Published online Nov 7, 2018. doi: 10.3748/wjg.v24.i41.4635
Peer-review started: August 7, 2018
First decision: August 30, 2018
Revised: September 2, 2018
Accepted: October 5, 2018
Article in press: October 5, 2018
Published online: November 7, 2018
Processing time: 92 Days and 12.8 Hours
Esophageal cancer (EC) presents a high mortality rate, mainly due to its aggressive nature. Squamous cell carcinoma is the most common histological type worldwide, though, a continuous increase in esophageal adenocarcinomas has been noted in the past decades. Common risk factors associated with EC include smoking, alcohol consumption, gastroesophageal reflux disease, Barrett’s esophagus and obesity. In an effort to overcome chemotherapy resistance in oncology, it was discovered that histone acetylation/deacetylation equilibrium is altered in carcinogenesis, leading to changes in chromatin structure and altering expression of genes important in the cell cycle, differentiation and apoptosis. Based on this knowledge, histone acetylation was addressed as a potential novel chemotherapy drug target to repress cancer cell proliferation. There are four classes of histone deacetylases (HDACs) inhibitors with a variety of different mechanisms of actions that render them possible anti-cancer drugs. They arrest the cell cycle, inhibit differentiation and angiogenesis and induce apoptosis. They do not necessarily act on histone proteins, since they can also exert indirect anti-cancer effects, by modifying various cellular proteins. In addition, HDACs have also been associated with increased chemotherapy resistance. Based on the literature, HDACs have been associated with EC, with surveys revealing that increased expression of certain HDACs correlates with advanced TNM stages, tumor grade, metastatic potential and decreased 5-year overall and disease-free survival. The aim of this survey is to elucidate the molecular identity and mechanism of action of HDAC inhibitors as well as verify their potential utility as anti-cancer agents in esophageal cancer.
Core tip: Esophageal cancer (EC) remains one of the most lethal malignancies, mainly due to its aggressive nature. In an effort to overcome chemotherapy resistance, it was discovered that histone acetylation/deacetylation equilibrium is altered in carcinogenesis, leading to changes in chromatin structure and altering expression of genes important in the cell cycle, differentiation and apoptosis. Therefore, histone acetylation was addressed as a potential novel chemotherapy drug target. Based on the literature, histone deacetylases (HDACs) have been associated with EC, with surveys elucidating that increased expression of certain HDACs correlates with advanced TNM stages, tumor grade, metastatic potential and decreased 5-year overall and disease-free survival.
- Citation: Schizas D, Mastoraki A, Naar L, Spartalis E, Tsilimigras DI, Karachaliou GS, Bagias G, Moris D. Concept of histone deacetylases in cancer: Reflections on esophageal carcinogenesis and treatment. World J Gastroenterol 2018; 24(41): 4635-4642
- URL: https://www.wjgnet.com/1007-9327/full/v24/i41/4635.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i41.4635
Esophageal cancer (EC) remains one of the most lethal malignancies worldwide, mainly due to its aggressive nature and the eight most common malignancy of the gastrointestinal (GI) tract[1]. It is also often diagnosed in late stages, making a curative approach less likely. The 5-year survival rate ranges from 15%-25% and disease outcome is strongly associated with early diagnosis[2]. Squamous cell carcinoma (SCC) is described as the most common histological type worldwide, though in many countries a continuous increase in esophageal adenocarcinomas has been reported. The incidence of EC is 2-4 times higher in males compared to females[3]. There is a slight difference in the predisposing parameters associated with each subtype of esophageal carcinoma, with smoking and alcohol consumption being the most important risk factors for SCC and gastroesophageal reflux disease, Barrett’s esophagus and obesity being implicated in adenocarcinomas[3]. Well defined molecular pathways and targets involved in esophageal carcinogenesis include tissue inhibitors of metalloproteinase (TIMP) 3 and 4 and vascular endothelial growth factor receptor (VEGFR). Expression of human epidermal growth factor receptor 2 (HER2)/neu and c-kit is also high in EC, with slightly higher rates of expression in adenocarcinomas rather than SCCs[4].
During the last decades there has been a lot of effort in overcoming chemotherapy resistance in tumor cells. This has led to the investigation of more cellular compounds implicated in gene expression and transcription processes. Among the findings, it was discovered that histone acetylation/deacetylation equilibrium is affected in carcinogenesis, leading to modified chromatin structure and therefore changes in gene expression[5]. It is common knowledge that in eukaryotic cells, DNA is tightly developed around a histone core, forming the nucleosome, which is the basic DNA structure. Further coiling of the nucleosomes leads to the formation of the chromosomes. Histone can undergo various alterations including acetylation, phosphorylation, methylation and ubiquitination affecting chromosomal stability and gene expression[6,7]. Uncoiling promotes gene expression, providing access of transcription factors in the DNA. On the contrary, heterochromatin represses gene transcription and is associated with hypoacetylated histones. Based on the above, histone acetylation was addressed as a potential chemotherapy drug target to repress cancer cell proliferation. Histone deacetylase (HDAC) function in human cells is to counteract the action of acetyltransferases, providing an equilibrium in histone acetylation. In cancer cells, absence of balance between acetyltransferases and HDACs provokes significant modifications in chromatin structure altering expression of genes important in the cell cycle, differentiation and apoptosis[8].
The aim of this review article is, at first, to elucidate the molecular identity and mechanism of action of HDAC inhibitors as well as verify their potential utility as anti-cancer agents. More importantly, we will also describe and critically review the relevant literature of HDAC implication in esophageal carcinoma.
Histone acetylation is sustained in all cells by the functional equilibrium between two categories of enzymes: Histone acetyltransferases (HATs) and HDACs. Based on the HDACs’ homology to their yeast analogues, they are divided in four classes. Class I are nuclear HDACs 1, 2, 3 and 8. Members of class IIa include HDACs 4, 5, 7 and 9, with HDACs 6 and 10 belonging to Class IIb and being located both in the nucleus and the cytoplasm. Class IV includes only HDAC 11. Class I, II and IV are Zn2+-dependent and work by editing histones. Class III HDACs have a different mechanism of action and are NAD+ dependent homologues of the yeast sirtuin proteins[9,10]. HDACs have recently been implicated in normal cell cycle regulation and differentiation as their pivotal role in tight gene control has been verified.
The increased function of HDACs has been associated with many neurodegenerative diseases, as well as normal aging and heart failure[11]. In addition, HDACs have been implicated in many different types of cancer, including HDAC1 that can be highly expressed in prostate, gastric, lung, esophageal, colon and breast cancer, HDAC2 in colorectal, cervical and gastric cancer cells and HDAC3 in colorectal, gastric and prostate cancer[5]. In some malignant lesions, increased expression of certain HDACs has also been correlated with worst prognosis and decreased survival.
Chromatin structure and coiling is a process very dynamic, by nature. Lysine residues’ acetylation is a mechanism of major significance in the relaxation of DNA structures and gene transcription. In cancer cells, the loss of balance between HATs and HDACs causes changes in chromatin structure and alters expression of genes basic in the cell cycle, differentiation and apoptosis. For instance, in cells that provoke hyperacetylation of histones, due to HDAC repression, chromatin is less condensed and favors an increased access of transcription factors to the DNA sequence promoter regions, increasing expression of many genes, that may potentially include oncogenes, leading to tumorigenesis[8]. On the other hand, in cells that favor hypoacetylation, through HDAC overexpression, there is an increase in cellular proliferation through proteins affecting the cell cycle. Many HDACs, including HDAC4, that is implicated in increased proliferation of EC cellular pathways, help cells overpass the G1-S checkpoint. Flow cytometry of these cell structures treated with HDAC inhibitors has shown an increase in the G1 population with a concomitant decrease in the S population of cells. The main mechanism of action of HDAC4 is through an increase in cyclin dependent kinases 2 and 4 and an augmentation in phosphorylated retinoblastoma protein. Altogether, these alterations help cells progress to the S phase of the cell cycle[12]. Another mechanism that may alter normal cell cycle control, in cells overexpressing HDACs, is through HDAC-mediated inhibition of p21 protein, that normally arrests cell cycle in G1-S phase[13].
Inhibiting histone deacetylation can cause changes in expression of many genes involved in cell differentiation, apoptosis, angiogenesis, motility, inflammation and metabolism. One of the most important ways that HDAC inhibitors affect the cell cycle is the increase provided in the expression of the gene coding for p21 protein that acts as a signal regulator for the cell to go through G1 and proceed to the S phase. In cases it is over-expressed, it functions closely with protein p53, arresting the cell cycle in G1 phase and inhibiting cellular differentiation[14].
As far as apoptosis is concerned, HDAC inhibitors have been proposed to activate both the intrinsic and extrinsic pathways. In carcinogenesis, evasion of apoptosis remains one of the most important mechanisms contributing to cell immortality. Many cancer cells have increased levels of antiapoptotic factors, including BCL-XL[15]. Incubation of such cells in an HDAC inhibitor leads to a downregulation of related factors and upregulation of pre-apoptotic ones[16]. HDAC inhibitors can also induce apoptosis even when caspases enzymes are inhibited, showing the ability to accelerate cell death even in apoptosis-resistant cells, possibly through the activation of autophagy[5]. At this point, it is important to understand that HDAC overexpression occurs only in cancer cells. As a result, HDAC inhibitors neither affect cell cycle nor induce apoptosis in cells with normal HAT/HDAC expression[17].
HDACs have also been found to remove acyl groups from other non-histone proteins, like transcription factors and important regulatory proteins of cellular proliferation, differentiation and apoptosis[8]. After understating the implication of histones in chromatin structure and gene expression, there were a lot of questions concerning the accumulation of these non-histone proteins and whether they do influence the anti-cancer action of these novel agents. To answer that, DNMT3B, a methyltransferase that abnormally methylates and suppresses tumor suppressor genes and LSD1, which has been implicated in sustaining cancer cell proliferation were assessed[18]. Results after HDAC inhibition were indicative of a significant decrease in both DNMT3B and LSD1, showing potential additional anti-cancer mechanisms of action of these drugs[10]. Therefore, HDAC inhibition can affect gene transcription in cancer cells in a direct fashion with changes in the histone core, or in an indirect modality through the removal of acyl groups from other important regulatory proteins. Using HDAC inhibitors can induce a state of growth repression, arrest differentiation and initiate apoptosis[8].
Furthermore, HDAC over-expression in cancer cells has been associated with increased macropinocytosis highly related with cellular migration and substantial metastatic potential. HDAC6 activity, precisely, has been shown to increase macropinocytosis. Supporting the above information, HDAC6 inhibition led to a decrease in cellular migration and endocytosis, through actin remodeling processes, reducing the metastatic capacity of tumor cells[19]. Zhu et al[20] taking advantage of this HDAC associated increased macropinocytosis, linked an HDAC inhibitor (cinnamic acid) to neutral red (NR), a molecule well known for achieving huge intracellular concentrations through pinocytosis. The resulting HDAC inhibition’s cytotoxicity was augmented by increased drug accumulation in cancer cells through macropinocytosis. This study displayed for the first time an important alternative pathway for drug distribution to cancer cells, exploiting HDAC-induced macropinocytosis[20].
Moreover, HDACs can also play a major role in chemotherapy resistance. One hypothesis that can explain this finding is based on the changes on chromatin structure. Altering chromatin’s coiling status can prevent DNA targeting drugs from accessing their target molecule, reducing the extent of DNA damage and improving cellular survival. Taking the above into consideration, it is understandable why HDAC inhibitors have been shown to increase chemotherapy cell vulnerability[21]. Results from different clinical trials have shown that combination treatment of an HDAC inhibitor with chemotherapy regimens or ionizing radiation leads to an additive/synergistic effect, partly explained through hyper-acetylation mediated DNA relaxation and increased drug penetration[5].
Last but not least, HDAC inhibition increases reactive oxygen species (ROS), causing DNA and membrane damage in cancer cells. At the same time, it affects angiogenesis, a process of utmost importance in cancer cell survival, by decreasing the levels of circulating VEGF and hypoxia inducible factor-1 alpha (HIF-1α), arresting cellular proliferation[22]. Additionally, one important feature of all HDAC inhibitors in tumors is the formation of more differentiated, less aggressive cellular colonies[23].
However, despite the increasing amount of information we possess on the field of HDACs and their inhibitors as anti-cancer drugs, there are many cellular interactions that are not yet thoroughly investigated. HDAC inhibition leads to activation of various cellular pathways and acetylation of many different cellular proteins. It is interesting to identify these pathways and describe their interactions with HDACs, since gene analysis in cells treated with an HDAC inhibitor has shown an increase in gene proteins associated with cell cycle processes and cell to cell adhesions[23]. In that direction, Yar Saglam et al[24] studied the potential association of an HDAC inhibitor with EF24 that inhibits NF-κB pathway, based on the knowledge that class I HDAC inhibitors cause augmentation in genes regulated by this pathway, leading to an increase in cellular proliferation. Interestingly, their results verified that combination treatment further decreased cellular proliferation than the administration of class I HDAC inhibitor alone[24].
HDAC inhibitors include four categories of drugs that are not structurally equal and also show different affinity for the various categories of HDACs. The 4 classes include the short chain fatty acids (butyrate and valproic acid), hydroxamates (i.e., trichostatin A-TSA), benzamides (i.e., MS275) and cyclic tetrapeptides (i.e., FK228). Hydroxamates are the only type of HDAC inhibitors that possess a pan-HDAC inhibition activity, with the rest showing a more selective inhibition[25]. At the moment, United States Food and Drug Administration (FDA) has approved three HDAC inhibitors as anti-cancer medication, including Romodepsin (FK228), Vorinistat (SAHA) and Belinostat approved in 2014, with many others undergoing clinical trials[26]. The most sensitive biomarkers used in measuring HDAC inhibition effect are H3 and H4[27].
Novel HDAC inhibitors can have several heterogenous mechanisms of action. Their epigenetic activation of many genes through histone modification is well described. Henderson et al[16] developed AR-42, a hydroxamate combined phenylbutyrate, that is one of the newest inhibitors under clinical trials with many histone-independent actions. It mediates Akt dephosphorylation, inhibits gp130/Stat3 pathway, inactivates mechanisms implicated in DNA repair, suppresses kit activation and initiates proteasomal degradation of topoisomerase II[16]. Also, it has a unique effect on tumor cachexia, inhibiting cancer-induced muscle atrophy and weight loss that is important, since cachexia is prevalent among cancer patients and also increases morbidity and mortality[16]. The variety of mechanisms also underlines the need for further in vitro investigation of the HDAC inhibitors and the necessity for a better understanding of all the interconnected cellular pathways.
Table 1 summarizes the mechanisms of action of HDACs in carcinogenesis.
| Increased expression of genes associated with the cell cycle, increasing proliferation, by overpassing the G1-S checkpoint, either by increasing CDK-2 and -4[12], or by inhibiting p21 protein that arrests cell cycle[13]. |
| Inhibition of both the intrinsic and extrinsic apoptotic pathways[15]. |
| Acyl group removal from other non-histone proteins, like transcription factors and regulatory proteins, indirectly affecting cellular proliferation[8]. |
| Increased macropinocytosis, a process closely related to cellular migration and increased metastatic potential[19]. |
| Increased chemotherapy resistance in tumor cells[21]. |
| Protection from cellular damage caused by ROS[22]. |
| Inhibition of angiogenesis by decreasing VEGF and HIF-1α[22]. |
| Reduction of E-cadherin and increased vimentin expression, increasing metastatic potential[12]. |
| Increased HSP function, conferring stability to oncogenic proteins[33]. |
| Decreased function of DNA repair enzymes[39]. |
As mentioned above, EC is one of the most fatal carcinomas of the GI tract, mainly due to its late diagnosis and aggressive nature. Thus, there is an increasing need for tools and biomarkers that will be able to predict prognosis after resection, resistance to chemotherapy, or help develop new combination regimens to increase patients’ survival. As in many other cancers, loss of normal equilibrium between histone acetyltransferases and deacetylases play a significant role in esophageal carcinogenesis. DNMT1 and class I HDAC1, 2 and 3 are often increased in EC[28]. To our knowledge, information about the implication of HDACs and possible use of HDAC inhibitors in EC have not been gathered before in one article.
Available data on HDAC expression in esophageal adenocarcinomas are limited in the literature. Class I HDACs seem to be commonly implicated, with HDAC1 and HDAC2 being separately studied. Langer et al[29] suggested that increased HDAC1 expression (detected in 46% of relevant specimens) did not correlate with T, N or M stage and neither with tumor grade. On the other hand, HDAC2 overexpression (found in 70% of the specimens) showed a statistically significant correlation with increased lymphatic spreading of the tumor (N stage) and lower tumor differentiation (higher grade). Nevertheless, correlation between HDAC1 or 2 expression and survival analysis has not been verified[29]. Furthermore, increased HDAC2 expression showed a non-significant tendency for better chemotherapy response, compared to HDAC1, but it cannot be used as a biomarker for chemotherapy cell vulnerability[29].
In esophageal SCC (ESCC) increased interest has been expressed in understanding the implication of HDAC4 in carcinogenesis. Among 86 paired ESCC patients, Zeng et al[12] reported a significant increase in HDAC4 expression in cancer cells compared to paired normal tissue samples. In an effort to assess HDAC4’s clinical significance in these patients, high expression was statistically significantly associated with unfavorable clinicopathologic characteristics, including higher grade cancer and more advanced T, N and TNM stages[12]. Furthermore, patients with high HDAC4 expression showed a 5-year overall survival of 20.15% with a 5-year progression free survival of 30.92%, that were significantly shorter compared to patients with low HDAC4 expression that presentd 68.75% and 53.59% relevant rates respectively[27]. From this study, HDAC4 was found to be, together with TNM stage, a significant predictor of overall survival and a marginally significant parameter of progression free survival in patients with ESCC, suggesting its potential use as a biomarker for ESCC patients’ prognosis. HDAC4 inhibition was also associated with increased E-cadherin and decreased vimentin expression, inhibiting epithelial to mesenchymal transition (EMT) and EC cell metastatic potential[12].
Xue et al[30] used RNA interference screening to identify the HATs/HDACs ratio implicated in EC. In their study they included genes, that when inhibited, they could cause a decrease in cancer cell viability by ≥ 20%. Therefore, HDAC1 was recognized to be implicated in tumorigenesis[30]. HAT1 was also the first acetyltransferase to be implicated in EC, and since then it has been linked to more malignancies, though its contribution to esophageal carcinogenesis and associated value as target therapy still remains unknown[30]. Since, HDAC1 had already been addressed, Xue and his colleagues studied HAT1. Silencing of HAT1, led to an increase in the population of cells arrested in G2 phase and a decrease in cyclinB1, that works as the checkpoint from G2 to M phase[30]. Immunohistochemical analysis from 167 EC specimens, showed a positive expression in 67% (104 specimens) with higher expression in primary tumor site compared to normal esophageal tissue. Higher expression was associated with poorer tumor differentiation, but, nevertheless, a worse survival prognosis was not found in a statistical significant correlation[30].
In carcinogenesis, heat shock proteins, like HSP90, are cellular regulators providing stability to multiple oncogenic proteins maintaining a high proliferation rate and cancer expansion[31]. In esophageal carcinomas, investigations have already proved that cancer cells present increased HSP90 expression, when compared to normal esophageal tissue[32]. Tao et al[33] studied the association of HDAC6 with esophageal carcinogenesis, noticing an increased expression in comparison to normal tissue. When HDAC6 was inhibited with RNA interference it was documented that cancer cells had reduced motility and invasion capacity, through increased acetylation of a-tubulin[33]. In addition to the HDAC6-tubulin interaction, a new co-relation between HDAC6 and HSP90 was investigated. HDAC6 downregulation led to increased acetylation of HSP90, inhibiting stability of oncogenic proteins, like EGFR, and repressing tumor growth, providing a novel target in EC[33]. Combination treatment with HDAC6/HSP90 inhibition showed the most potent arrest in cellular mitotic activity than any of the drugs administered alone[33].
In accordance with the above studies, Hu et al[34] addressed the possibility that selenium anti-cancer effect could be due to its interactions with enzymes implicated in histone acetylation. Selenium levels have been inversely associated with the risk for cancer development in humans. The exact mechanism of action of selenium anti-cancer effect is not well understood[35]. Studies though, have proven the significance of selenium in decreasing ESCC’s prevalence[36]. Moreover, selenomethionine can help protect against mild esophageal squamous cell dysplasia. Hu et al[34] concluded that methylseleninic acid (MSA) inhibits cellular growth of ESCC, through inactivation of HDAC activity, affecting gene expression through histone modification. An upregulation of general control nonrepressed protein 5 (GCN5) was also noted, increasing histone acetylation. After incubation of mice with MSA, they noted a 33% decrease in weight of MSA-treated tumors and a significant decrease in tumor growth rate, when at the same time the weight of affected mice remained the same. Apoptosis was greatly influenced, as concluded from the increase in caspase 3, one of the catalytic enzymes in the common apoptotic pathway[34]. In MSA-treated cancer cells, an increase in krüppel-like factor 4 (KLF4) was also suggested. KLF4, played an important role in mediating MSA-induced apoptosis, though further evaluation is essential for HDAC-KLF4 molecular interactions and potential novel targets[34].
As mentioned above, class I HDACs 1,2 and 3 are commonly overexpressed in esophageal carcinomas. Though, this overexpression does not only affect malignant cells. Increased levels of HDACs 1,2 and 3 have also been found in tissue surrounding the neoplasm, as well as normal esophageal tissue[37]. Considering this information, it is awkward to address HDAC inhibitors as potential therapeutic agents in patients with EC. Though, their unique cancer cell selectivity for inducing apoptosis makes these drugs suitable for treating EC.
In order to estimate the vulnerability of EC cells to HDAC inhibitors, combination treatments with azacytidine have been tested in patients with both SCCs and adenocarcinomas. Results confirmed a decrease in cell viability. The combination therapeutic approach provided better outcomes compared to either HDAC inhibitor or azacytidine alone. Interestingly, combined treatment in a cell line consisting of normal esophageal cells, showed increased concentrations of the drugs inside the cell, at the same level that was observed in cancer cell lines, though there was no decrease in viability, supporting the hypothesis that these novel drugs have a cancer-specific effect[17]. Interpreting this specificity can be very hard, since the level of HDAC activity in both cell lines was decreased. Nevertheless, in the cancer cell lines an increased number of DNA double strand breaks and apoptosis mediators was noted, leading to the assumption that benign cells survive due to their resistance to these effects[17]. This can be attributed to the diminished function of DNA repair enzymes in malignant cells; enzymes that in normal cells can repair the damage and escape apoptosis[38].
HDAC inhibitors have also been detected to enhance radiosensitivity in EC. This can be an interesting therapeutic advantage, since radiation therapy is included in the treatment of EC, especially of ESCC. Apart from all the aforementioned mechanisms of action of HDAC inhibitors, the synergistic effect with radiation involves mainly their ability to inhibit repair of double strand DNA breaks, through increased acetylation of the repair enzymes involved[39]. Valproic acid, apart from being one of the most potent drugs in treating seizure disorders, it also possesses HDAC inhibition activity, the exact mechanism of which still remains unclear. Based on this, Makita et al[39] initiated valproic acid pretreatment in cancer cell lines and monitored an increase in H2AX, the most sensitive marker for irradiation induced DNA breaks. Valproic acid was found to prolong H2AX levels after irradiation compared to patients treated with radiation only. Therefore, it was suggested as a conjugate treatment for patients with EC, in order to enhance the local cytotoxicity of radiation therapy[39].
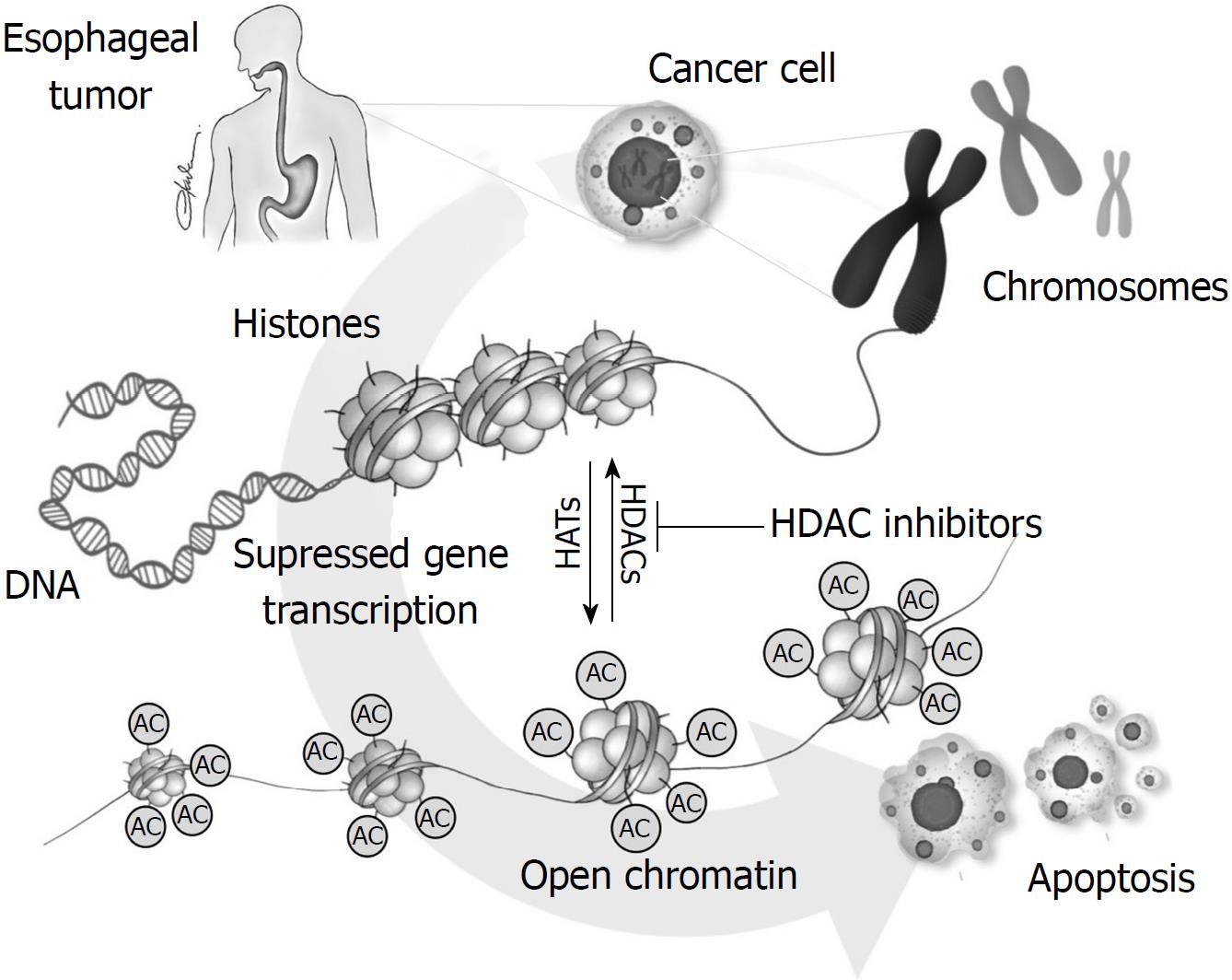
Table 2 summarizes the results of the studies reporting the association of HDAC involvement in esophageal cancer. Figure 1 illustrates the role of HDACs in esophageal carcinogenesis.
| Ref. | Year | Number of pathology samples | HDACs/HATs studied | Correlation with clinicopathological data |
| Langer et al[29] | 2010 | 180 EC samples | HDAC1 and 2 | Increased N stage and Tumor Grade correlated with increased HDAC2 expression |
| Xue et al[30] | 2014 | 167 EC samples | HDAC1 and HAT1 | Increased Tumor Grade correlated with increased HAT1 expression |
| Zeng et al[12] | 2016 | 86 ESCC samples | HDAC4 | Increased T, N, M, TNM stage, Tumor Grade and increased metastatic potential correlated with increased HDAC4 expression Worse 5-yr OS and DFS was noted in patients with increased HDAC4 expression |
| Tao et al[33] | 2018 | Study performed on cell lines of EC | HDAC6 | Increased cell motility and invasion capacity and increased HSP-90 activity, conferring oncogenic protein stability, correlated with increased HDAC6 expression |
In the past decade there has been an increasing amount of research in the field of HDAC inhibition. Right now, we have extensive studies that have proven HDAC inhibition’s efficacy, particularly in oncology. Despite the large amount of information that is available, concerning HDAC and their implication in carcinogenesis, more research is necessary to contribute to the understanding of differences between the HDAC functions in the normal and cancer cell, as well as their interactions with other cellular targets. Aspects of their action in various cellular pathways still remain unclear. HDAC inhibitors’ ability to have synergistic or additive effect with other chemotherapeutic regimens could prove very useful in the wide utilization of these novel drugs in many different cancer types. Better understanding of the HDAC enzymes, will help guide pharmaceutical research and clinical trials in developing the best possible drug combination regimens, based on the type of cancer and HDAC expression. In addition to the above, further investigation is needed in order to be able to better answer questions regarding the interaction of HDAC expression in cancer cells and the effect on patient’s prognosis, association with clinicopathological parameters and chemotherapy response.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Sandhu DS S- Editor: Ma RY L- Editor: A E- Editor: Yin SY
| 1. | Rakovich G, Ouellette D, Beauchamp G. An unusual tumor of the esophagus. J Thorac Cardiovasc Surg. 2010;139:e91-e93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 2. | Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1956] [Cited by in RCA: 1960] [Article Influence: 163.3] [Reference Citation Analysis (5)] |
| 3. | Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21:7933-7943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 554] [Cited by in RCA: 746] [Article Influence: 74.6] [Reference Citation Analysis (15)] |
| 4. | Maurer J, Schöpp M, Thurau K, Haier J, Köhler G, Hummel R. Immunohistochemical analysis on potential new molecular targets for esophageal cancer. Dis Esophagus. 2014;27:93-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 5. | Ali SR, Humphreys KJ, McKinnon RA, Michael MZ. Impact of Histone Deacetylase Inhibitors on microRNA Expression and Cancer Therapy: A Review. Drug Dev Res. 2015;76:296-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Kothapalli N, Sarath G, Zempleni J. Biotinylation of K12 in histone H4 decreases in response to DNA double-strand breaks in human JAr choriocarcinoma cells. J Nutr. 2005;135:2337-2342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Li Z, Zhu WG. Targeting histone deacetylases for cancer therapy: from molecular mechanisms to clinical implications. Int J Biol Sci. 2014;10:757-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 8. | Damaskos C, Garmpis N, Valsami S, Kontos M, Spartalis E, Kalampokas T, Kalampokas E, Athanasiou A, Moris D, Daskalopoulou A. Histone Deacetylase Inhibitors: An Attractive Therapeutic Strategy Against Breast Cancer. Anticancer Res. 2017;37:35-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 143] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 9. | Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 805] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 10. | Cai MH, Xu XG, Yan SL, Sun Z, Ying Y, Wang BK, Tu YX. Depletion of HDAC1, 7 and 8 by Histone Deacetylase Inhibition Confers Elimination of Pancreatic Cancer Stem Cells in Combination with Gemcitabine. Sci Rep. 2018;8:1621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 11. | Mielcarek M, Zielonka D, Carnemolla A, Marcinkowski JT, Guidez F. HDAC4 as a potential therapeutic target in neurodegenerative diseases: a summary of recent achievements. Front Cell Neurosci. 2015;9:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 12. | Zeng LS, Yang XZ, Wen YF, Mail SJ, Wang MH, Zhang MY, Zheng XF, Wang HY. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging (Albany NY). 2016;8:1236-1249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 13. | Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548-13558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 418] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 14. | Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014-10019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 878] [Cited by in RCA: 884] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 15. | Bai J, Sui J, Demirjian A, Vollmer CM Jr, Marasco W, Callery MP. Predominant Bcl-XL knockdown disables antiapoptotic mechanisms: tumor necrosis factor-related apoptosis-inducing ligand-based triple chemotherapy overcomes chemoresistance in pancreatic cancer cells in vitro. Cancer Res. 2005;65:2344-2352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 102] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 16. | Henderson SE, Ding LY, Mo X, Bekaii-Saab T, Kulp SK, Chen CS, Huang PH. Suppression of Tumor Growth and Muscle Wasting in a Transgenic Mouse Model of Pancreatic Cancer by the Novel Histone Deacetylase Inhibitor AR-42. Neoplasia. 2016;18:765-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Ahrens TD, Timme S, Hoeppner J, Ostendorp J, Hembach S, Follo M, Hopt UT, Werner M, Busch H, Boerries M. Selective inhibition of esophageal cancer cells by combination of HDAC inhibitors and Azacytidine. Epigenetics. 2015;10:431-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 18. | Peralta-Arrieta I, Hernández-Sotelo D, Castro-Coronel Y, Leyva-Vázquez MA, Illades-Aguiar B. DNMT3B modulates the expression of cancer-related genes and downregulates the expression of the gene VAV3 via methylation. Am J Cancer Res. 2017;7:77-87. [PubMed] |
| 19. | Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol Cell Biol. 2007;27:8637-8647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 169] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 20. | Zhu BY, Shang BY, Du Y, Li Y, Li L, Xu XD, Zhen YS. A new HDAC inhibitor cinnamoylphenazine shows antitumor activity in association with intensive macropinocytosis. Oncotarget. 2017;8:14748-14758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 21. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3299] [Cited by in RCA: 3405] [Article Influence: 189.2] [Reference Citation Analysis (0)] |
| 22. | Chien W, Lee DH, Zheng Y, Wuensche P, Alvarez R, Wen DL, Aribi AM, Thean SM, Doan NB, Said JW. Growth inhibition of pancreatic cancer cells by histone deacetylase inhibitor belinostat through suppression of multiple pathways including HIF, NFkB, and mTOR signaling in vitro and in vivo. Mol Carcinog. 2014;53:722-735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Mishra VK, Wegwitz F, Kosinsky RL, Sen M, Baumgartner R, Wulff T, Siveke JT, Schildhaus HU, Najafova Z, Kari V. Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4- and MYC-dependent manner. Nucleic Acids Res. 2017;45:6334-6349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 24. | Yar Saglam AS, Yilmaz A, Onen HI, Alp E, Kayhan H, Ekmekci A. HDAC inhibitors, MS-275 and salermide, potentiates the anticancer effect of EF24 in human pancreatic cancer cells. EXCLI J. 2016;15:246-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 25. | Walkinshaw DR, Yang XJ. Histone deacetylase inhibitors as novel anticancer therapeutics. Curr Oncol. 2008;15:237-243. [PubMed] |
| 26. | Koutsounas I, Giaginis C, Theocharis S. Histone deacetylase inhibitors and pancreatic cancer: are there any promising clinical trials? World J Gastroenterol. 2013;19:1173-1181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 27. | Shi B, Xu W. The development and potential clinical utility of biomarkers for HDAC inhibitors. Drug Discov Ther. 2013;7:129-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 28. | Zhao SL, Zhu ST, Hao X, Li P, Zhang ST. Effects of DNA methyltransferase 1 inhibition on esophageal squamous cell carcinoma. Dis Esophagus. 2011;24:601-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 29. | Langer R, Mutze K, Becker K, Feith M, Ott K, Höfler H, Keller G. Expression of class I histone deacetylases (HDAC1 and HDAC2) in oesophageal adenocarcinomas: an immunohistochemical study. J Clin Pathol. 2010;63:994-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 30. | Xue L, Hou J, Wang Q, Yao L, Xu S, Ge D. RNAi screening identifies HAT1 as a potential drug target in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:3898-3907. [PubMed] |
| 31. | Workman P, Burrows F, Neckers L, Rosen N. Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann NY Acad Sci. 2007;1113:202-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 422] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 32. | Wu X, Wanders A, Wardega P, Tinge B, Gedda L, Bergstrom S, Sooman L, Gullbo J, Bergqvist M, Hesselius P. Hsp90 is expressed and represents a therapeutic target in human oesophageal cancer using the inhibitor 17-allylamino-17-demethoxygeldanamycin. Br J Cancer. 2009;100:334-343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 33. | Tao H, Chen YY, Sun ZW, Chen HL, Chen M. Silence of HDAC6 suppressed esophageal squamous cell carcinoma proliferation and migration by disrupting chaperone function of HSP90. J Cell Biochem. 2018;119:6623-6632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Hu C, Liu M, Zhang W, Xu Q, Ma K, Chen L, Wang Z, He S, Zhu H, Xu N. Upregulation of KLF4 by methylseleninic acid in human esophageal squamous cell carcinoma cells: Modification of histone H3 acetylation through HAT/HDAC interplay. Mol Carcinog. 2015;54:1051-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 35. | Vinceti M, Filippini T, Del Giovane C, Dennert G, Zwahlen M, Brinkman M, Zeegers MP, Horneber M, D'Amico R, Crespi CM. Selenium for preventing cancer. Cochrane Database Syst Rev. 2018;1:CD005195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 36. | Mark SD, Qiao YL, Dawsey SM, Wu YP, Katki H, Gunter EW, Fraumeni JF Jr, Blot WJ, Dong ZW, Taylor PR. Prospective study of serum selenium levels and incident esophageal and gastric cancers. J Natl Cancer Inst. 2000;92:1753-1763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 183] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Nakagawa M, Oda Y, Eguchi T, Aishima S, Yao T, Hosoi F, Basaki Y, Ono M, Kuwano M, Tanaka M. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep. 2007;18:769-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci USA. 2010;107:14639-14644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 39. | Makita N, Ninomiya I, Tsukada T, Okamoto K, Harada S, Nakanuma S, Sakai S, Makino I, Kinoshita J, Hayashi H. Inhibitory effects of valproic acid in DNA double-strand break repair after irradiation in esophageal squamous carcinoma cells. Oncol Rep. 2015;34:1185-1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |