Published online Aug 14, 2018. doi: 10.3748/wjg.v24.i30.3426
Peer-review started: March 28, 2018
First decision: May 9, 2018
Revised: May 24, 2018
Accepted: June 22, 2018
Article in press: June 22, 2018
Published online: August 14, 2018
Processing time: 139 Days and 16 Hours
To construct a long non-coding RNA (lncRNA) signature for predicting hepatocellular carcinoma (HCC) prognosis with high efficiency.
Differentially expressed lncRNAs (DELs) between HCC specimens and peritumor liver specimens were identified using the edgeR package to analyze The Cancer Genome Atlas (TCGA) LIHC dataset. Univariate Cox proportional hazards regression was performed to obtain the DELs significantly associated with overall survival (OS) in a training set. These OS-related DELs were further analyzed using a stepwise multivariate Cox regression model. Those lncRNAs fitted in the multivariate Cox regression model and independently associated with overall survival were chosen to build a prognostic risk formula. The prognostic value of this formula was then validated in the test group and the entire cohort and further compared with two previously identified prognostic signatures for HCC. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed to explore the potential biological functions of the lncRNAs in the signature.
Based on lncRNA expression profiling of 370 HCC patients from the TCGA database, we constructed a 5-lncRNA signature (AC015908.3, AC091057.3, TMCC1-AS1, DCST1-AS1 and FOXD2-AS1) that was significantly associated with prognosis. HCC patients with high-risk scores based on the expression of the 5 lncRNAs had significantly shorter survival times compared to patients with low-risk scores in both the training and test groups. Multivariate Cox regression analysis demonstrated that the prognostic value of the 5 lncRNAs was independent of clinicopathological parameters. A comparison study involving two previously identified prognostic signatures for HCC demonstrated that this 5-lncRNA signature showed improved prognostic power compared with the other two signatures. Functional enrichment analysis indicated that the 5 lncRNAs were potentially involved in metabolic processes, fibrinolysis and complement activation.
Our present study constructed a 5-lncRNA signature that improves survival prediction and can be used as a prognostic biomarker for HCC patients.
Core tip: In the present study, we developed a 5-long non-coding RNA (lncRNA) signature for predicting the prognosis of hepatocellular carcinoma (HCC) patients based on The Cancer Genome Atlas database. The signature was reproducible and robust in another independent large-scale HCC cohort, supporting its utility and effectiveness. In addition, the prognostic value of the 5-lncRNA signature was independent of clinicopathological variables. When compared with two previously identified signatures for HCC survival prediction, this 5-lncRNA signature showed superior prognostic power. Our study indicates that the 5-lncRNA signature could improve survival prediction and could be used as a prognostic biomarker for HCC patients.
- Citation: Zhao QJ, Zhang J, Xu L, Liu FF. Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma. World J Gastroenterol 2018; 24(30): 3426-3439
- URL: https://www.wjgnet.com/1007-9327/full/v24/i30/3426.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i30.3426
Hepatocellular carcinoma (HCC) is the sixth most commonly diagnosed cancer in the world[1]. According to previous epidemiologic studies, the incidence of HCC varies strikingly worldwide and is particularly high in eastern Asian countries, including China, and sub-Saharan Africa[2,3]. The 5-year overall survival rate for HCC is lower than 20%[4], and the ratio of its mortality to morbidity is 0.95[1]. Because of its poor prognosis, HCC ranks as the second leading cause of cancer-related deaths worldwide[1]. An estimated 782500 new liver cancer cases and 745500 deaths occurred worldwide in 2012, among which 50% occurred in China[3]. There are multiple risk factors related to HCC, including hepatitis B or C viral infection, chronic alcohol abuse, nonalcoholic fatty liver disease and smoking[5,6]. Although treatment for HCC, including surgical resection, has improved over the past decades, the overall survival rate for this disease remains devastatingly high due to its high recurrence rate (50%-70% at 5 years)[7-9]. Because HCC is a heterogeneous disease with substantially variable clinical outcomes, the search for effective biomarkers to predict recurrence and prognosis is indispensable. To date, no widely accepted molecular biomarkers for HCC aggressiveness are available. In the past 40 years, serum alpha fetoprotein (AFP) levels have been utilized for the diagnosis of HCC and for predicting its response to therapy. However, AFP levels can be influenced by tumor size and cancer stage, and they are not reliable in clinical applications[10]. In addition, the American Association for the Study of Liver Diseases concluded that the use of AFP levels lacks sufficient sensitivity and specificity to effectively monitor or diagnose HCC[11].
With the development of high-throughput sequencing technologies, it has become easy to acquire whole genome profiles for specific cancers and develop more reliable prognostic signatures. Long non-coding RNAs (lncRNAs) are mRNA-like transcripts of more than 200 nucleotides (nt) with little or no protein-coding capacity[12,13]. In the past, they were previously thought to be redundant segments of the genome, but in recent decades, emerging studies have indicated the importance of lncRNAs in cellular physiological and pathological processes[14,15]. Increasing evidence suggests that dysregulated lncRNAs are associated with various human diseases, particularly the initiation and progression of various human cancers[16,17]. Prognostic lncRNA signatures have been examined in many cancer types, including renal cancer, glioblastoma, colorectal cancer, lymphoma, and others[18-21]. For HCC, most of the published gene signatures associated with prognosis have focused on mRNAs and microRNAs[22-25]. To the best of our knowledge, very few lncRNA signatures have been developed for HCC prognosis prediction[26]. Thus, it is necessary to identify a more effective lncRNA signature for HCC prognosis. In the present study, we aimed to construct a lncRNA signature capable of predicting HCC prognosis with high efficiency.
In this work, we analyzed a cohort of 370 HCC patients from The Cancer Genome Atlas (TCGA) to identify a potential lncRNA signature for predicting the survival of HCC patients. We identified a five-lncRNA prognostic signature from the TCGA dataset and determined that its prognostic value was independent from clinical factors. The identification of prognostic lncRNAs suggests the potential roles of lncRNAs in HCC pathogenesis and progression.
Level 3 RNA-seq data (HTSeq-counts) from 374 HCC tumor specimens and 50 peritumoral liver specimens and their corresponding clinicopathological information were downloaded from the TCGA project (https://cancergenome.nih.gov/) on June 2, 2017. Because TCGA data are a community resource project, additional ethical approval was not acquired, and the present study adhered to TCGA publication guidelines and data access policies. After excluding the data without complete survival information, a total of 370 HCC patients with complete follow up data were enrolled in our study and then randomly divided into a training set (n = 184) and test set (n = 186) using SPSS software (version 24.0). The clinicopathological parameters of the HCC patients in each group are listed in Table 1.
| Variables | Training group (n = 184) | Test group (n = 186) | Entire group (n = 370) | |
| Age, yr | < 60 | 84 | 85 | 169 |
| ≥ 60 | 100 | 101 | 201 | |
| Sex | Male | 129 | 120 | 24 |
| Female | 55 | 66 | 121 | |
| Weight, kilograms | < 70 | 92 | 85 | 177 |
| ≥ 70 | 78 | 89 | 167 | |
| NA | 14 | 12 | 26 | |
| Child-Pugh grade | A | 109 | 107 | 216 |
| B | 9 | 12 | 21 | |
| C | 0 | 1 | 1 | |
| NA | 66 | 66 | 132 | |
| Fibrosis ishak score | 0 | 32 | 42 | 74 |
| 1-5 | 38 | 31 | 69 | |
| 6 | 36 | 33 | 69 | |
| NA | 78 | 80 | 158 | |
| Vascular tumor invasion | None | 100 | 106 | 206 |
| Micro | 45 | 46 | 91 | |
| Macro | 8 | 9 | 17 | |
| NA | 31 | 25 | 56 | |
| Serum FAP level, ng/mL | < 100 | 102 | 90 | 192 |
| ≥ 100 | 37 | 48 | 85 | |
| NA | 45 | 48 | 93 | |
| Tumor grade | 1 | 37 | 18 | 55 |
| 2 | 82 | 95 | 177 | |
| 3 + 4 | 61 | 72 | 133 | |
| NA | 4 | 1 | 5 | |
| Pathologic stage | I | 92 | 79 | 171 |
| II | 43 | 42 | 85 | |
| III + IV | 37 | 53 | 90 | |
| NA | 12 | 12 | 24 | |
Only lncRNAs with a description in NCBI or Ensemble were selected for further study in this paper. We obtained the expression profiles of 6929 lncRNAs from the RNA-seq data of the TCGA LIHC cohort. Differentially expressed lncRNAs (DELs) between the HCC specimens and peritumor liver specimens were identified with the edgeR package, using an adjusted P < 0.05 and log2 |fold change| > 1. The expression level of each lncRNA was log2 transformed for the downstream analyses.
Univariate Cox proportional hazards regression was performed to obtain the DELs that were significantly associated with the overall survival (OS) of HCC patients in the training group. After acquiring survival-related lncRNAs (P < 0.01), we excluded those not expressed in at least 10% of the samples. The remaining OS-related lncRNAs were then adjusted sing the stepwise multivariate Cox regression model. Finally, those lncRNAs fitted in the multivariate Cox regression model and independently associated with OS were chosen. A prognostic risk formula was established based on a linear combination of the expression level of these lncRNAs multiplied by the regression coefficient derived from the multivariate Cox regression model as previously described[18-21].The subjects in each dataset were classified into a high-risk group and low-risk group according to the median risk score of the risk formula derived from the training set.
Univariate Cox proportional hazards regression was performed to obtain survival-related DELs, and the stepwise multivariate Cox regression model was performed for further selection. Overall survival analyses in the high-risk and low-risk groups were performed using Kaplan-Meier survival curves and a log-rank test. Receiver operating curve analyses were performed to assess the specificity and sensitivity of the prognosis prediction. The above analyses were performed using R (version 3.3.1). To verify the independence of the prognostic value of the 5-lncRNA signature and clinicopathological parameters, univariate and multivariate Cox regression analyses were performed using SPSS software (version 24.0). In the comparison study, Kaplan-Meier survival analysis and receiver operating curve (ROC) analysis were also performed using SPSS (version 24.0).
To identify co-expressed lncRNA-mRNA pairs, we performed Person correlation analyses with R (version 3.3.1) for each of the five lncRNAs with protein-coding genes based on the RNA-seq data of the TCGA LIHC cohort. The protein-coding genes with a correlation coefficient > 0.5 and a P < 0.01 were considered to be significantly correlated genes. For functional enrichment analysis, the correlated protein-coding genes were subjected to gene ontology (GO) and Kyoto Encyclopediaof Genes and Genomes (KEGG) pathway analyses using DAVID Bioinformatics Resources (version 6.8)[27,28]. Significant functional categories were identified and limited to GO terms in the “Biological Process” (GOTERM-BP-DIRECT) and KEGG pathway categories, using the human whole genome as the background. Significantly enriched GO terms with similar functions were visualized using the EnrichmentMap plugin in Cytoscape (version 3.5.1)[29].
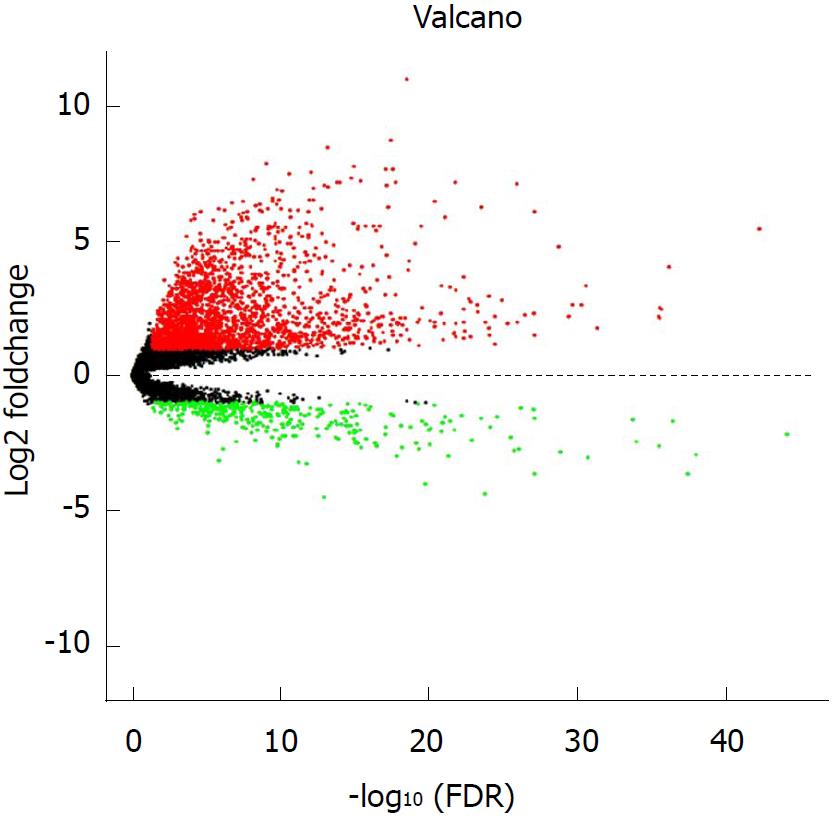
Using the edgeR package, we identified a total of 2593 lncRNAs differentially expressed (log2|fold change| > 1 and adjusted P < 0.05) between 374 HCC tumor specimens and 50 peritumor liver specimens, including 2240 upregulated and 353 downregulated lncRNAs (Figure 1). A total of 370 HCC samples with complete survival information were subjected to further analyses. For the training set, univariate Cox proportional hazards regression analyses revealed 82 lncRNAs significantly correlated with OS (P < 0.01) among the 2593 differentially expressed lncRNAs. Among the 82 OS-related lncRNAs, we further excluded those expressed in less than 10% of the HCC specimens, and the remaining 30 lncRNAs were subjected to further selection.
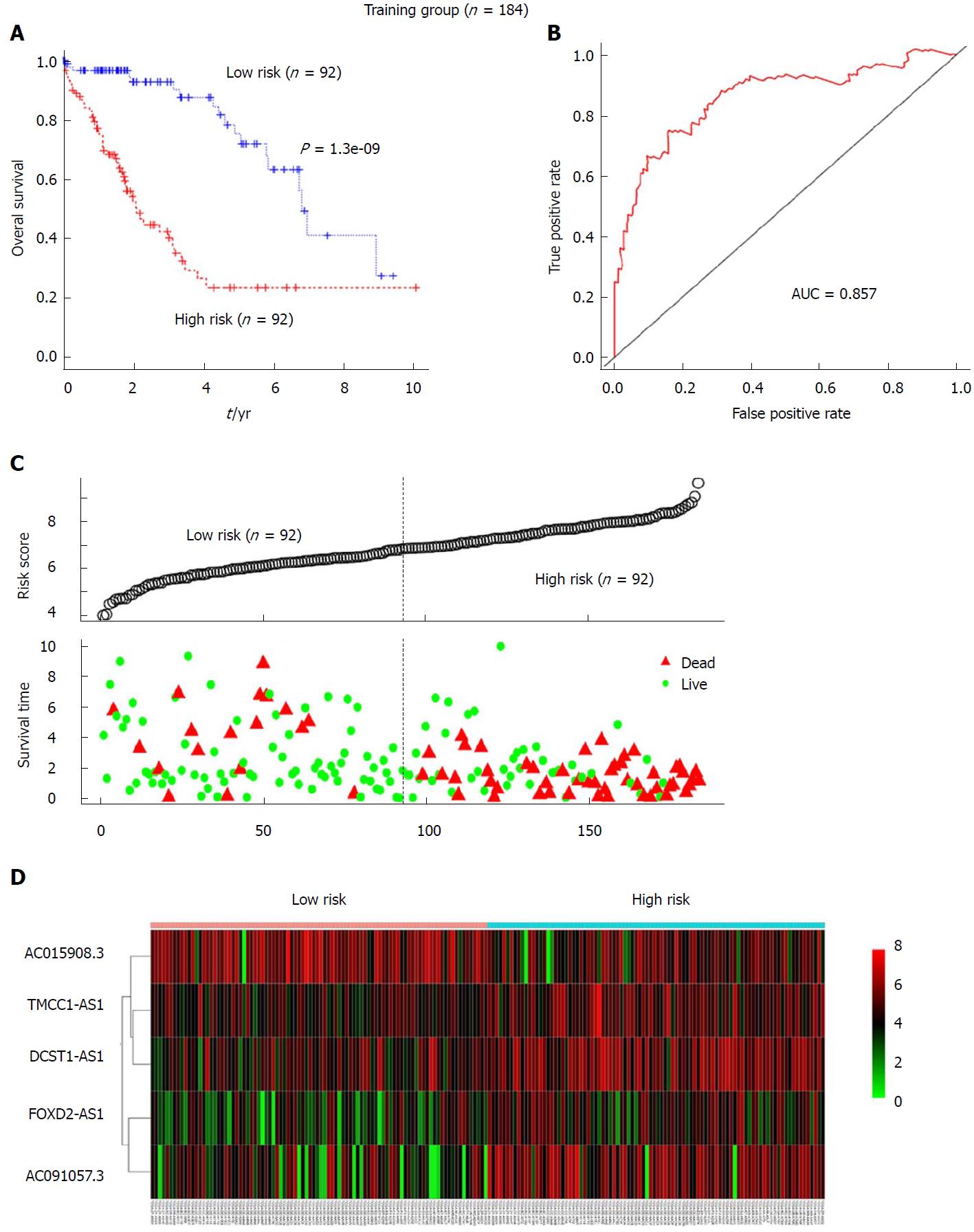
Stepwise multivariable Cox proportional hazards regression analyses were performed to identify the optimal prognostic lncRNAs among the 30 candidate lncRNAs. Based on this model, a final 5 lncRNAs were found to be significantly and independently related to prognosis. We then constructed a prognostic signature based on the expression levels of these 5 lncRNAs and their coefficients derived from the multivariable Cox model. The formula is as follows: risk score = (-0.1900 × the expression level of AC015908.3) + (0.1764 × the expression level of FOXD2-AS1) + (0.3588 × the expression level of AC091057.3) + (0.5615 × the expression level of TMCC1-AS1) + (0.4877 × the expression level of DCST1-AS1). Detailed information for the 5 lncRNAs is listed in Table 2. The risk score for each patient in the training group was calculated using the formula. The training set was then divided into a high-risk group (n = 92) and a low-risk group (n = 92) according to the median risk score. Kaplan-Meier analysis revealed that the high-risk group had a significantly poorer prognosis than that of the low-risk group (P = 1.3e-09, log-rank test, Figure 2A). The median survival time for the high-risk group and the low-risk group was 2.096 and 6.811 years, respectively. Additionally, the 3- and 5-year survival rates of the high-risk group were 40% and 23.5%, whereas the corresponding survival rates were 90% and 71.8%, respectively, in the low-risk group. To evaluate the performance of the 5-lncRNA signature for predicting the prognosis of HCC patients, a time-dependent ROC analysis was conducted. The area under the ROC curve (AUC) for the 5-lncRNA signature was 0.857, which indicated good performance (Figure 2B). The risk scores of patients in the training group were also ranked, and survival status was plotted for each patient on a dot plot (Figure 2C). The mortality for patients in the high-risk group was much higher than that in the low-risk group. A heat map displays the expression profiles of these five lncRNAs in the samples from the training group; the expression profiles are ranked according to risk score (Figure 2D). Among the 5 lncRNAs, AC015908.3 showed a negative coefficient derived for the multivariate Cox regression model and seemed to be a protective factor, as its high expression predicts a low risk. The other 4 lncRNAs with positive coefficients, including FOXD2-AS1, AC091057.3, TMCC1-AS1 and DCST1-AS1, seemed to be risk factors and all were upregulated in the high-risk group compared to the low-risk group within the training set.
| Gene ID | Gene symbol | Coefficient | Hazard ratio | P value |
| ENSG00000264016 | AC015908.3 | -0.1900 | 0.7792 | 0.000305 |
| ENSG00000237424 | FOXD2-AS1 | 0.1764 | 1.2865 | 0.007317 |
| ENSG00000269974 | AC091057.3 | 0.3588 | 1.4682 | 0.000375 |
| ENSG00000271270 | TMCC1-AS1 | 0.5615 | 1.5417 | 0.000287 |
| ENSG00000232093 | DCST1-AS1 | 0.4877 | 1.3909 | 0.001632 |
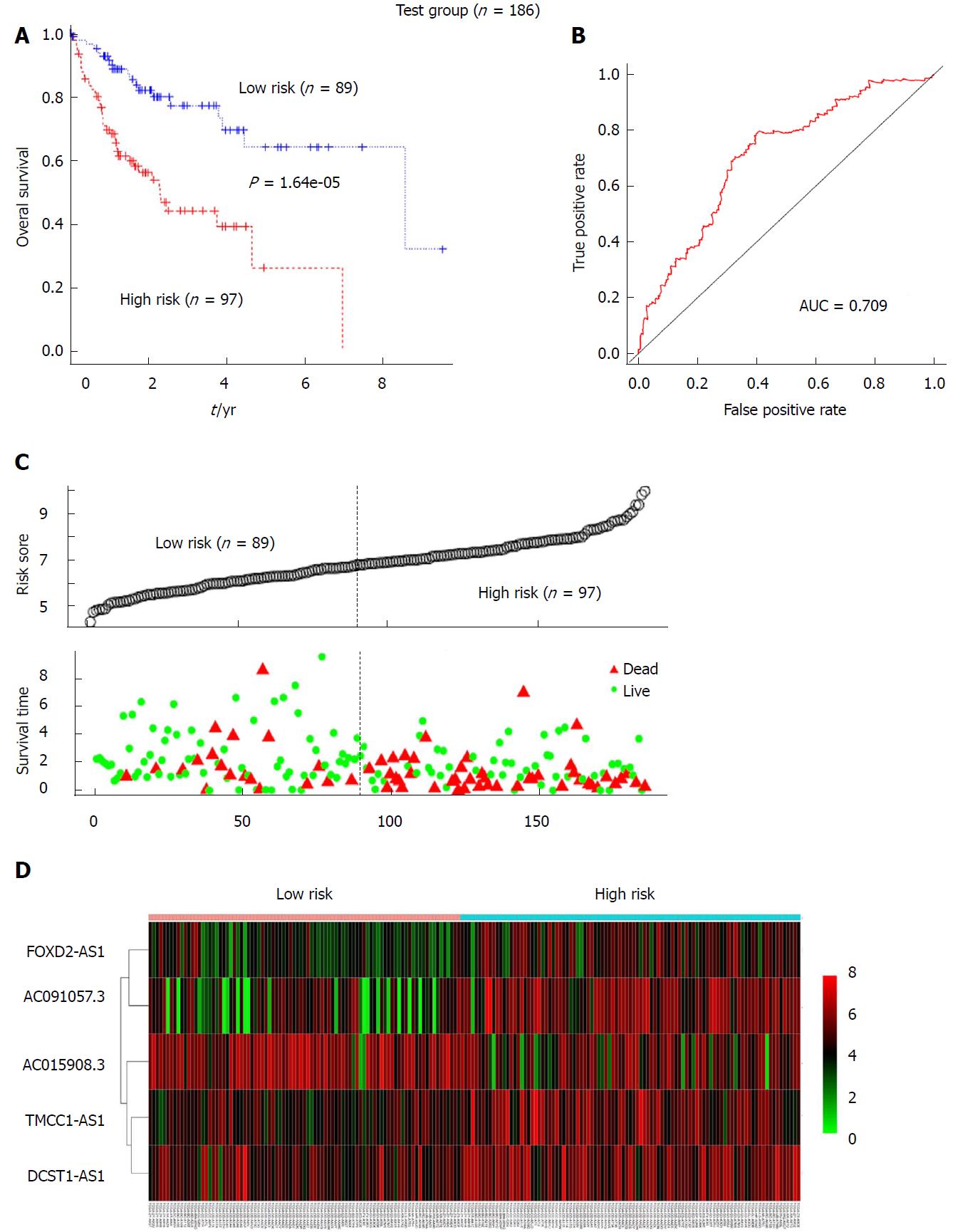
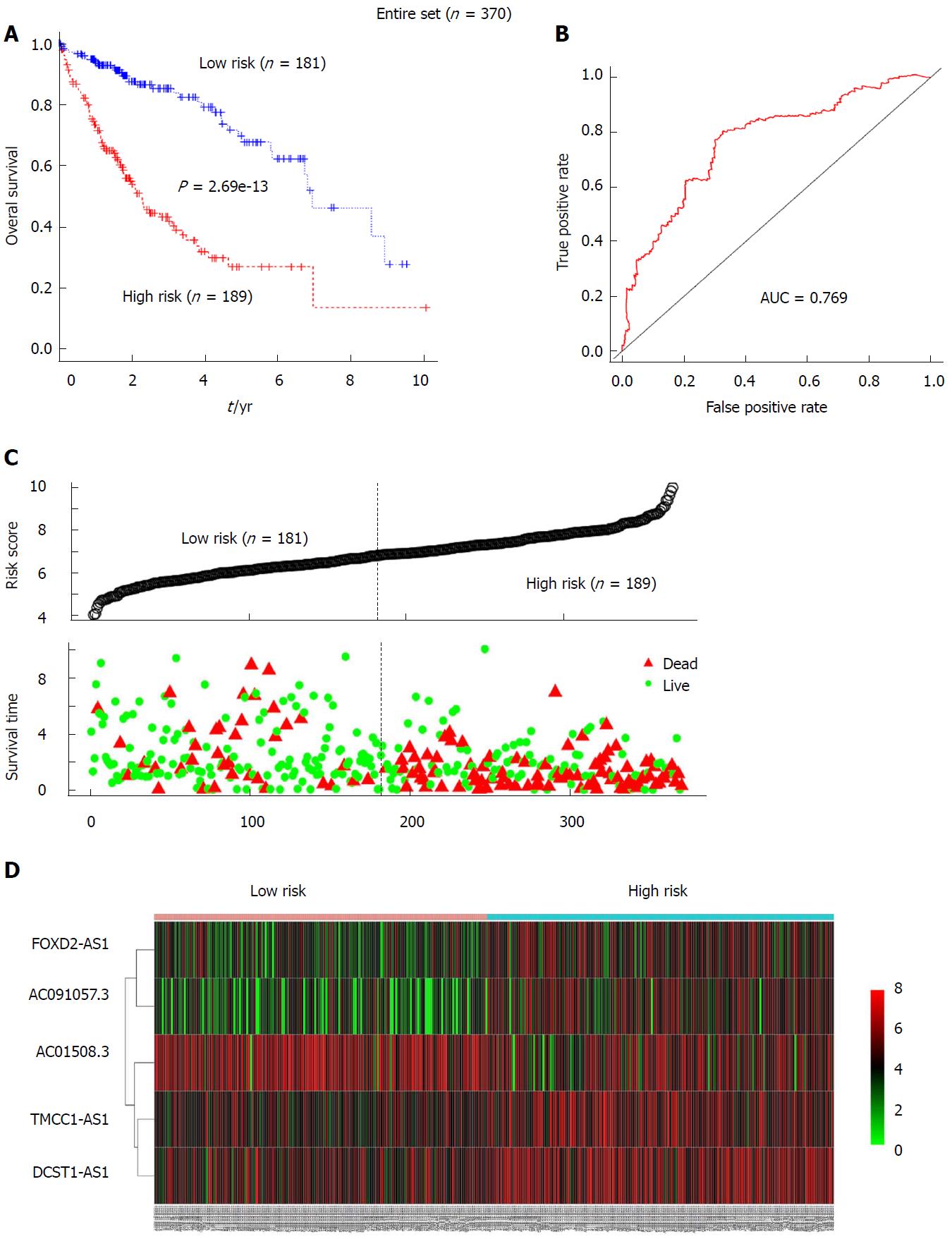
To further verify the prognostic value of the 5-lncRNA signature for HCC patients, risk scores for patients in the test group were calculated according to the constructed formula based on the expression of the 5 lncRNAs. The test group was also divided into high-risk (n = 97) and low-risk (n = 89) groups using the same cutoff as for the training group. Kaplan-Meier analysis revealed that the survival rate of the high-risk subgroup was much lower than that of the low-risk subgroup in the test set (median OS: 2.293 years vs 8.562 years; log-rank P = 1.64e-05) (Figure 3A). For the entire set, a similar result was obtained by Kaplan-Meier analysis. Among the entire cohort, the median survival of the high-risk group (n = 189) was 2.197 years, which was significantly lower than the median OS of 6.937 years for the low-risk group (n = 181) (P = 2.69e-13, Figure 4A). The AUC for the 5-lncRNA-based risk score of overall survival was 0.709 and 0.769 for the test group (Figure 3B) and the entire group (Figure 4B), respectively, with both showing robust utility. In addition, ranked risk scores and survival status for each subject were plotted for the test group (Figure 3C) and the entire set (Figure 4C). Heatmaps display the expression profiles of the five lncRNAs for each subject in the test group (Figure 3D) and the entire cohort (Figure 4D), which were ranked according to risk score.
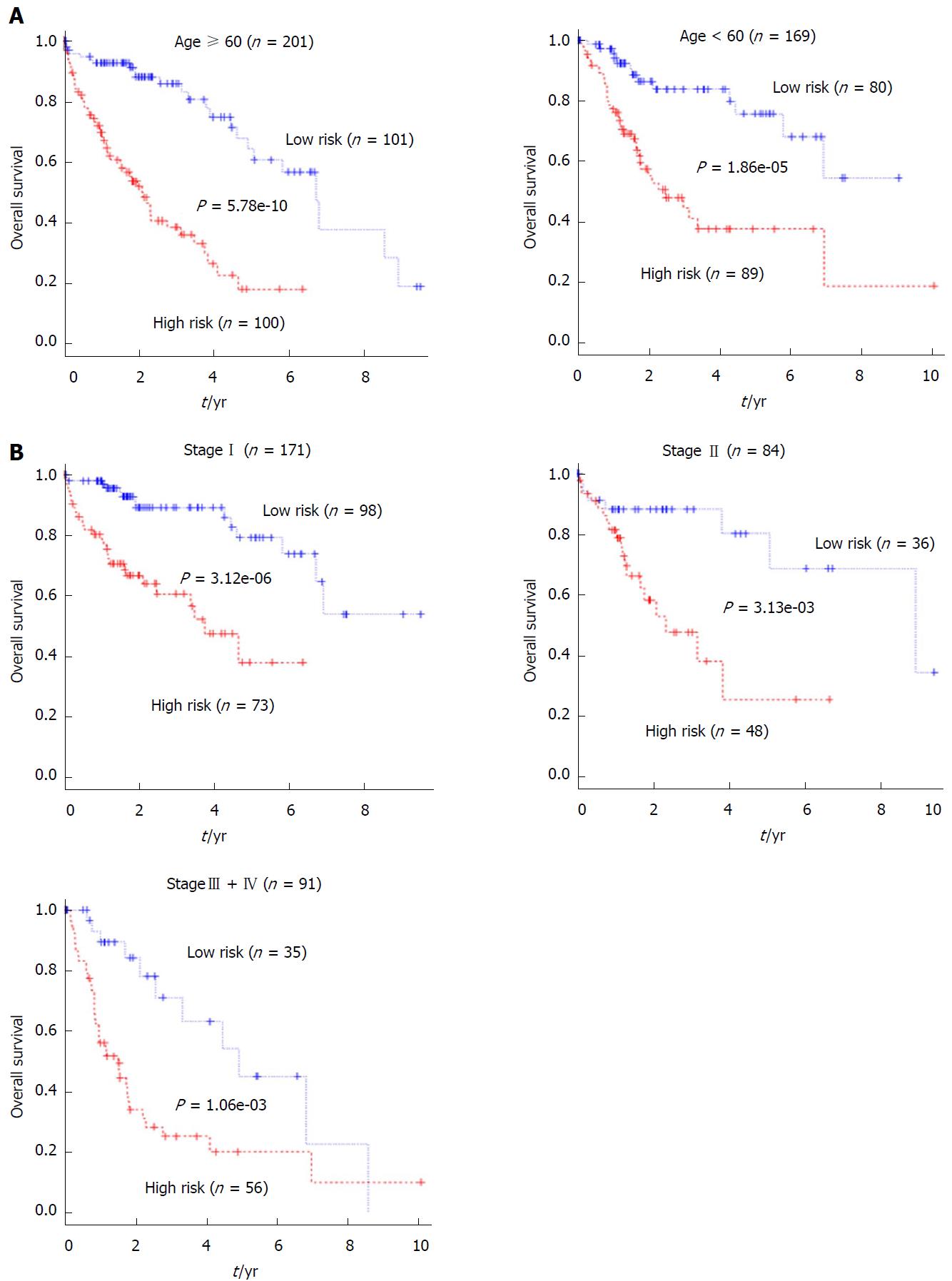
Univariate and multivariate Cox regression analyses were performed with the 5-lncRNA-based risk score and clinicopathological factors, including age, gender, weight, Child-Pugh grading, fibrosis extent, vascular tumor invasion, serum FAP levels, tumor grade and pathological stage as explanatory variables and overall survival as the dependent variable. The univariate Cox regression demonstrated that the 5-lncRNA signature-based risk score and pathologic stage were able to effectively predict the prognosis of HCC patients. In addition, in the training set and the entire set, patient age seemed to be related to survival, although this did not reach significance (Table 3). In contrast, none of the other clinicopathological parameters were associated with prognosis in either set. Multivariate Cox regression analysis revealed that after adjusting for other factors, age (only for the entire set), pathologic stage and the 5-lncRNAsignature were the only factors significantly associated with overall survival (Table 2). Patients from the entire cohort were then stratified by age (Figure 5A) and pathological stage (Figure 5B). Each subgroup was then divided into a high-risk and low-risk group based on the 5-lncRNA risk score median derived from the training group. Kaplan-Meier analysis revealed that for all of the subgroups, the high-risk group had significantly poorer survival than the low-risk group. All of these results strongly suggest that the prognostic value of the 5-lncRNA-based risk score is independent of clinicopathological factors.
| Variables | Univariate analysis | Multivariate analysis | ||||
| HR | 95%CI of HR | P value | HR | 95%CI of HR | P value | |
| Training set (n = 184) | ||||||
| Risk score | 2.718 | 2.093-3.530 | < 0.0001 | 2.830 | 2.091-3.831 | < 0.0001 |
| Age | 1.018 | 0.998-1.038 | 0.073 | |||
| Sex (Male/Female) | 1.022 | 0.619-1.690 | 0.931 | |||
| Weight | 1.006 | 0.993-1.108 | 0.397 | |||
| Child-Pugh grade | 0.845 | 0.253-2.827 | 0.195 | |||
| Fibrosis ishak score | 0.685 | 0.756-1.283 | 0.910 | |||
| Vascular invasion (yes/no) | 0.834 | 0.480-1.449 | 0.519 | |||
| FAP | 0.805 | 0.932-1.056 | 0.805 | |||
| Tumor grade | ||||||
| (G1 + G2/G3 + G4) | 1.133 | 0.680-1.887 | 0.632 | |||
| Pathologic stage | 1.900 | 1.098-3.288 | 0.022 | |||
| I/II | 0.604 | 0.303-1.203 | 0.152 | |||
| I/III + IV | 0.298 | 0.163-0.543 | < 0.0001 | |||
| Test set (n = 186) | ||||||
| Risk score | 1.603 | 1.270-2.024 | < 0.0001 | 1.568 | 1.196-2.055 | 0.001 |
| Age | 1.006 | 0.987-1.026 | 0.522 | |||
| Sex (Male/Female) | 1.420 | 0.856-2.356 | 0.174 | |||
| Weight | 1.000 | 0.984-1.105 | 0.960 | |||
| Child-Pugh grade | 0.475 | 0.195-1.157 | 0.101 | |||
| Fibrosis ishak score | 1.640 | 0.706-3.813 | 0.250 | |||
| Vascular invasion (yes/no) | 1.069 | 0.613-1.866 | 0.614 | |||
| FAP | 1.035 | 0.981-1.093 | 0.210 | |||
| Tumor grade | ||||||
| (G1 + G2/G3 + G4) | 1.092 | 0.655-1.818 | 0.736 | |||
| Pathologic stage | 2.103 | 1.193-3.707 | 0.010 | |||
| I/II | 0.865 | 0.426-1.757 | 0.688 | |||
| I/III + IV | 0.445 | 0.250-0.793 | 0.006 | |||
| Entire set (n = 370) | ||||||
| Risk score | 1.957 | 1.646-2.327 | < 0.0001 | 2.011 | 1.638-2.469 | < 0.0001 |
| Age | 1.013 | 0.999-1.027 | 0.068 | 1.016 | 1.000-1.032 | 0.048 |
| Sex (Male/Female) | 1.166 | 0.817-1.664 | 0.396 | |||
| Weight | 0.998 | 0.988-1.007 | 0.615 | |||
| Child-Pugh grade (A/B + C) | 0.620 | 0.306-1.256 | 0.184 | |||
| Fibrosis ishak score | 1.232 | 0.742-2.045 | 0.365 | |||
| Vascular invasion | 0.962 | 0.650-1.424 | 0.846 | |||
| (yes/no) | ||||||
| Serum AFP level | 1.023 | 0.980-1.068 | 0.306 | |||
| Tumor grade | ||||||
| (G1 + G2/G3 + G4) | 1.119 | 0.780-1.604 | 0.542 | |||
| Pathologic stage | 2.017 | 1.359-2.993 | 0.027 | |||
| I/II | 0.648 | 0.398-1.056 | 0.082 | |||
| I/III + IV | 0.351 | 0.236-0.524 | < 0.0001 | |||
Two HCC-related prognostic signatures have recently been developed and reported, including a 3-gene signature by Binghua Li and a 4-lncRNA signature by Zhonghao Wang that were both derived from the TCGA dataset[25,26]. To compare the prognostic value of the 5-lncRNA signature developed in our present study (hereafter referred to as 5LncSig) with the existing 3-gene signature by Binghua Li (hereafter referred to as 3GeneSig) and the 4-lncRNA signature by Zhonghao Wang (hereafter referred to as ZhongSig), we calculated the risk scores of each patient in the entire cohort based on formulae derived from each of these signatures. The 3GeneSig and ZhongSig both successfully and significantly predicted prognosis in the entire TCGA LIHC cohort (Figure 6A and B). Furthermore, comparison of the Kaplan-Meier curves revealed that patients in the high-risk group predicted by 5LncSig showed a dramatically poorer prognosis than those in the low-risk groups predicted by the 3GeneSig and ZhongSig (Figure 6A and B), and patients in the low-risk group predicted by 5LncSig had a much better prognosis than those in the high-risk group predicted by the other two signatures (Figure 6A and B). To compare the sensitivity and specificity of the 5LncSig for prognosis prediction with the other two existing signatures, we performed time-dependent ROC analysis. The AUC of overall survival for the 3GeneSig and the ZhongSig was 0.701 and 0.721, respectively (Figure 6C), both lower than that of the 5LncSig (0.769). Thus, the prognostic power of 5LncSig, developed in the present study, was superior to that of the previously developed 3-gene and 4-lncRNA signatures.
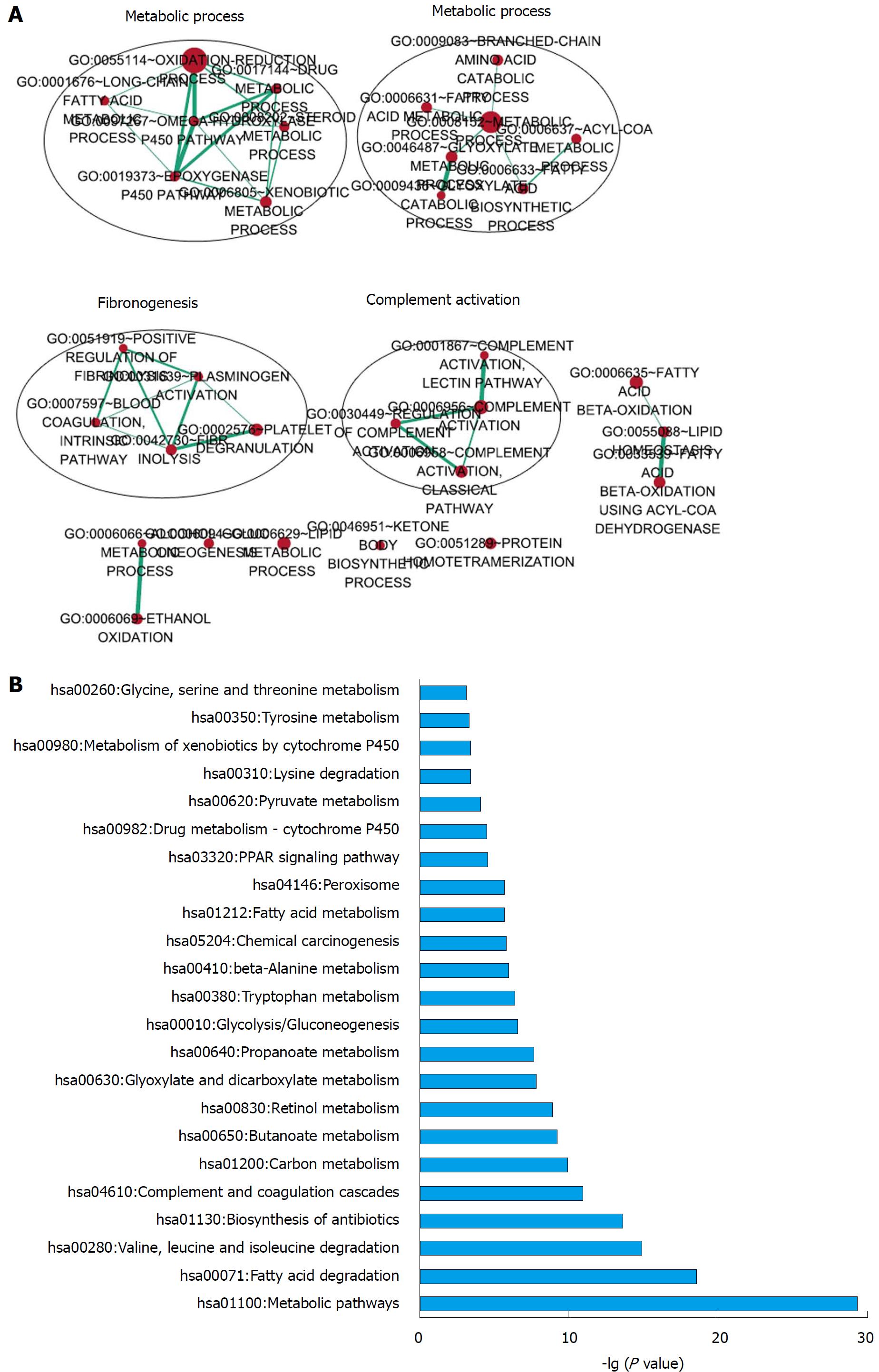
To explore the functional implications of these 5 lncRNAs, we performed Pearson correlation analyses between the 5 lncRNAs and protein-coding genes based on their expression levels in the TCGA LIHC cohort. The protein-coding genes that correlated with at least 1 of the 5 lncRNAs (Pearson coefficient > 0.5, P < 0.01) were considered to be correlated genes. We chose the 200 correlated genes with the highest Pearson coefficients for further analysis. Functional enrichment analysis revealed that these genes were primarily enriched in 32 GO terms (Benjamin P value < 0.1, Figure 7A) and 23 KEGG pathways (P < 0.001, Figure 7B). Further analysis revealed that these enriched GO functional terms are mostly involved in metabolic processes, fibrinolysis and complement activation (Figure 7A).
HCC is a heterogeneous disease with differential prognoses and a high mortality. Until now, no biomarkers have been shown to effectively predict the survival of HCC patients, and thus, finding effective biomarkers for HCC is crucial.
Previous investigations of gene regulation and disease pathogenesis have mainly focused on protein-coding genes, which account for only a very small proportion (2%) of transcribed genes in eukaryotic species[13]. Recent developments in genome and transcriptome sequencing technologies have profoundly expanded our knowledge of non-coding RNAs, which are much more abundant than canonical protein-coding mRNAs[30,31]. Multiple studies indicate that lncRNAs act not only as intermediaries between DNA and protein but also as important regulators of diverse cellular functions. lncRNAs have been shown to regulate the expression and function of protein-coding genes at the chromatin, transcriptional and post-transcriptional levels[31]. Many studies have revealed the contribution of lncRNAs in cancer development, indicating their potential as novel biomarkers for cancer diagnosis and prognosis[32-35].
LncRNA signatures for prognostic prediction prognoses have been developed for many cancers including renal cancer, glioblastoma, and colorectal cancer, among others[18-21]. Regarding HCC, the existing gene signatures for survival prediction have focused mostly on mRNAs and microRNAs. Several potential lncRNA biomarkers associated with the progression and prognosis of HCC have been identified, such as TSLNC8, HOXD-AS1 and CACS2[36-38]. These lncRNAs are thought to impact HCC progression through their regulation of tumor cell proliferation, EMT, apoptosis and migration. Although many of these lncRNAs are closely associated with the prognosis and survival of HCC patients, their prognostic value has been tested only in small-scale studies; they have not yet been validated in a large clinical cohort. Until now, relatively few comprehensive lncRNA signatures for the prediction of HCC survival have been constructed[26]. TCGA is an open-access database including samples from hundreds of patients with various malignancies. In the present study, we downloaded the RNA sequencing data of the TCGA LIHC cohort and acquired lncRNA expression profiles for HCC patients in the dataset. Using univariate and stepwise multivariate Cox regression analyses, we developed a prognostic formula for HCC based on the expression of 5 lncRNAs including AC015908.3, AC091057.3, TMCC1-AS1, DCST1-AS1 and FOXD2-AS1. In the training set, HCC patients with high-risk scores based on the 5-lncRNA signature had a significantly reduced survival time compared to those with low-risk scores. The prognostic value of the 5-lncRNA signature for HCC patients was further validated in the test group and the entire group, with robust and reproducible predictive indices. The results of these analyses suggest that the prognostic value of the 5-lncRNA-based risk model is robust and reliable for predicting survival in HCC patients.
In the present study, when adjusted using multivariate Cox regression analyses, age (only for the entire set), pathological stage and the 5-lncRNA signature were shown to independently predict the survival of HCC patients. The results of stratification analyses demonstrated that the prognostic value of the 5-lncRNA signature remained significant and robust in HCC subgroups stratified by age and pathological stage. In an attempt to further validate its prognostic value in other HCC cohorts, we downloaded data from several GEO datasets. Unfortunately, most of the 5 lncRNA probes could not be found. There are many existing prognostic signatures for HCC, and we therefore compared our 5-lncRNA signature with two recently developed signatures: a 3-gene signature and a 4-lncRNA signature. The results indicate that the predictive performance of the 5-lncRNA signature was superior to that of the other two signatures for HCC overall survival.
To the best of our knowledge, the functions of these 5 lncRNAs have not been reported. Functional enrichment analysis revealed that the protein-coding genes that were significantly correlated with these 5lncRNAs are enriched for metabolic processes, fibrinolysis and complement activation. KEGG pathway analysis revealed that these genes are enriched in pathways related to metabolism. These results suggest that the 5 lncRNAs may participate in the initiation and progression of HCC through these pathways. However, further studies are needed to investigate and validate the functions of these 5 lncRNAs.
In conclusion, our present study developed a 5-lncRNA signature for predicting the prognosis of HCC patients. The signature was reproducible and robust in a second independent large-scale HCC cohort, supporting its value and effectiveness. In addition, the prognostic value of the 5-lncRNA signature was independent of clinicopathological variables. Our study indicates that the 5-lncRNA signature could improve survival prediction and could be used as a prognostic biomarker for HCC patients.
Hepatocellular carcinoma (HCC) is the sixth most commonly diagnosed cancer in the world. Although treatment for HCC, including surgical resection, has improved over the past decades, its overall survival rate remains devastatingly high due to its high rate of recurrence. Because HCC is a heterogeneous disease with substantially variable clinical outcomes, the search for effective biomarkers to predict recurrence and prognosis is crucial.
Recent studies have demonstrated the importance of long non-coding RNAs (lncRNAs) in physiological and pathological cellular processes. Increasing evidence suggests that lncRNA dysregulation is associated with various human diseases, particularly the initiation and progression of various human cancers. For patients with HCC, most of the existing prognostic signatures have focused on mRNAs or microRNAs, and only a few lncRNA signatures have been developed. In the present study, we aimed to construct a lncRNA signature for the prediction of HCC prognosis with high efficiency.
To construct a lncRNA signature for the prediction of HCC prognosis with high efficiency.
Differentially expressed lncRNAs (DELs) between HCC specimens and peritumor liver specimens were acquired from the The Cancer Genome Atlas (TCGA) LIHC dataset using the edgeR package. Univariate Cox proportional hazards regression was performed to identify the DELs that were significantly associated with overall survival for the training set. The stepwise multivariate Cox regression model was applied. Those lncRNAs fitted in the multivariate Cox regression model and independently associated with overall survival were chosen to build a prognostic risk formula. The prognostic value of this formula was validated in the test group and the full cohort and further compared with two previously developed prognostic signatures for HCC.
We identified a five-lncRNA prognostic signature from the TCGA dataset and determined that its prognostic value was independent from clinicopathological factors. The signature was reproducible and robust in another independent large-scale HCC cohort, supporting its utility and effectiveness.
This study constructed a 5-lncRNA signature that improves survival prediction, and can be used as a prognostic biomarker for HCC patients.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Namisaki T, Tsui SK S- Editor: Wang JL L- Editor: Filipodia E- Editor: Huang Y
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20515] [Article Influence: 2051.5] [Reference Citation Analysis (20)] |
| 2. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21370] [Article Influence: 2137.0] [Reference Citation Analysis (3)] |
| 3. | Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15 Suppl 4:5-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 737] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 4. | Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1738] [Cited by in RCA: 1735] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 5. | Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2-S6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 880] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 6. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4267] [Article Influence: 237.1] [Reference Citation Analysis (2)] |
| 7. | Joliat GR, Allemann P, Labgaa I, Demartines N, Halkic N. Treatment and outcomes of recurrent hepatocellular carcinomas. Langenbecks Arch Surg. 2017;402:737-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 1107] [Article Influence: 100.6] [Reference Citation Analysis (1)] |
| 9. | Zheng J, Kuk D, Gönen M, Balachandran VP, Kingham TP, Allen PJ, D’Angelica MI, Jarnagin WR, DeMatteo RP. Actual 10-Year Survivors After Resection of Hepatocellular Carcinoma. Ann Surg Oncol. 2017;24:1358-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 10. | Kudo M. Surveillance, diagnosis, treatment, and outcome of liver cancer in Japan. Liver Cancer. 2015;4:39-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 146] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 11. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6573] [Article Influence: 469.5] [Reference Citation Analysis (1)] |
| 12. | Schmitt AM, Chang HY. Long Noncoding RNAs: At the Intersection of Cancer and Chromatin Biology. Cold Spring Harb Perspect Med. 2017;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 13. | Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 516] [Cited by in RCA: 572] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 14. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4442] [Article Influence: 277.6] [Reference Citation Analysis (0)] |
| 15. | Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391-6400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 463] [Cited by in RCA: 525] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 16. | Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655-4669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 290] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 17. | Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1127] [Cited by in RCA: 1340] [Article Influence: 95.7] [Reference Citation Analysis (0)] |
| 18. | Shi D, Qu Q, Chang Q, Wang Y, Gui Y, Dong D. A five-long non-coding RNA signature to improve prognosis prediction of clear cell renal cell carcinoma. Oncotarget. 2017;8:58699-58708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J. An Immune-Related Six-lncRNA Signature to Improve Prognosis Prediction of Glioblastoma Multiforme. Mol Neurobiol. 2018;55:3684-3697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 20. | Zeng JH, Liang L, He RQ, Tang RX, Cai XY, Chen JQ, Luo DZ, Chen G. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget. 2017;8:16811-16828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 21. | Sun J, Cheng L, Shi H, Zhang Z, Zhao H, Wang Z, Zhou M. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci Rep. 2016;6:27842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 22. | Lin Z, Xu SH, Wang HQ, Cai YJ, Ying L, Song M, Wang YQ, Du SJ, Shi KQ, Zhou MT. Prognostic value of DNA repair based stratification of hepatocellular carcinoma. Sci Rep. 2016;6:25999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 23. | Lu M, Kong X, Wang H, Huang G, Ye C, He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:8775-8784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 24. | Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 25. | Li B, Feng W, Luo O, Xu T, Cao Y, Wu H, Yu D, Ding Y. Development and Validation of a Three-gene Prognostic Signature for Patients with Hepatocellular Carcinoma. Sci Rep. 2017;7:5517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 26. | Wang Z, Wu Q, Feng S, Zhao Y, Tao C. Identification of four prognostic LncRNAs for survival prediction of patients with hepatocellular carcinoma. PeerJ. 2017;5:e3575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 27. | Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10350] [Cited by in RCA: 11170] [Article Influence: 657.1] [Reference Citation Analysis (0)] |
| 28. | Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24735] [Cited by in RCA: 27978] [Article Influence: 1748.6] [Reference Citation Analysis (0)] |
| 29. | Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1397] [Cited by in RCA: 1682] [Article Influence: 112.1] [Reference Citation Analysis (0)] |
| 30. | Chen JA, Conn S. Canonical mRNA is the exception, rather than the rule. Genome Biol. 2017;18:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 823] [Cited by in RCA: 955] [Article Influence: 79.6] [Reference Citation Analysis (0)] |
| 32. | Bi M, Yu H, Huang B, Tang C. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in gastric cancer cells through regulating CDKN1A. Gene. 2017;626:337-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 33. | Hua F, Liu S, Zhu L, Ma N, Jiang S, Yang J. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/β-catenin signaling pathway in cervical cancer. Biomed Pharmacother. 2017;92:1128-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 34. | Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16:129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 35. | Spurlock CF 3rd, Shaginurova G, Tossberg JT, Hester JD, Chapman N, Guo Y, Crooke PS 3rd, Aune TM. Profiles of Long Noncoding RNAs in Human Naive and Memory T Cells. J Immunol. 2017;199:547-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 36. | Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, Hu Z, Yu T, Ding J, Li J. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67:171-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 174] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 37. | Lu S, Zhou J, Sun Y, Li N, Miao M, Jiao B, Chen H. The noncoding RNA HOXD-AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Mol Cancer. 2017;16:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 38. | Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, Dou C, Xu M, Liu Q, Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 201] [Article Influence: 25.1] [Reference Citation Analysis (0)] |