a | F: 5’- ACACTCCAGCTGGGTAGCAGCACATAATGG-3’ |
| R: 10 × miscript universal primer (QIAGEN) | |
| RNU6A | F: 5’- CTCGCTTCGGCAGCACA-3’ |
| R: 5’- AACGCTTCACGAATTTGCGT -3’ | |
| Cdc42 | F: 5’- TTATGACAGATTACGACCGC -3’ |
| R: 5’- CCAACAAGCAAGAAAGGAGT -3’ | |
| ZO-1 | F: 5’- AACAAGCCAGCAGAGACCTC -3’ |
| R: 5’- CTTCATACATGGGGACGCGA -3’ | |
| E-cadherin | F: 5’- TCCAGTGAACAACGATGGCA -3’ |
| R: 5’- AATGTACTGCTGCTTGGCCT -3’ | |
| GAPDH | F: 5’- CCTCCTCACAGTTGCCATGT -3’ |
| R: 5’- CTGGTTGAGCACAGGGTACT -3’ |
To determine whether Cdc42 was a direct target of miR-15a, a dual luciferase assay was performed using 293T cells. Mutant Cdc42 3′-UTR reporters were created by mutating the seed region of the predicted miR-15a target site. Approximately 1 × 105 cells were plated in 24-well plates. After 24 h, hsa-mir-15a mimic plasmid and its negative control (miR-NC) were cotransfected with either Cdc42-WT or Cdc42-MUT luciferase reporter vector; 48 h after transfection, a Dual Luciferase Reporter Assay System (Promega, Madison, WI, United States) was used to examine the relative luciferase activities. Relative luciferase activities were normalized to the value with Cdc42-MUT and miR-NC cotransfection.
Caco-2 cells (5 × 104) were passaged in 35-mm confocal dishes, washed in phosphate buffer solution (PBS), and fixed with 4% paraformaldehyde for 20 min. Cells were permeabilized with PBS-T for 10 min, blocked with 4% bovine serum albumin at room temperature for 30 min, incubated with primary antibody solution at 4 °C overnight, and secondary antibody solution at room temperature for 1 h, and DAPI solution for 5 min. Immunofluorescence images were captured using a Leica confocal microscope and analyzed by Photoshop version 7.0.
Antibodies were used as follows: anti-ZO-1 at 1:400 (Cell Signaling Technology, #13663), anti-E-cadherin at 1:200 (Cell Signaling Technology, #3195), and secondary antibody was Dnk pAb to Rb IgG (Alexa Fluor 647, Abcam).
All experiments were performed in triplicate. All data are expressed as the mean ± standard deviation (SD). The statistical analysis of differences was performed using t test of variance and Pearson’s r test of correlation using GraphPad Prism version 6.0. P < 0.05 was considered statistically significant.
We searched the online microRNAs target database Target Scan and found that Cdc42 was a candidate target gene of miR-15a (Figure 1A). We then constructed firefly luciferase reporter vectors containing either wild-type Cdc42 3’UTR (Cdc42-WT) or mutated Cdc42 3’UTR with a modified miR-15a binding site (Cdc42-MUT). We transfected 293T cells with either Cdc42-WT or Cdc42-MUT, along with plasmid containing miR-15a mimics (miR-15a) or its negative control plasmid (miR-NC). At 48 h after transfection, we used a dual-luciferase reporter assay to examine the relative luciferase activities. The relative luciferase activities were decreased in cells cotransfected with Cdc42-WT and miR-15a mimics compared to cells cotransfected with Cdc42-MUT and miR-15a mimics (Figure 1B, P < 0.05), confirming that Cdc42 was a direct target of miR-15a.
TNF−α induces miR-15a upregulation and Cdc42 downregulation in Caco-2 cells
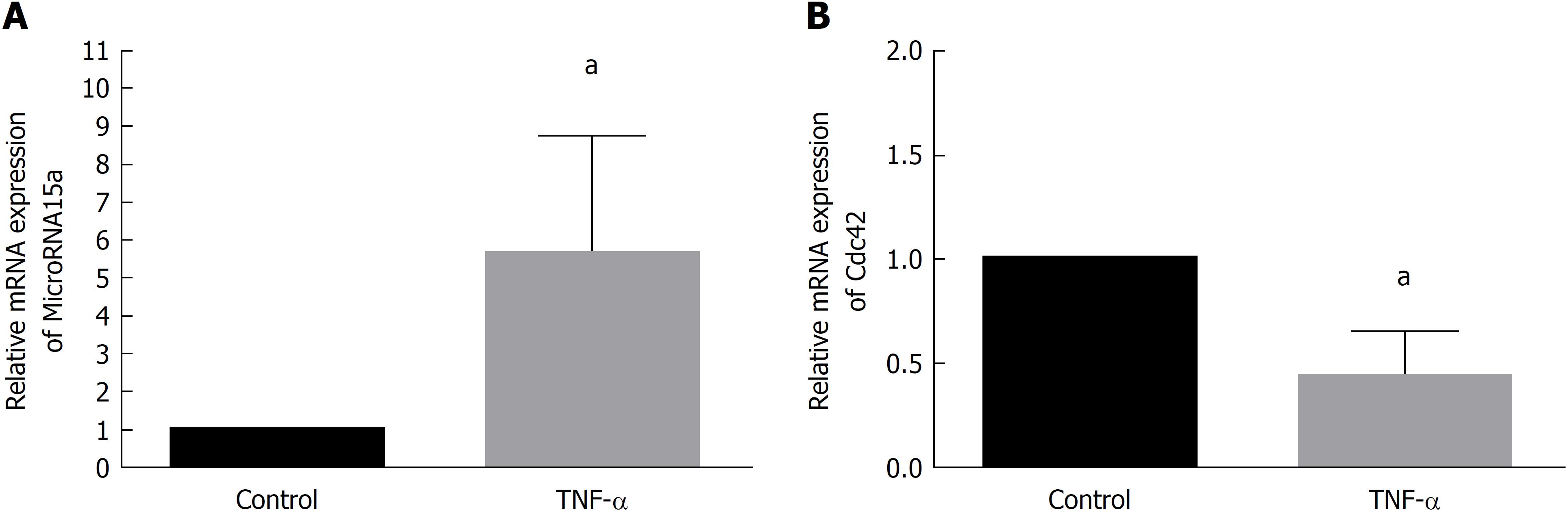
We cultured Caco-2 cells and passaged 1 × 105 cells in 35-mm dishes. After 24 h, we cultured cells with medium containing 35 ng/mL TNF-α (TNF-α group) or regular medium (control group) for 60 h and then harvested. qRT-PCR demonstrated that miR-15a levels increased upon TNF-α treatment compared to the control group (Figure 2A), while Cdc42 levels decreased in the TNF-α group compared to the control group (Figure 2B). These results suggest that TNF-α induced miR-15a upregulation and Cdc42 downregulation, and high TNF-α expression has been found in most pediatric patients[25].
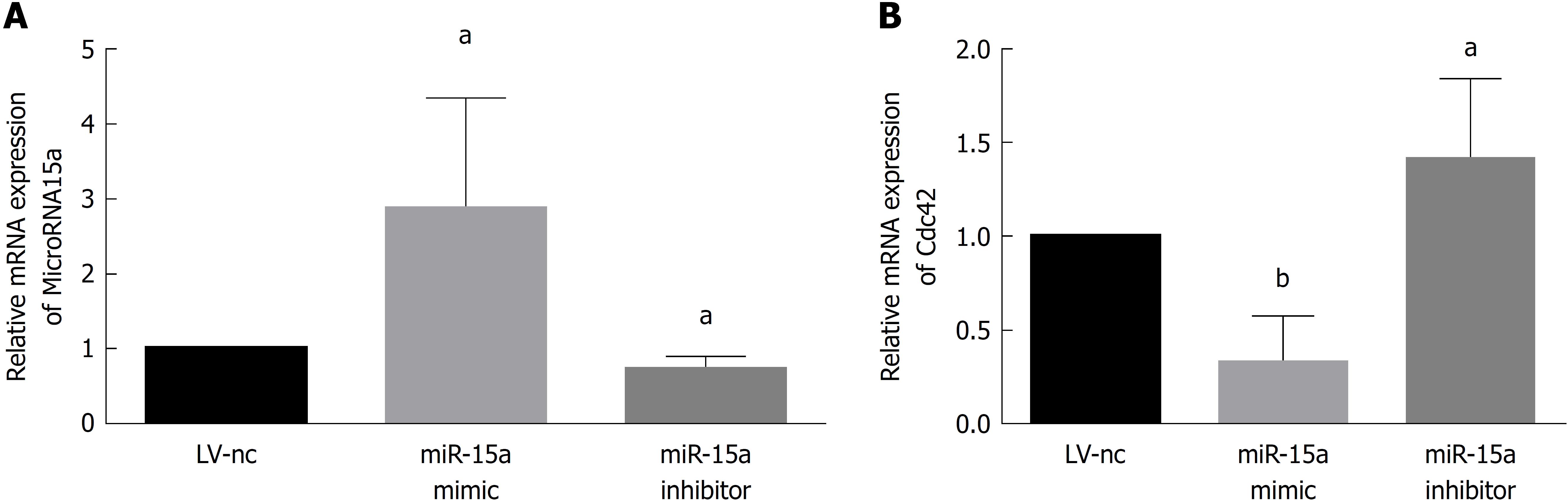
We transfected Caco-2 cells with miR-15a mimic lentivirus and its negative control (miR15a-NC) or miR-15a inhibitor lentivirus and its negative control (LV-NC) and found no difference in miR-15a and Cdc42 expression between the two negative control groups. In the miR-15a mimic group, miR-15a was overexpressed compared to the negative control group (Figure 3A). In the miR-15a inhibitor group, miR-15a expression was decreased compared to the negative control group (Figure 3A). Cdc42 was downregulated in the miR-15a mimic group and upregulated in the miR-15a inhibitor group (Figure 3B). These results demonstrate that miR-15a negatively regulates Cdc42 in Caco-2 cells.
MiR-15a overexpression downregulates epithelial junctions, while overexpressed Cdc42 rescues low epithelial junction protein expression induced by miR-15a upregulation or high TNF-α expression
We found that in the TNF-α group, the epithelial junction proteins E-cadherin and ZO-1 were decreased compared to the control (WT) group (Figure 4A-D). In the miR-15a mimic group, E-cadherin and ZO-1 were also decreased compared with the WT group (Figure 4A-D). We found no difference in E-cadherin and ZO-1 expression between the WT and miR15a-NC groups.
We transfected Caco-2 cells with miR-15a inhibitor lentivirus or its negative control (LV-NC). After passage, we added 35 ng/mL TNF-α to stimulate the cells for 60 h (miR-15a inhibitor + TNF-α or LV-NC + TNF-α). E-cadherin and ZO-1 expression did not change in the miR-15a inhibitor + TNF-α cells compared to the WT group (Figure 4A-D). E-cadherin and ZO-1 expression was similar in the LV-NC + TNF-α group to the WT + TNF-α group.
We also transfected Caco-2 cells with Cdc42 lentivirus (Lv-Cdc42) or its negative control (Lv-Cdc42-NC). Cdc42 was overexpressed in the Lv-Cdc42 group compared to the Lv-Cdc42-NC group, but there were no differences between the WT and Lv-Cdc42-NC groups. Expression of E-cadherin and ZO-1 was not different in the Lv-Cdc42-NC group compared to the WT cells or the miR15a-NC group.
We then used 35 ng/mL TNF-α to stimulate Lv-Cdc42 (Lv-Cdc42 + TNF-α) and Lv-Cdc42-NC cells (Lv-Cdc42-NC + TNF-α) for 60 h and found that E-cadherin and ZO-1 expression decreased in the Lv-Cdc42-NC + TNF-α cells compared to the WT cells (Figure 4A-D), while it increased in the Lv-Cdc42 + TNF-α cells compared to the miR-15a mimic or Lv-Cdc42-NC + TNF-α cells (Figure 4A-D).
Taken together, these results demonstrate that overexpressed TNF-α can disrupt the intestinal epithelial barrier. miR-15a overexpression can damage the intestinal epithelia similarly to TNF-α overexpression. Increased Cdc42 expression can reverse the damage caused by miR-15a overexpression.
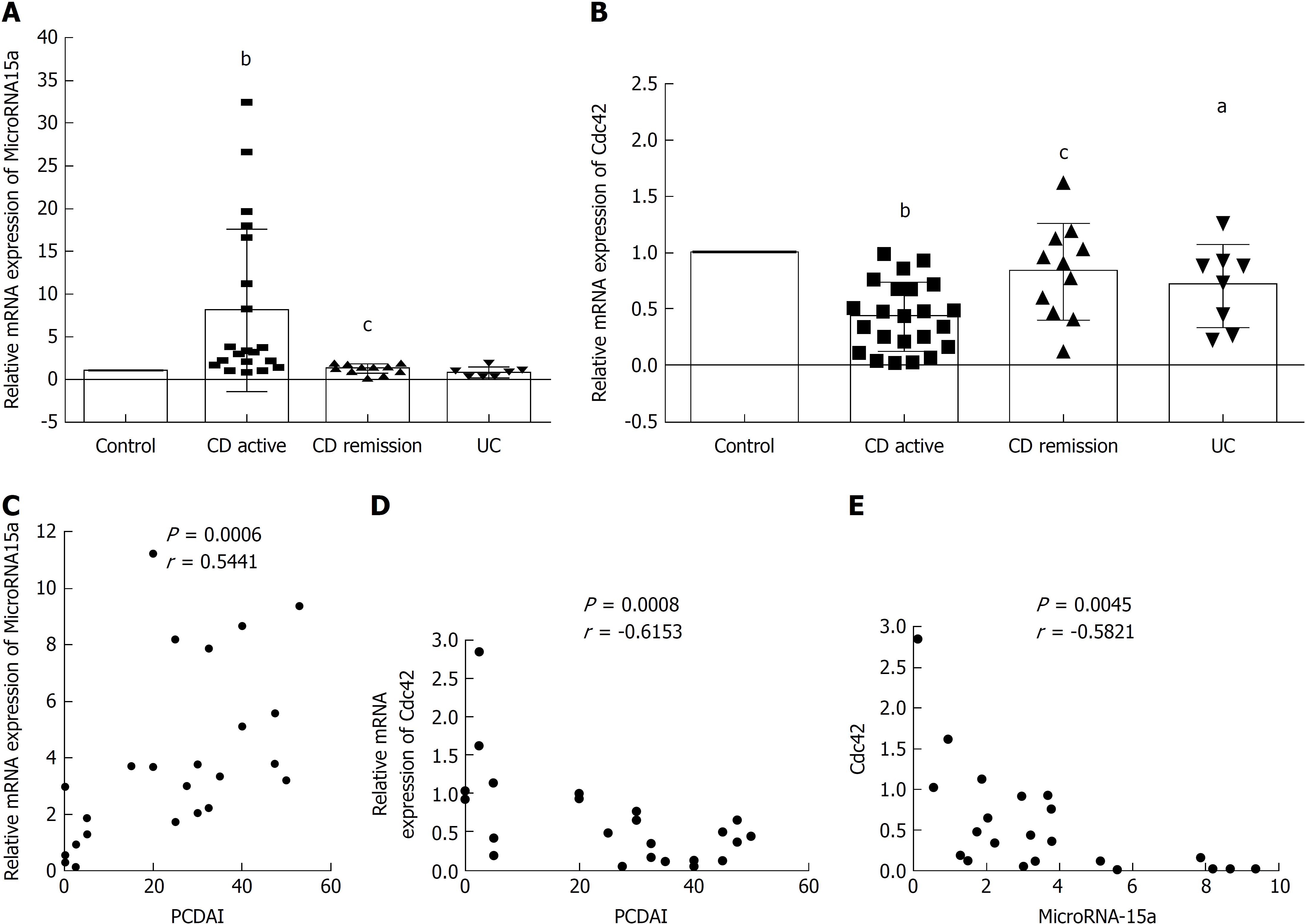
We investigated the expression of miR-15a and Cdc42 in 33 pediatric IBD patients (26 CD and seven UC) and 21 controls from our hospital (Table 2). PCDAI or PUCAI was calculated as follows: CD active group (PCDAI ≥ 10, CD AC group), CD remission group (PCDAI < 10, CD RE group), UC active group (PUCAI ≥ 10 group); 19 in CD AC group, seven in CD RE group, and seven in UC group (Table 3). Ileocecal tissues were collected by endoscopy from all patients. qRT-PCR was used to evaluate miR-15a and Cdc42 gene expression in these tissues, and the CD AC group, CD RE group, and UC group were compared with the age- and sex-matched control group (Table 3). qRT-PCR showed that miR-15a expression was increased in the CD AC group compared to the control group (Figure 5A), while it was decreased in the CD RE group compared to the CD AC group (Figure 5A). Cdc42 was decreased in the CD AC group compared to the control group (Figure 5B), while it was increased in the CD RE group compared to the CD AC group (Figure 5B). Cdc42 expression was decreased in the UC group compared to control group (Figure 5B). miR-15a expression did not significantly change between the UC and control groups (Figure 5A).
| Matched group | Patient no. | Group | Sex | Age, yr | Disease | PCDAI/PUCAI |
| 1 | 1 | Control | M | 3 | JP | |
| 2 | Control | M | 3 | JP | ||
| 3 | CD AC | M | 3 | CD | 52.5 | |
| 4 | CD AC | M | 2.5 | CD | 25 | |
| 2 | 5 | Control | F | 7 | JP | |
| 6 | Control | F | 7 | JP | ||
| 7 | CD RE | M | 8 | CD | 0 | |
| 8 | CD RE | F | 7 | CD | 2.5 | |
| 9 | UC | F | 7 | UC | 30 | |
| 3 | 10 | Control | M | 8 | JP | |
| 11 | Control | M | 9 | JP | ||
| 12 | CD AC | M | 7.5 | CD | 27.5 | |
| 13 | UC | M | 8 | UC | 20 | |
| 14 | UC | M | 8 | UC | 20 | |
| 4 | 15 | Control | F | 9 | JP | |
| 16 | CD AC | F | CD | 40 | ||
| 5 | 17 | Control | M | 10 | JP | |
| 18 | Control | M | 10 | JP | ||
| 19 | Control | M | 10 | JP | ||
| 20 | CD AC | M | 9.6 | CD | 20 | |
| 21 | CD AC | M | 10 | CD | 30 | |
| 22 | CD RE | M | 10 | CD | 5 | |
| 6 | 23 | Control | M | 11 | JP | |
| 24 | Control | M | 11 | JP | ||
| 25 | CD AC | M | 11 | CD | 32.5 | |
| 26 | CD AC | M | 12.2 | CD | 50 | |
| 27 | CD AC | M | 11 | CD | 45 | |
| 28 | CD AC | F | 11 | CD | 15 | |
| 7 | 29 | Control | F | 12 | JP | |
| 30 | Control | F | 12 | JP | ||
| 31 | Control | F | 12 | JP | ||
| 32 | CD AC | F | 12 | CD | 25 | |
| 33 | CD AC | F | 12 | CD | 35 | |
| 34 | CD AC | F | 12 | CD | 32.5 | |
| 35 | CD AC | F | 12 | CD | 47.5 | |
| 36 | CD RE | M | 13.1 | CD | 0 | |
| 37 | UC | M | 13.1 | UC | 35 | |
| 38 | UC | F | 12 | UC | 20 | |
| 8 | 39 | Control | F | 13 | JP | |
| 40 | CD AC | F | 13 | CD | 45 | |
| 41 | CD AC | F | 12.8 | CD | 32.5 | |
| 9 | 42 | Control | F | 14 | JP | |
| 43 | CD AC | F | 14 | CD | 20 | |
| 44 | UC | F | 14 | UC | 40 | |
| 45 | CD RE | F | 14 | CD | 2.5 | |
| 10 | 46 | Control | M | 14 | JP | |
| 47 | CD RE | M | 15.1 | CD | 0 | |
| 48 | CD AC | M | 14 | CD | 30 | |
| 49 | CD AC | M | 14 | CD | 47.5 | |
| 11 | 50 | Control | F | 16 | JP | |
| 51 | CD RE | F | 15.8 | CD | 5 | |
| 12 | 52 | Control | M | 5 | JP | |
| 53 | Control | M | 5 | JP | ||
| 54 | UC | M | 6.1 | UC | 35 |
We also used Pearson’s r test to analyze the correlation between miR-15a, Cdc42 and PCDAI. We found that miR-15a was positively correlated with PCDAI (Figure 5C), Cdc42 was negatively correlated with PCDAI (Figure 5D), and miR-15a was negatively correlated with Cdc42 (Figure 5E).
Altogether, these results demonstrate that miR-15a negatively regulates Cdc42 in pediatric IBD patients, and both are correlated with PCDAI. PCDAI is one of the indicators of the severity of CD.
IBD is a multifactorial disease, characterized by chronic inflammation of the gastrointestinal tract. MicroRNAs can offer a potential missing link between the genetic, en–vironmental and immunological factors involved in the pathogenesis of IBD[26,27]. They play an important role in the development of IBD. MicroRNAs mediate post-transcriptional gene expression by binding to the 3’-UTRs of target genes, thus inhibiting protein translation or inducing gene degradation. Altered expression of microRNAs has been linked to different cell functions, including signal transduction, differentiation, proliferation and apoptosis[28-30]. In this study, we focused on miR-15a and found that miR-15a can bind the 3’-UTR of Cdc42, suggesting that Cdc42 is a target gene of miR-15a. Dual-luciferase reporter assay confirmed that Cdc42 is a miR-15a target gene. Our results were similar to other studies[21-23]. Cdc42 is a Rho family small GTPase that is known to control various cellular functions including adhesion, migration, transcription and growth[10,11]. It has a special role in intestinal stem cell division, survival and differentiation in mice[10,14]. It was recently reported that Cdc42 is critical for intestinal epithelial stem cell differentiation into a functional intestinal barrier[10,14]. A number of studies in IBD patients have shown that intestinal barrier function is disrupted in active and quiescent disease states.
High TNF-α expression has been found in most pediatric patients[25]. TNF-α has an important role in microRNA expression. Therefore, we used TNF-α to stimulate Caco-2 cells and found that TNF-α upregulated miR-15a and downregulated Cdc42. This result demonstrated that TNF-α has an important role in miR-15a expression and that miR-15a-Cdc42 signaling participated in the development of IBD. Therefore, we used lentiviruses to up- or downregulate miR-15a expression in Caco-2 cells. We found that in the miR-15a mimic group, Cdc42 was downregulated, but upregulated in cells infected with a miR-15a inhibitor. These results show that miR-15a negatively regulates Cdc42.
We also found that after TNF-α stimulation, E-cadherin and ZO-1 expression decreased in Caco-2 cells. These belong to the intestinal epithelial junction proteins. The change in epithelial junction proteins can disrupt the epithelial barrier. A number of studies in IBD patients has demonstrated that intestinal barrier function is disrupted. We also found that expression of E-cadherin and ZO-1 also decreased in miR-15a mimic Caco-2 cells, similar to cells with high TNF-α expression. However, E-cadherin and ZO-1 expression did not decrease in the miR-15a inhibitor + TNF-α group compared to the WT group.
We transfected Caco-2 cells with Cdc42 lentivirus (Lv-Cdc42) or its negative control (Lv-Cdc42-NC), and then stimulated both with TNF-α. We found that in Lv-Cdc42 cells, E-cadherin and ZO-1 expression increased compared to Lv-Cdc42-NC + TNF-α or miR-15a mimic cells, suggesting that Cdc42 overexpression can reverse epithelial junction expression caused by miR-15a upregulation. These results demonstrated that miR15a negatively regulates intestinal epithelial junctions through Cdc42 in Caco-2 cells. The miR-15a-Cdc42 signaling pathway is involved in many other biological processes, and our study confirmed it to be important in the development of gastrointestinal inflammation.
We investigated miR-15a and Cdc42 expression in pediatric IBD patients and found that miR-15a was increased and Cdc42 was decreased in the CD AC group compared to the control group. In contrast, miR-15a was decreased and Cdc42 increased in the CD RE group compared to the CD AC group. We analyzed the correlation between miR-15a, Cdc42 and PCDAI and found that miR-15a was positively correlated with PCDAI, Cdc42 negatively correlated with PCDAI, and miR-15a negatively correlated with Cdc42. These results demonstrated that miR-15a negatively regulates Cdc42 in children with IBD, and both are correlated with PCDAI.
This study had some limitations. First, we found a correlation between miR-15a, Cdc42 and PCDAI, but the R values were not high, possibly due to the small number of patients. Second, we only included seven UC patients, which may be one of the reasons why we did not find altered expression of miR-15a in UC patients.
This study shows that miR-15a negatively regulates intestinal epithelial junctions through Cdc42, but we did not study how Cdc42 regulates intestinal epithelial junctions. This will be studied in our future research.
In conclusion, miR-15a negatively regulates intestinal epithelial junctions through Cdc42 in Caco-2 cells and pediatric IBD. This is believed to be the first study of the miR-15a-Cdc42 signaling pathway in pediatric IBD patients. Additionally, this study provides evidence that Cdc42 is a miR-15a target gene, and this will provide a critical clue for understanding IBD development.
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder with an increasing prevalence. Pediatric IBD has severe phenotypes, but the etiology of IBD remains unknown. A number of studies have shown that microRNAs participate in the development of IBD and that miR-15a has an altered expression level in IBD patients, but the underlying mechanisms have not been fully elucidated. Cell division cycle (Cdc)42 is a Rho family small GTPase, and it has been reported that Cdc42 is critical for intestinal epithelial stem cell differentiation into a functional intestinal barrier. MicroRNA-Cdc42 signaling plays a role in some biological processes, and Target Scan identified a putative seed region of miR-15a in the wild-type 3’-UTR of Cdc42. Thus, we hypothesized that miR-15a regulates Cdc42 to disrupt the intestinal barrier in IBD development.
The main goal of this study is to find the role of microRNA-Cdc42 signaling in pediatric IBD development. Our study found that miR-15a negatively regulates intestinal epithelial junctions through Cdc42 in Caco-2 cells and in pediatric IBD. This finding will provide a critical clue for understanding IBD development in the future.
The objectives of this study were to determine whether Cdc42 is regulated by miR-15a in the development of pediatric IBD. Through our study, we found that miR-15a negatively regulates intestinal epithelial junctions through Cdc42 in Caco-2 cells and in pediatric IBD. This study provides a new clue for understanding the development of IBD and will help researchers study the etiology of IBD and provide new methods for IBD treatment in the future.
We cultured cells, used tumor necrosis factor (TNF)-α to stimulate cells, employed plasmids and lentiviruses to change miR-15a and Cdc42 expression, performed dual-luciferase assay, quantitative reverse transcription polymerase chain reaction and immunofluorescence. Statistical analysis was performed using t test of variance and Pearson’s r test of correlation using Graphpad Prism version 6.0.
We demonstrated that miR-15a negatively regulates intestinal epithelial junctions through Cdc42 in Caco-2 cells and pediatric IBD. This is believed to be the first study of the miR-15a-Cdc42 signaling pathway in pediatric IBD patients.
This study had some limitations. First, we found a correlation between miR-15a, Cdc42 and Pediatric Crohn’s Disease Activity Index, but the R values were not high, possibly due to the small number of patients. Second, we only included seven ulcerative colitis (UC) patients, which may be one of the reasons why we did not find altered miR-15a expression in UC patients. Last, we showed that miR-15a negatively regulates intestinal epithelial junctions through Cdc42, but we did not study how Cdc42 regulates intestinal epithelial junctions. This will be studied in our future research.
MiR-15a negatively regulates intestinal epithelial junctions through Cdc42 in pediatric IBD. This is believed to be the first study of the miR-15a - Cdc42 signaling pathway in pediatric IBD patients. Additionally, this study provides evidence that Cdc42 is a miR-15a target gene, and this will provide a critical clue for understanding IBD development, and may also provide new methods for future IBD treatments.
It is better get more patients, particularly UC patients. We will study how Cdc42 regulates intestinal epithelial junctions in our future research.
The authors are grateful to Xiao-Long Dong for his skillful technical assistance and Hua-Qing Zhong for bioinformatics support.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: de Souza HSP, Sebastian S S- Editor: Ma RY L- Editor: Filipodia E- Editor: Yin SY
| 1. | Wang XQ, Zhang Y, Xu CD, Jiang LR, Huang Y, Du HM, Wang XJ. Inflammatory bowel disease in Chinese children: a multicenter analysis over a decade from Shanghai. Inflamm Bowel Dis. 2013;19:423-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (1)] |
| 2. | Chandel S, Prakash A, Medhi B. Current scenario in inflammatory bowel disease: drug development prospects. Pharmacol Rep. 2015;67:224-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Yang L, Yan Y. Protein kinases are potential targets to treat inflammatory bowel disease. World J Gastrointest Pharmacol Ther. 2014;5:209-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (1)] |
| 4. | Raisch J, Darfeuille-Michaud A, Nguyen HT. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol. 2013;19:2985-2996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 130] [Cited by in RCA: 158] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 5. | Rebane A, Akdis CA. MicroRNAs: Essential players in the regulation of inflammation. J Allergy Clin Immunol. 2013;132:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 6. | Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624-1635.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 404] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 7. | Iborra M, Bernuzzi F, Invernizzi P, Danese S. MicroRNAs in autoimmunity and inflammatory bowel disease: crucial regulators in immune response. Autoimmun Rev. 2012;11:305-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 8. | Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, Ando T, Oyamada Y, Isozaki Y, Hongo H. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol. 2010;25 Suppl 1:S129-S133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 9. | Béres NJ, Szabó D, Kocsis D, Szűcs D, Kiss Z, Müller KE, Lendvai G, Kiss A, Arató A, Sziksz E. Role of Altered Expression of miR-146a, miR-155, and miR-122 in Pediatric Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:327-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J Cell Biol. 2008;183:625-633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 282] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 11. | Jin D, Durgan J, Hall A. Functional cross-talk between Cdc42 and two downstream targets, Par6B and PAK4. Biochem J. 2015;467:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 12. | Elvers M. RhoGAPs and Rho GTPases in platelets. Hamostaseologie. 2016;36:168-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Yang L, Zheng Y. Cdc42: a signal coordinator in hematopoietic stem cell maintenance. Cell Cycle. 2007;6:1445-1450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Sakamori R, Das S, Yu S, Feng S, Stypulkowski E, Guan Y, Douard V, Tang W, Ferraris RP, Harada A. Cdc42 and Rab8a are critical for intestinal stem cell division, survival, and differentiation in mice. J Clin Invest. 2012;122:1052-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 15. | Schlegel N, Meir M, Spindler V, Germer CT, Waschke J. Differential role of Rho GTPases in intestinal epithelial barrier regulation in vitro. J Cell Physiol. 2011;226:1196-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Mehta S, Nijhuis A, Kumagai T, Lindsay J, Silver A. Defects in the adherens junction complex (E-cadherin/ β-catenin) in inflammatory bowel disease. Cell Tissue Res. 2015;360:749-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Groschwitz KR, Hogan SP. Intestinal barrier function: molecular regulation and disease pathogenesis. J Allergy Clin Immunol. 2009;124:3-20; quiz 21-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1248] [Article Influence: 78.0] [Reference Citation Analysis (2)] |
| 18. | McCole DF. IBD candidate genes and intestinal barrier regulation. Inflamm Bowel Dis. 2014;20:1829-1849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 19. | Landy J, Ronde E, English N, Clark SK, Hart AL, Knight SC, Ciclitira PJ, Al-Hassi HO. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol. 2016;22:3117-3126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 294] [Cited by in RCA: 369] [Article Influence: 41.0] [Reference Citation Analysis (4)] |
| 20. | Förster C. Tight junctions and the modulation of barrier function in disease. Histochem Cell Biol. 2008;130:55-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 392] [Cited by in RCA: 454] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 21. | Wang J, Xu R, Wu J, Li Z. MicroRNA-137 Negatively Regulates H2O2-Induced Cardiomyocyte Apoptosis Through CDC42. Med Sci Monit. 2015;21:3498-3504. [PubMed] |
| 22. | Zhang H, Fu Y, Shi Z, Su Y, Zhang J. miR-17 is involved in Japanese Flounder (Paralichthys olivaceus) development by targeting the Cdc42 mRNA. Comp Biochem Physiol B Biochem Mol Biol. 2016;191:163-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Zheng L, Zhang Y, Lin S, Sun A, Chen R, Ding Y, Ding Y. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int J Clin Exp Pathol. 2015;8:10534-10544. [PubMed] |
| 24. | Levine YY, Koletzko J, Turner D. [ESPGHAN revised Porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents]. Zhonghua Er Ke Za Zhi. 2016;54:728-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 25. | Mouzaoui S, Djerdjouri B, Makhezer N, Kroviarski Y, El-Benna J, Dang PM. Tumor necrosis factor-α-induced colitis increases NADPH oxidase 1 expression, oxidative stress, and neutrophil recruitment in the colon: preventive effect of apocynin. Mediators Inflamm. 2014;2014:312484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology. 2013;145:293-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 231] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 27. | Lin J, Welker NC, Zhao Z, Li Y, Zhang J, Reuss SA, Zhang X, Lee H, Liu Y, Bronner MP. Novel specific microRNA biomarkers in idiopathic inflammatory bowel disease unrelated to disease activity. Mod Pathol. 2014;27:602-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 28. | Chen WX, Ren LH, Shi RH. Implication of miRNAs for inflammatory bowel disease treatment: Systematic review. World J Gastrointest Pathophysiol. 2014;5:63-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 29. | Papaconstantinou I, Stamatis K, Tzathas C, Vassiliou I, Giokas G, Gazouli M. The role of variations within microRNA in inflammatory bowel disease. Eur J Gastroenterol Hepatol. 2013;25:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 30. | Whiteoak SR, Felwick R, Sanchez-Elsner T, Fraser Cummings JR. MicroRNAs in inflammatory bowel diseases: paradoxes and possibilities. Inflamm Bowel Dis. 2015;21:1160-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |