Published online Oct 7, 2018. doi: 10.3748/wjg.v24.i37.4263
Peer-review started: May 30, 2018
First decision: July 18, 2018
Revised: August 9, 2018
Accepted: August 24, 2018
Article in press: August 24, 2018
Published online: October 7, 2018
Processing time: 123 Days and 1.3 Hours
To identify functional proteins involved in pancreatic-duodenal homeobox-1 (PDX1)-mediated effects on gastric carcinogenesis.
A PDX1-overexpressed model was established by transfecting gastric cancer cell line SGC7901 with pcDNA3.1(+)-PDX1 vector (SGC-PDX1). Transfection with empty pcDNA3.1 vector (SGC-pcDNA) served as control. Comparative protein profiles of the two groups were analyzed by two-dimensional electrophoresis based-proteomics (2DE gel-based proteomics). The differential proteins identified by 2DE were further validated by qRT-PCR and immunoblotting. Finally, co-immunoprecipitation was used to determine any direct interactions between PDX1 and the differential proteins.
2DE gel proteomics identified seven differential proteins in SGC-PDX1 when compared with those in SGC-pcDNA. These included four heat shock proteins (HSPs; HSP70p1B, HSP70p8, HSP60, HSP27) and three other proteins (ER60, laminin receptor 1, similar to epsilon isoform of 14-3-3 protein). Immunoblotting validated the expression of the HSPs (HSP70, HSP60, HSP27). Furthermore, their expressions were lowered to 80%, 20% and 24%, respectively, in SGC-PDX1, while PDX1 exhibited a 9-fold increase, compared to SGC-pcDNA. However, qRT-PCR analysis revealed that mRNA levels of the HSPs were increased in SGC-PDX1, suggesting that the expression of the HSPs was post-translationally regulated by the PDX1 protein. Finally, co-immunoprecipitation failed to identify any direct interaction between PDX1 and HSP70 proteins.
This study demonstrates the potential involvement of HSPs in PDX1-mediated effects on the genesis of gastric cancer.
Core tip: Using a pcDNA3.1(+)-pancreatic-duodenal homeobox-1 (PDX1) vector, a PDX1-overexpressed model was built. Seven differential proteins were identified in SGC-PDX1 by two-dimensional electrophoresis gel proteomics compared with those in SGC-pcDNA. Four heat shock proteins were identified and confirmed by immunoblotting. qRT-PCR analysis further revealed that the expression of the HSPs was post-translationally regulated by the PDX1 protein. This study suggests the potential involvement of HSPs in PDX1 mediated effects on gastric carcinogenesis.
- Citation: Ma J, Wang BB, Ma XY, Deng WP, Xu LS, Sha WH. Potential involvement of heat shock proteins in pancreatic-duodenal homeobox-1-mediated effects on the genesis of gastric cancer: A 2D gel-based proteomic study. World J Gastroenterol 2018; 24(37): 4263-4271
- URL: https://www.wjgnet.com/1007-9327/full/v24/i37/4263.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i37.4263
Homeobox genes greatly contribute to the pattern formation of embryos by encoding many homeodomain transcriptional regulators[1,2]. The human gene of pancreatic-duodenal homeobox-1 (PDX1) is located at chromosomal locus 13q12.1[3]. In adults, PDX1 is expressed in Brunner’s glands of the duodenum, pancreatic β cells and gastric pyloric gland cells. As one of the key homeodomain transcription regulators, PDX1 plays an important role in the development of the digestive system, including antrum, duodenum, and pancreas[4,5]. PDX1 is also a biomarker of pancreatic stem cells and regulates normal islet function[6]. In addition, PDX1 is expressed in the distal stomach and is involved in the secretion of hormones, such as somatostatin, serotonin, and gastrin[4]. Dysregulation of PDX1 may lead to pancreatic and gastric carcinogenesis. For example, overexpression of PDX1 was shown to be associated with the development of pancreatic cancer as it promoted proliferation, invasion, and colony formation in cancer cells[7]. Moreover, we earlier reported downregulation of PDX1 in gastric cancer, which suggests its potential role as a tumor suppressor. Further, overexpression of PDX1 induced apoptosis and inhibited proliferation, clone formation, and migration of gastric cancer cells. In addition, stable transfection with PDX1 was shown to suppress development of gastric cancer in vivo[8]. We have also shown that silencing of PDX1 in gastric cancer is likely caused by promoter hypermethylation and histone hypoacetylation[9]. However, the downstream mechanism by which PDX1 mediates gastric tumorigenesis remains elusive.
Proteins are the fundamental molecules that perform cellular functions. Changes in the protein expression profile under pathological conditions may reflect potential pathogenic mechanisms. Proteomic approaches represent a powerful tool to explore the underlying mechanism of tumorigenesis by characterizing the cellular events related to tumor development, angiogenesis, and progression. A number of proteomic approaches have been used to investigate the pathogenesis of gastric cancer[10-12]. Among these, 2D gel electrophoresis is a conventional approach and has been the principal step in the development of proteomics. Subsequent to 2D gel electrophoresis, protein expression profiles have been elucidated by computational image analysis and identified by mass spectrometry[13]. Although a number of different proteomics techniques have evolved in recent years, 2DE-based proteomics is still widely used to identify cancer-associated proteins[14].
To clarify the role of downstream mediators of PDX1 in gastric tumorigenesis, we established a PDX1-overexpressed gastric cancer cell model by transfection. After validation of the PDX1 overexpressed model, we used a 2D gel-based proteomic approach to determine the differentially expressed proteins as compared to that in the empty vector transfected gastric cancer cells. The expressions of identified candidate proteins were further assessed by real-time qRT-PCR and/or immunoblotting. Finally, co-immunoprecipitation was used to examine whether a differential protein, HSP70 has direct interactions with PDX1 protein. Our results will greatly extend our understanding of the mechanisms of PDX1 mediated effects on gastric tumorigenesis.
Human gastric cancer cell line SGC7901 was cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 μg/mL penicillin. The cells were maintained in a constant temperature incubator at 37 °C and 5% CO2. The cells were harvested and passaged as required.
The vector of pcDNA3.1(+)-PDX1 was established according to the methods previously published[15]. The SGC7901 cells were cultured in 6-well plates and transfected with pcDNA3.1(+)-PDX1 vector (SGC-PDX1) or pcDNA3.1(+) control vector (SGC-pcDNA) using liposome transfection reagent (Lipofectamine™ 2000 Transfection Reagent, ThermoFisher Scientific, United States). After six hours, fresh complete medium was used to replace the transfection medium and the cells of each group were aliquoted in triplicate. After 24-h incubation, the transfected SGC7901 cells were harvested for protein extraction. Initially, the culture plates were placed on ice, followed by addition of 250 μL lysis buffer for 10 min. The lysed cells were scraped off and sonicated for 10 min. Subsequently, the cell lysates were centrifuged at 20000 g for 30 min, and the supernatant containing cell proteins was collected. The whole cell lysate proteins were purified using a Cleanup kit (ProteoPrep® Total Extraction Sample Kit, Sigma-Aldrich, United States) to remove salt and lipid impurities. Protein concentration was determined by bicinchoninic acid (BCA) method. Finally, 600 μg protein from each sample was used for 2D gel electrophoresis.
2D gel electrophoresis was performed according to the method described elsewhere[16]. The first-dimensional isoelectric focusing of protein sample was conducted on Immobiline™ pH 3-10 IPG linear strips (Amersham, Pharmacia Biotech Inc.), and focused by an IPGphor electrophoresis system (Ettan IPGphor Isoelectric Focusing System, Amersham Biosciences, United States) by following the manufacturer’s instructions. During second dimensional separation, focused strips were subjected onto the top of 12.5% SDS-PAGE gradient gels and overlaid with 1% agarose gel buffer. The gels were run under 20 °C at a speed of 15 mA/gel in stacking gel and 30 mA/gel in resolving gel until bromophenol blue front was within 0.5-1 cm of the gel bottom. The 2D gels were then stained with silver and scanned with UMax Powerlook 2110XL (GE Amersham). Image analysis was conducted using Image Master 2D platinum 5.0 software (Amersham pharmcia, Biotech) to identify differentially expressed protein profiles.
The differential protein spots between the two groups were excised using an automated Spot Handling Workstation (Amersham pharmcia, Biotech) and were subjected to discoloration and dehydration. The proteins were digested by trypsin and the peptides were obtained using peptide extraction buffer. The peptide suspension was vacuum-dried and resuspended in D/W. The resuspended peptide extracts were then spotted on a matrix-assisted laser-desorption ionization (MALDI) target. MALDI-MS analysis was conducted through a MALDI-time-of-flight (MALDI-TOF) mass spectrometer (Applied Biosystems, United States) and peptide mass mapping was performed by searching the NCBInr database.
Total RNA from the cells was isolated using a Mini-RNease RNA extract kit (Qiagen, Germany) according to the manufacturer’s instructions. cDNA was reverse-transcribed from total RNA using a Thermoscript RT-PCR system (Gibco BRL, Gaithersburg, MD, United States). Applied Biosystems Sequence Detection System 7900 (Applied Biosystems, United States) was used to perform the qRT-PCR analysis. The reaction was performed using 10 μL mixture of 300 ng cDNA templates, 500 nmol of each primer and Power SYBR GREEN PCR Master Mix (Applied Biosystems, United States) as previously reported[8]. The generated melting curves and CT values were used to calculate the copy numbers of PDX1 or HSPs mRNA. Online tool was utilized to design the PCR primers and the corresponding sequences, which are shown in Table 1.
| Gene name | Forward | Reverse | Product size (bp) |
| PDX1 | 5’-ATCTCCCCATACGAAGTGCC-3’ | 5’-CGTGAGCTTTGGTGGATTTCAT-3’ | 92 |
| GAPDH | 5’-ATGGGGAAGGTGAAGGTCG-3’ | 5’-GGGGTCATTGATGGCAACAATA-3 | 108 |
| HSPA2 | 5’-CACCACCTATTCGTGCGTC-3’ | 5’-TTTCCGTCCAATCAGCCTCTT-3’ | 196 |
| HSPA6 | 5’-CAAGGTGCGCGTATGCTAC-3’ | 5’-GCTCATTGATGATCCGCAACAC-3’ | 224 |
| HSPA8 | 5’-GGAGGTGGCACTTTTGATGTG-3’ | 5’-CAAGCAGTACGGAGGCGTCT-3’ | 200 |
| HSPA1B | 5’-TTTGAGGGCATCGACTTCTACA-3 | 5’-CCAGGACCAGGTCGTGAATC-3’ | 148 |
| HSPA1L | 5’-CTACTGCCAAGGGAATCGCC-3’ | 5’GCCGATCAGACGTTTAGCATC-3’ | 227 |
| HSP27 | 5’-GGACGAGCATGGCTACATCT-3’ | 5’-CTTTACTTGGCGGCAGTCTC-3’ | 237 |
| HSP60 | 5’-CACCGTAAGCCTTTGGTCAT-3’ | 5’-CCCTCTTCTCCAAACACTGC-3’ | 188 |
As described in our previous study[8], the whole cell lysate protein was mixed with SDS-PAGE sample buffer and boiled for five minutes. Prepared protein samples were separated by SDS-PAGE electrophoresis and electrotransferred onto polyvinylidene fluoride membranes. After blocking, the membranes were blotted with primary antibodies against PDX1, HSP27, HSP60, and HSP70 (Santa Cruz Biotechnology). The membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies and visualized using an enhanced chemiluminescence system (Amersham, Piscataway, NJ, United States).
SGC7901 cells were co-transfected with both pcDNA3.1(+)-PDX1 and pcDNA3.1(+)-HSP70 plasmids and cultured for 24 h. The cells were subsequently washed with PBS and lysed using lysis buffer (1 mmol/L phenylmethylsulfonyl fluoride, 5 mmol/L 2-mercaptoethanol, 2 mmol/L MgCl2, 20 mmol/L HEPES, 150 mmol/L NaCl, 10 μg/mL leupeptin and 10 μg/mL aprotinin). The lysates were transferred onto protein A beads (Thermo Fisher Scientific) and incubated overnight with primary antibodies for PDX1 or HSP70 (Santa Cruz Biotechnology) at 4 °C. The beads were washed twice using lysis buffer to remove unbound proteins. To elute the bound proteins, the beads were then resuspended in sample buffer and boiled at 95 °C for three minutes. The collected unbound and bound proteins were stored at -80 °C for further use during immunoblotting.
Data are presented as mean ± SD, and the differences in continuous variables were assessed by Mann-Whitney U test (Student t test). P-values < 0.05 were considered statistically significant and all statistical tests were two-tailed.
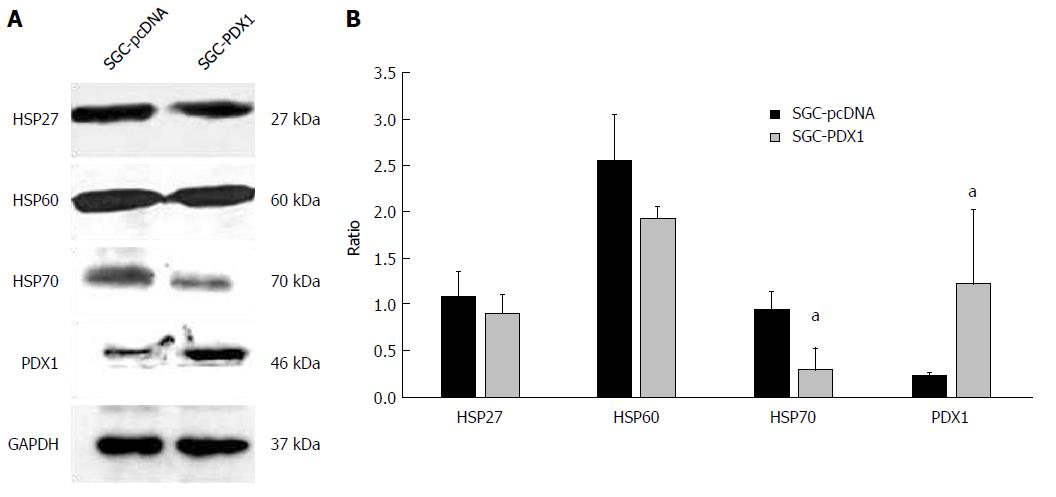
To study the downstream mechanisms of PDX1 in gastric carcinogenesis, we established a PDX1 overexpressed model of gastric cancer cell line SGC7901 using a vector of pcDNA3.1(+)-PDX1 (SGC-PDX1). The whole cell lysate proteins were extracted and subjected to immunoblotting to confirm the establishment of PDX1 overexpressed model. As shown in Figure 1A, the expression of PDX1 in pSGC-PDX1 cells was significantly increased compared to that in the SGC-pcDNA cells. These results indicated the successful establishment of a PDX1 overexpressed model, which was further used for 2D gel-based proteomic analysis.
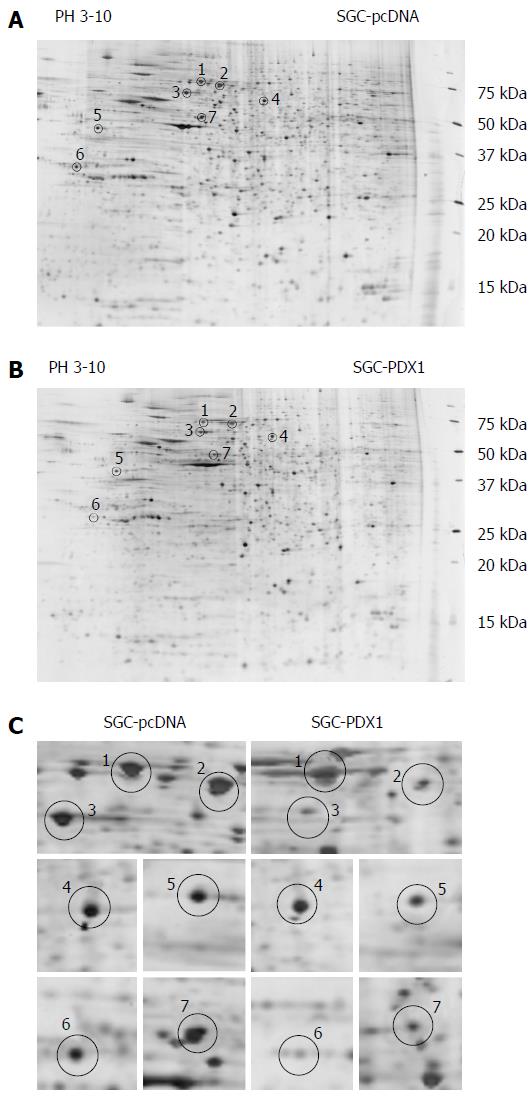
The comparative protein profile of PDX1 overexpressed model and control group was determined using 2D PAGE coupled with MALDI-TOF-MS. The 2-DE maps were displayed between a pH range of 3-10. Figure 2A and 2B shows the representative 2-DE maps of SGC-PDX1 and SGC-pcDNA. Compared with SGC-pcDNA control cells, seven proteins were found to be downregulated about two-fold in SGC-PDX1 cells (Figure 2C). The seven differentially expressed protein spots were excised from replicate 2D-gels and the corresponding peptides were obtained by proteolytic digestion. Extracted peptides were subjected to MALDI-TOF-MS analysis and then the database search was performed. The seven differentially expressed proteins were identified as, heat shock protein (70 kDa) and its isoforms, heat shock protein (60 kDa), heat shock protein (27 kDa), glucose regulated protein (58 kDa), laminin receptor 1, epsilon isoform of 14-3-3 protein. The detailed information of the seven differentially expressed proteins is shown in Table 2.
| Spot No. | Rank protein name | Accession No. | Protein score | Protein score CI% | Protein MW |
| S1 | Heat shock 70 kDa protein 8 isoform 2 (homo sapiens) | gi 24234686 | 595 | 100 | 53598.4 |
| HSPA8 protein (homo sapiens) | gi 48257068 | 546 | 100 | 54804.2 | |
| Heat shock 70 kDa protein 2 (homo sapiens) | gi 3287 9973 | 290 | 100 | 70263.0 | |
| Heat shock 70 kDa protein 6 (HSP70B’) (homo sapiens) | gi 55960611 | 81 | 100 | 71440.4 | |
| S2 | DNAK-type molecular chaperone HSPA1L-human | gi 2119712 | 559 | 100 | 70110.0 |
| HSPA1A protein (homo sapiens) | gi 14414588 | 558 | 100 | 70294.1 | |
| Heat shock 10 kDa protein 1-like (homo sapiens) | gi 55961919 | 372 | 100 | 70730.5 | |
| Heat shock 70 kDa protein 1-like (homo sapiens) | gi 21759781 | 361 | 100 | 70748.4 | |
| Heat shock 70 kDa protein 1B (homo sapiens) | gi 55962554 | 296 | 100 | 52199.8 | |
| S3 | Chaperonin 60 (Hsp60) (homo sapiens) | gi 6996447 | 375 | 100 | 61187.4 |
| S4 | ER-60 protein (homo sapiens) | gi 2245365 | 223 | 100 | 57146.9 |
| Glucose regulated protein 58 kDa (Bos taurus) | gi 27805905 | 113 | 100 | 57293.0 | |
| S5 | Ribosomal protein SA (laminin receptor 1) (homo sapiens) | gi 47125390 | 204 | 100 | 32933.5 |
| S6 | PREDICTED: Similar to epsilon isoform of 14-3-3 protein | gi 57091321 | 53 | 37.851 | 29326.5 |
| S7 | Heat shock 70 kDa protein 1B (homo sapiens) | gi 55962554 | 67 | 97.468 | 52199.8 |
Through literature search and review, we found that the heat shock proteins (HSPs) were shown to play an important role in the regulation of tumorigenesis, including proliferation, invasion, and colony formation of cancer cells[17-19]. This finding suggested that the HSPs might also be involved in gastric carcinogenesis because they also showed differential protein expressions in SGC-PDX1 as revealed by 2D gel-based proteomics. Therefore, we selected the heat shock proteins for further evaluation.
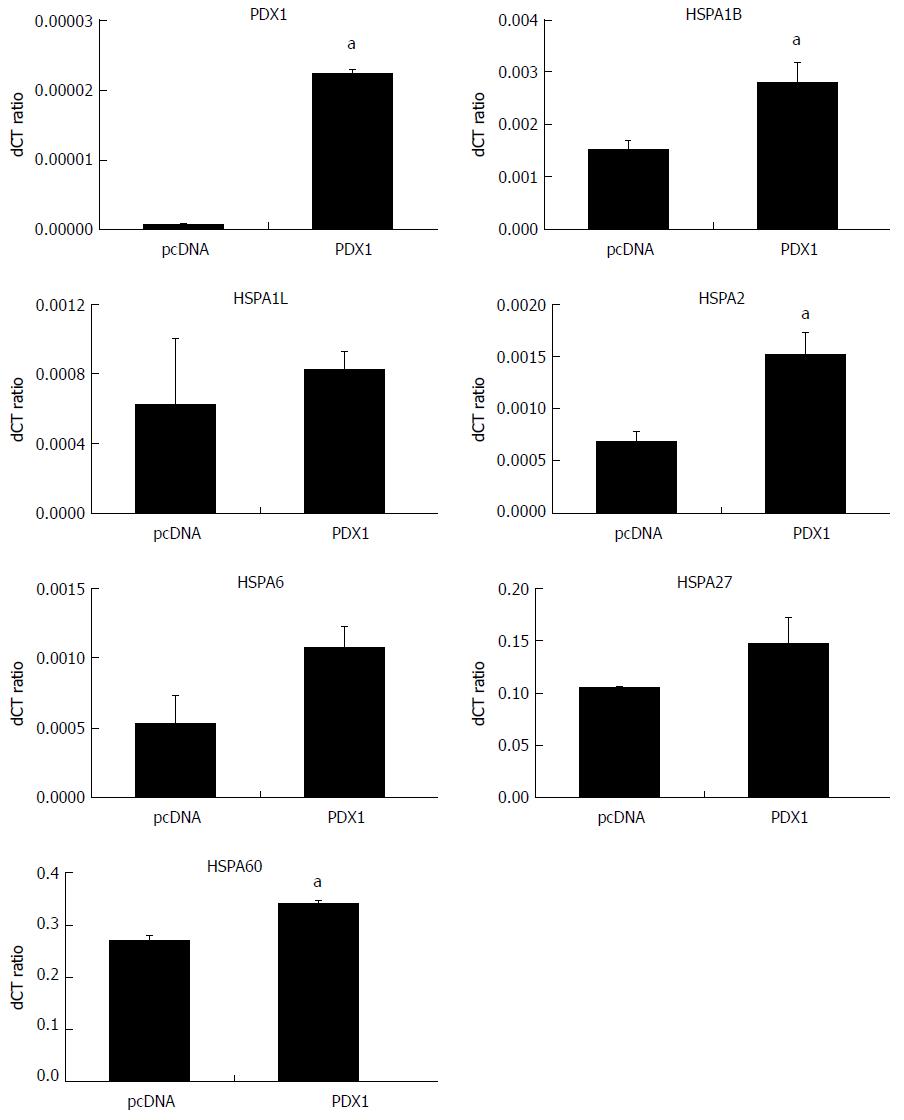
SGC7901 cells were maintained under the same culture conditions and transfected with corresponding vectors as mentioned earlier. Total RNA and whole cell proteins were extracted for qRT-PCR and immunoblotting, respectively. The expression of PDX1 mRNA was significantly upregulated in SGC-PDX1 cells as compared to that in the control group (Figure 3). This indicated the successful transfection of SGC7901 cells with the PDX1 gene. In addition, the mRNA levels of the HSP70 isoforms (HSPA1B, HSPA1L, HSPA2, HSPA6), HSP27, and HSP60 were found to have increased after PDX1 overexpression (Figure 3) in SGC7901 cells. Immunoblotting confirmed that the expression of the PDX1 protein was significantly increased in SGC-PDX1 cells. Moreover, the expression of HSP70, HSP27, and HSP60 were downregulated in SGC-PDX1 cells (reduced to 80%, 24%, and 20%, respectively), which was consistent with the proteomic results (Figure 1). However, these results demonstrated the opposite patterns of mRNA and protein expression of HSP70, HSP27, and HSP60 in SGC-PDX1 cells, which suggests that the expressions of these proteins were post-translationally regulated in SGC-PDX1 cells.
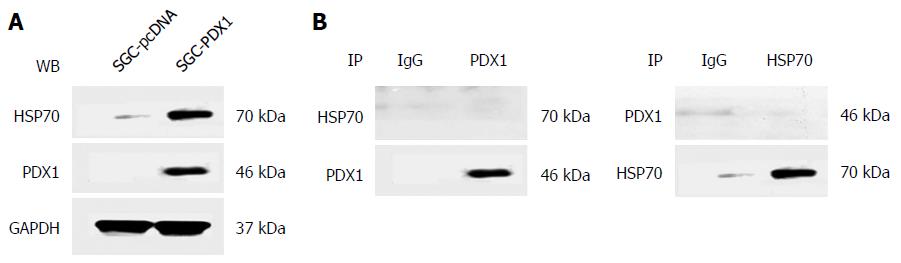
Results of immunoblotting showed that the difference in the expression of HSP70 between the two groups was much higher than that for the other two HSP proteins, which emphasized the significance of the HSP70 protein in PDX1-mediated effects on gastric tumorigenesis. Therefore, we further evaluated whether PDX1 and HSP70 proteins have direct interactions. SGC7901 cells were co-transfected with both pcDNA3.1(+)-PDX1 and pcDNA3.1(+)-HSP70 plasmids and cultured for 24 h. The whole cell lysate protein of co-transfected SGC7901 cells were then subjected to co-immunoprecipitation. As illustrated in Figure 4A, both PDX1 and HSP70 proteins were identified in the cell lysate proteins of co-transfected SGC7901 cells, which confirmed the input of PDX1 and HSP70 proteins in co-immunoprecipitation assay. The whole cell lysate proteins were then subjected to co-immunoprecipitation assay using precipitating antibodies against PDX1 and HSP70. The corresponding immunoprecipitated proteins were further detected by immunoblotting using primary antibodies against HSP70 and PDX1. Interestingly, no significant binding between PDX1 and HSP70 proteins was observed (Figure 4B), which implied that there was no direct interaction between PDX1 and HSP70 proteins.
Gastric cancer is a leading cause of cancer-associated mortality across the world[20]. Despite years of extensive research, the molecular mechanisms involved in the pathogenesis of gastric cancer are not completely understood. Recent advances in proteomics display its great potential for use in cancer diagnosis, prognostic assessment, and to understand the molecular mechanism of carcinogenesis. Proteomic approaches have been widely applied in research on gastric cancer[21-23]. Liu et al[24] used 2D gel-based proteomics to compare the differential serum protein profiles between gastric cancer patients and healthy controls, and identified several serum biomarkers for gastric cancer. Poon et al[22] also demonstrated that an exclusive serum proteomic fingerprint, identified by SELDI-based proteomics, can be used for noninvasive diagnosis of gastric cancer. In addition, many prognostic biomarkers of gastric cancer were identified by proteomic approaches, such as S100P[25] and S100A9[25] proteins. Chen et al[26] utilized a quantitative proteomic technique to identify the differentially expressed proteins in metastatic gastric cancer cells as compared to that in noninvasive gastric cancer cells. After downstream validation, they found that vimentin and galectin-1 were potential markers of gastric cancer metastasis and suggested their involvement in cell-cell and cell-ECM adhesion interactions.
PDX1 is a member of the homeobox family and plays an important role in the development of embryonic digestive system. A previous study demonstrated that PDX1 knockout mice showed abnormal growth of gastroduodenal junction, which increased the difficulty in emptying gastric contents and ultimately led to gastric retention[27]. Aberrant expression of the PDX1 gene has been associated with carcinogenesis. PDX1 was shown to function as a tumor promoter by enhancing the proliferation, invasion[28], and induction of acinar-to-ductal metaplasia[29] in pancreatic cancer. Hence, there is a growing interest to develop a novel therapy for pancreatic cancer by targeting PDX1. Interestingly, PDX1 was also found to be associated with the tumorigenesis of gastric cancer. Both Faller et al[30] and Sakai et al[31] have reported abnormal expression of PDX1 protein in pseudo-pyloric glandular metaplasia. In a previous study, we found that PDX1 protein was downregulated in Helicobacter pylori infection, incisural antralisation, and intestinal metaplasia[32]. We further identified significantly decreased PDX1 mRNA and PDX1 protein expression in gastric cancer cells. Downregulation of PDX1 in gastric cancer tissues might be caused by promoter hypermethylation and histone hypoacetylation[9]. We also observed that the transient overexpression of the PDX1 gene could inhibit proliferation and induce apoptosis of gastric cancer cells. Moreover, stable overexpression of the PDX1 gene suppressed colony formation, wound healing, migration of gastric cancer cells, and decreased tumor incidence in nude mice; these findings imply that PDX1 may act as a tumor suppressor in the context of gastric cancer[8].
In the present study, we sought to clarify the downstream mediators of PDX1-mediated effects on gastric cancer tumorigenesis. We used 2D gel-based proteomics to identify differentially expressed proteins between PDX1-overexpressed gastric cancer cells and the control empty vector transfected gastric cells. Seven differentially expressed proteins were identified by proteomic analysis, which included three heat shock proteins. The three HSPs proteins, HSP70, HSP60, and HSP27, were significantly decreased in gastric cells after PDX1 overexpression. The results of immunoblotting analysis were consistent with those of proteomics analysis. However, the mRNA levels of HSP70, HSP60, and HSP27 were significantly increased after PDX1 overexpression. These results indicated that overexpression of PDX1 could downregulate HSP70, HSP60, and HSP27 proteins through post-translational regulation pathways, such as mRNA degradation, glycosylation, degradation, or phosphorylation.
HSPs are a group of chaperone proteins that play dual roles in tumorigenesis[33]. HSP70, HSP60, and HSP27 proteins were reported to be upregulated in gastric cancer and associated with tumor progression and poor prognosis[34-36]. Our immunoblotting analysis revealed that HSP70 proteins showed the greatest differential expression in PDX1-overexpressed gastric cells, which emphasized its significance in PDX1 mediated effects on gastric cancer tumorigenesis. Besides, HSP70 protein was shown to serve as a tumor promoter in gastric cancer by reducing apoptosis[37,38]. Therefore, PDX1 might affect the apoptosis of gastric cancer cells by regulating the expression of HSP70. To further evaluate the interaction between PDX1 and HSP70, we performed a co-immunoprecipitation assay. However, no direct interactions could be found between the two proteins, which indicates that there may be some intermediate mediators that link PDX1 and HSP70 proteins in the regulation of gastric cancer tumorigenesis. Hence, further studies are warranted to elucidate the relationship between PDX1 and HSP70, along with a detailed mechanism regarding their collaboration to regulate gastric tumorigenesis.
In conclusion, our study showed that proteomics is a powerful tool to study the molecular mechanisms involved in the genesis of gastric cancer. In addition, the inhibition of PDX1 in gastric cancers may contribute to the upregulation of HSPs, especially HSP70.
As one of the homeobox genes that play critical roles in the pattern formation of embryos, pancreatic-duodenal homeobox-1 (PDX1) is widely expressed in Brunner’s glands of the duodenum, pancreatic β cells, and gastric pyloric gland cells. PDX1 plays a key role in the development of the digestive system, including antrum, duodenum, and pancreas. Downregulation of PDX1 has been observed in gastric cancer, which suggests its potential role in gastric tumorigenesis. Nevertheless, the downstream mechanisms that mediate the effect of PDX1 on gastric tumorigenesis are still poorly understood.
Although PDX1 has been found to be involved in gastric tumorigenesis, its downstream regulating mechanism is still unclear.
To clarify the differential protein profile in PDX1-overexpressed gastric cell line and explore functional proteins involved in PDX1-mediated effects on gastric tumorigenesis.
A PDX1-overexpressed model was established using gastric cancer cell line SGC7901 (SGC-PDX1). As a method that is still widely used to identify cancer-associated proteins, 2DE-based proteomics was applied to determine the differential protein profile between SGC-PDX1 and SGC-pcDNA. The differential proteins were then subjected to qRT-PCR and immunoblotting for further confirmation. Finally, direct interactions between PDX1 and the identified differential proteins were evaluated by co-immunoprecipitation.
Seven proteins were found to be differentially expressed in SGC-PDX1 using 2-DE proteomics. Immunoblotting confirmed that the three differential HSPs (HSP70, HSP60, HSP27) were downregulated in SGC-PDX1. However, qRT-PCR analysis identified increased HSP mRNA in SGC-PDX1, which indicates that PDX1 may post-translationally regulate the expression of the HSPs. Further study is warranted to elucidate the relationship of HPS70 and PDX1, as co-immunoprecipitation did not identify direct interaction between them.
HSPs are involved in PDX1-mediated effects on the genesis of gastric cancer and the interaction between HSP70 and PDX1 is indirect.
This study demonstrates the involvement of HSPs in PDX1-mediated effects on gastric tumorigenesis. Further study is warranted to elucidate the downstream regulating mechanism of PDX1 on HSPs.
HSP70 plasmid was kindly provided by Dr. BCY Wong, Hong Kong University, Hong Kong.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Aurello P, Christodoulidis G, Matowicka-Karna J S- Editor: Wang XJ L- Editor: Filipodia E- Editor: Yin SY
| 1. | Melton DA. Pattern formation during animal development. Science. 1991;252:234-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 123] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 2. | Alappat S, Zhang ZY, Chen YP. Msx homeobox gene family and craniofacial development. Cell Res. 2003;13:429-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 184] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 3. | Brooke NM, Garcia-Fernàndez J, Holland PW. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 310] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 4. | Larsson LI, Madsen OD, Serup P, Jonsson J, Edlund H. Pancreatic-duodenal homeobox 1 -role in gastric endocrine patterning. Mech Dev. 1996;60:175-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983-995. [PubMed] |
| 6. | Edlund H. Pancreatic organogenesis--developmental mechanisms and implications for therapy. Nat Rev Genet. 2002;3:524-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 348] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 7. | Liu T, Gou SM, Wang CY, Wu HS, Xiong JX, Zhou F. Pancreas duodenal homeobox-1 expression and significance in pancreatic cancer. World J Gastroenterol. 2007;13:2615-2618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 8. | Ma J, Chen M, Wang J, Xia HH, Zhu S, Liang Y, Gu Q, Qiao L, Dai Y, Zou B. Pancreatic duodenal homeobox-1 (PDX1) functions as a tumor suppressor in gastric cancer. Carcinogenesis. 2008;29:1327-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Ma J, Wang JD, Zhang WJ, Zou B, Chen WJ, Lam CS, Chen MH, Pang R, Tan VP, Hung IF. Promoter hypermethylation and histone hypoacetylation contribute to pancreatic-duodenal homeobox 1 silencing in gastric cancer. Carcinogenesis. 2010;31:1552-1560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Hu HD, Ye F, Zhang DZ, Hu P, Ren H, Li SL. iTRAQ quantitative analysis of multidrug resistance mechanisms in human gastric cancer cells. J Biomed Biotechnol. 2010;2010:571343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Lin LL, Huang HC, Juan HF. Discovery of biomarkers for gastric cancer: a proteomics approach. J Proteomics. 2012;75:3081-3097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 12. | Chen J, Kähne T, Röcken C, Götze T, Yu J, Sung JJ, Chen M, Hu P, Malfertheiner P, Ebert MP. Proteome analysis of gastric cancer metastasis by two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-mass spectrometry for identification of metastasis-related proteins. J Proteome Res. 2004;3:1009-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 13. | Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci USA. 2000;97:9390-9395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 946] [Cited by in RCA: 845] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 14. | Rogowska-Wrzesinska A, Le Bihan MC, Thaysen-Andersen M, Roepstorff P. 2D gels still have a niche in proteomics. J Proteomics. 2013;88:4-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 15. | Ma J, Liu QH, Sha WH, Wang QY. The construction and expression of the PDX1 vector (in Chinese). Guangdong Medical Journal. 2010;31:3030-3031. |
| 16. | Görg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665-3685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1331] [Cited by in RCA: 1184] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 17. | Arata S, Hamaguchi S, Nose K. Inhibition of colony formation of NIH 3T3 cells by the expression of the small molecular weight heat shock protein HSP27: involvement of its phosphorylation and aggregation at the C-terminal region. J Cell Physiol. 1997;170:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 18. | Yang Y, Rao R, Shen J, Tang Y, Fiskus W, Nechtman J, Atadja P, Bhalla K. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68:4833-4842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 19. | Park JW, Yeh MW, Wong MG, Lobo M, Hyun WC, Duh QY, Clark OH. The heat shock protein 90-binding geldanamycin inhibits cancer cell proliferation, down-regulates oncoproteins, and inhibits epidermal growth factor-induced invasion in thyroid cancer cell lines. J Clin Endocrinol Metab. 2003;88:3346-3353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 336] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 21. | Ryu JW, Kim HJ, Lee YS, Myong NH, Hwang CH, Lee GS, Yom HC. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci. 2003;18:505-509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 102] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 22. | Poon TC, Sung JJ, Chow SM, Ng EK, Yu AC, Chu ES, Hui AM, Leung WK. Diagnosis of gastric cancer by serum proteomic fingerprinting. Gastroenterology. 2006;130:1858-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | Bai Z, Ye Y, Liang B, Xu F, Zhang H, Zhang Y, Peng J, Shen D, Cui Z, Zhang Z. Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer. Int J Oncol. 2011;38:375-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 24. | Liu W, Liu B, Cai Q, Li J, Chen X, Zhu Z. Proteomic identification of serum biomarkers for gastric cancer using multi-dimensional liquid chromatography and 2D differential gel electrophoresis. Clin Chim Acta. 2012;413:1098-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Jia SQ, Niu ZJ, Zhang LH, Zhong XY, Shi T, Du H, Zhang GG, Hu Y, Su XL, Ji JF. Identification of prognosis-related proteins in advanced gastric cancer by mass spectrometry-based comparative proteomics. J Cancer Res Clin Oncol. 2009;135:403-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Chen YR, Juan HF, Huang HC, Huang HH, Lee YJ, Liao MY, Tseng CW, Lin LL, Chen JY, Wang MJ. Quantitative proteomic and genomic profiling reveals metastasis-related protein expression patterns in gastric cancer cells. J Proteome Res. 2006;5:2727-2742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447-2457. [PubMed] |
| 28. | Liu S, Ballian N, Belaguli NS, Patel S, Li M, Templeton NS, Gingras MC, Gibbs R, Fisher W, Brunicardi FC. PDX-1 acts as a potential molecular target for treatment of human pancreatic cancer. Pancreas. 2008;37:210-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 29. | Miyatsuka T, Kaneto H, Shiraiwa T, Matsuoka TA, Yamamoto K, Kato K, Nakamura Y, Akira S, Takeda K, Kajimoto Y. Persistent expression of PDX-1 in the pancreas causes acinar-to-ductal metaplasia through Stat3 activation. Genes Dev. 2006;20:1435-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 132] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 30. | Faller G, Kirchner T. Immunological and morphogenic basis of gastric mucosa atrophy and metaplasia. Virchows Arch. 2005;446:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Sakai H, Eishi Y, Li XL, Akiyama Y, Miyake S, Takizawa T, Konishi N, Tatematsu M, Koike M, Yuasa Y. PDX1 homeobox protein expression in pseudopyloric glands and gastric carcinomas. Gut. 2004;53:323-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 32. | Zhu S, Xia HH, Yang Y, Ma J, Chen M, Hu P, Gu Q, Liang Y, Lin H, Wong BC. Alterations of gastric homeoprotein expression in Helicobacter pylori infection, incisural antralisation, and intestinal metaplasia. Dig Dis Sci. 2009;54:996-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 33. | Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 663] [Cited by in RCA: 710] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 34. | Li XS, Xu Q, Fu XY, Luo WS. Heat shock protein 60 overexpression is associated with the progression and prognosis in gastric cancer. PLoS One. 2014;9:e107507. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 35. | Ge H, He X, Guo L, Yang X. Clinicopathological significance of HSP27 in gastric cancer: a meta-analysis. Onco Targets Ther. 2017;10:4543-4551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 36. | Bodoor K, Jalboush SA, Matalka I, Abu-Sheikha A, Waq RA, Ebwaini H, Abu-Awad A, Fayyad L, Al-Arjat J, Haddad Y. Heat Shock Protein Association with Clinico-Pathological Characteristics of Gastric Cancer in Jordan : HSP70 is Predictive of Poor Prognosis. Asian Pac J Cancer Prev. 2016;17:3929-3937. [PubMed] |
| 37. | Xiang TX, Li Y, Jiang Z, Huang AL, Luo C, Zhan B, Wang PL, Tao XH. RNA interference-mediated silencing of the Hsp70 gene inhibits human gastric cancer cell growth and induces apoptosis in vitro and in vivo. Tumori. 2008;94:539-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 38. | Arora N, Alsaied O, Dauer P, Majumder K, Modi S, Giri B, Dudeja V, Banerjee S, Von Hoff D, Saluja A. Downregulation of Sp1 by Minnelide leads to decrease in HSP70 and decrease in tumor burden of gastric cancer. PLoS One. 2017;12:e0171827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |