Published online Jul 21, 2018. doi: 10.3748/wjg.v24.i27.3021
Peer-review started: April 28, 2018
First decision: June 11, 2018
Revised: June 12, 2018
Accepted: June 27, 2018
Article in press: June 27, 2018
Published online: July 21, 2018
Processing time: 82 Days and 1.2 Hours
To assess the diagnostic accuracy of a new fecal test for detecting Helicobacter pylori (H. pylori), using13C-urea breath test as the reference standard, and explore bacterial antibiotic resistance.
We conducted a prospective two-center diagnostic test accuracy study. We enrolled consecutive people≥ 18 years without previous diagnosis of H. pylori infection, referred for dyspepsia between February and October 2017. At enrollment, all participants underwent 13C-urea breath test. Participants aged over 50 years were scheduled to undergo upper endoscopy with histology. Participants collected stool samples 1-3 d after enrollment for a new fecal investigation (THD fecal test). The detection of bacterial 23S rRNA subunit gene indicated H. pylori infection. We also used the index diagnostic test to examine mutations conferring resistance to clarithromycin and levofloxacin. Independent investigators analyzed index test and reference test standard results blinded to the other test findings. We estimated sensitivity, specificity, positive (PPV) and negative (NPV) predictive value, diagnostic accuracy, positive and negative likelihood ratio (LR), together with 95% confidence intervals (CI).
We enrolled 294 consecutive participants (age: Median 37.0 years, IQR: 29.0-46.0 years; men: 39.8%). Ninety-five (32.3%) participants had a positive13C-urea breath test. Twenty-three (7.8%) participants underwent upper endoscopy with histology, with a full concordance between 13C-urea breath test and histology in detecting H. pylori infection. Four (1.4%) out of the 294 participants withdrew from the study after the enrollment visit and did not undergo THD fecal testing. In the 290 participants who completed the study, the THD fecal test sensitivity was 90.2% (CI: 84.2%-96.3%), specificity 98.5% (CI:96.8%-100%), PPV 96.5% (CI: 92.6%-100%), NPV 95.6% (CI: 92.8%-98.4%), accuracy 95.9% (CI: 93.6%-98.2%), positive LR 59.5(CI: 19.3-183.4), negative LR 0.10 (CI: 0.05-0.18). Out of 83 infected participants identified with the THD fecal test, 34 (41.0%) had bacterial genotypic changes consistent with antibiotic-resistant H. pylori infection. Of these, 27 (32.5%) had bacterial strains resistant to clarithromycin, 3 (3.6%) to levofloxacin, and 4 (4.8%) to both antibiotics.
The THD fecal test has high performance for the non-invasive diagnosis of H. pylori infection while additionally enabling the assessment of bacterial antibiotic resistances.
Core tip: Existing studies on molecular tests for Helicobacter pylori (H. pylori) detection in stools show suboptimal quality. The THD fecal test is a newer method to detect bacterial DNA and mutations conferring antibiotic resistance. In this diagnostic test accuracy study involving unselected consecutive participants and blinded outcome assessment, we evaluated the diagnostic accuracy of the THD fecal test for detecting H. pylori, using the 13C-urea breath test as the reference standard. We found that the THD fecal test has high performance for the non-invasive diagnosis of H. pylori infection while additionally enabling the assessment of bacterial antibiotic resistances.
- Citation: Iannone A, Giorgio F, Russo F, Riezzo G, Girardi B, Pricci M, Palmer SC, Barone M, Principi M, Strippoli GF, Di Leo A, Ierardi E. New fecal test for non-invasive Helicobacter pylori detection: A diagnostic accuracy study. World J Gastroenterol 2018; 24(27): 3021-3029
- URL: https://www.wjgnet.com/1007-9327/full/v24/i27/3021.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i27.3021
Helicobacter pylori (H. pylori) infection occurs among 48.5% of the general population worldwide, with high geographic variability[1]. It is the leading cause of chronic/atrophic gastritis, peptic ulcer, gastric lymphoma, gastric carcinoma, and some extra-gastric disorders[2-4].Diagnostic approaches for H. pylori infection include invasive and non-invasive testing[4-6]. The 13C-urea breath test is established as a highly sensitive and specific test (96% sensitivity and 93% specificity) for the non-invasive diagnosis ofinfection[4,7]. A stool-based monoclonal antigen test has low acceptability in some contexts and needs local validation, despite high accuracy[4,8]. Invasive tests require upper endoscopy, limiting their application based on local practices and policies and to patients who have alarm symptoms[4,5,9]. Culture with antibiogram is only recommended after repeated treatment failures[4-6] due to its high false negative rate[10,11].
Molecular testing is a promising approach for diagnosing H. pylori infection and has the added advantage of identifying bacterial DNA mutations associated with antibiotic resistance. Molecular tests on gastric biopsy are commonly used only for research purposes due to the need for an invasive endoscopic procedure[12].Thus, the application of these tests on fecal samples is gaining interest. A recent meta-analysis identified the bacterial 23S ribosomal RNA subunit gene as the most accurate marker for diagnosis of infection using molecular tests on stool samples, with 82% (95%CI: 77%-86%) sensitivity and 99%(95%CI: 98%-100%) specificity[13]. The consistency of these results is limited by the inclusion of studies of suboptimal quality resulting from bias in participant selection and lack of blinded outcome assessment[14-19].
We aimed to assess the diagnostic accuracy of a new molecular test, theTHD fecal test[20], for the non-invasive detection of H. pylori DNA, using the 13C-urea breath test as the reference standard. We estimated the point prevalence of H. pylori DNA point mutations conferring resistance to clarithromycin and levofloxacin.
This study was conducted according to the Standards for Reporting of Diagnostic Accuracy (STARD) statement[21].
We performed a two-center cross-sectional study with prospective data collection. We included consecutive participantsexperiencing dyspeptic symptoms and without previous diagnosis of H. pylori infection, who were referred for diagnostic evaluation to the Gastroenterology Unit, University of Bari (Italy) or the National Institute of Gastroenterology “Saverio De Bellis”, CastellanaGrotte, Bari (Italy) between February and October 2017.
Participants were eligible if they were aged 18 years or older and had experienced dyspeptic symptoms, defined as the presence of one or more of: post-prandial fullness, early satiation, epigastric pain and epigastric burning for at least one (post-prandial fullness and early satiation) or three (epigastric pain and epigastric burning) days per week in the last three months with symptoms onset at least six months previously[22]. Exclusion criteria were treatment with proton pump inhibitors or 2-histamine receptor antagonists in the previous two weeks as well as use of antibiotics or bismuth salts in the previous four weeks, as these medications may increase false negative results of invasive and non-invasive current diagnostic tests for H. pylori infection by reducing the bacterial load[23-25]. Additional exclusion criteria were previous diagnosis of H. pylori infection and presence of chronic diarrhea, which can limit the accurate collection of stool samples for the THD fecal test. Potential participants were also excluded if they had alarm symptoms, including weight loss, dysphagia, gastrointestinal bleeding, an abdominal mass or iron deficiency anemia, which are an indication to perform upper endoscopy as a first-line diagnostic approach[4,5,9].
At the enrollment visit, eligible participants underwent the 13C-urea breath test (the reference standard) for the non-invasive investigation of H. pylori infection. According to current Italian Clinical guideline recommendations[5], participants older than 50 years were scheduled to undergo upper endoscopy with biopsy sampling for histology within one week. All participants were asked to provide stool samples collected 1 to 3 d after enrollment, using the THD device. These samples were used for the THD fecal test (index test). Independent investigators analyzed the index test and reference standard test results blinded to the other test findings, participants’ information and histology results. Pathologists performing histology examination were unaware of the results of the other two tests.
Thestudy was performed in agreement with the ethical guidelines of the Declaration of Helsinki and the protocol was approved by the local Ethics Committee (Ospedale Consorziale Policlinico, Bari, protocol number 74413). All participants gave written informed consent before inclusion in the study.
The technical details of H. pylori DNA extraction and analysis are reported in Appendix 1.
Within three days after the enrollment visit, participants collected and stored a stool sample using the THD fecal test equipment (THD Spa, Correggio, Reggio Emilia, Italy), which allows for obtaining an adequate stool-derived product to extract H. pylori DNA.
We pre-specified THD fecal test positivity as the identification of the H. pylori bacterial gene encoding the 23S ribosomal RNA subunit in the stool-derived product[20]. The detection of specific bacterial DNA point mutations indicated H. pylori resistance to clarithromycin and/or levofloxacin. In brief, we assessed A2142C, A2142G and A2143G point mutations in the 23S rRNA subunit gene for clarithromycin resistance, and C261A, C261G, G271A, A272G, G271T and A270T point mutations in the A-subunit of gyrase gene for levofloxacin resistance.
The technical details of the 13C-urea breath test are reported in Appendix 2.
At the enrollment visit, all participants underwent 13C-urea breath testing after overnight fasting. We used this test as the reference standard due to the non-invasiveness, high diagnostic accuracy (sensitivity of 96% and specificity of 93%), and consumer acceptance[4,7]. We pre-specified a 13C-urea breath test positivity for a difference between the baseline and 30 min breath sample that exceeded 4 parts per 1000 of 13carbon dioxide (13CO2)[26,27].
Participants older than 50 years were scheduled to undergo upper endoscopy with biopsy sampling for histology, according to current Italian guidelines[5]. Among these participants, a minimum of two biopsy samples from the gastric antrum (greater and lesser curvature, 3 cm proximal to the pyloric region) and two from the middle of the gastric body were collected for histologic examination[4].
We assessed the normal distribution of continuous variables using the Shapiro-Wilk test and expressed them as the mean and standard deviation (SD) or median and interquartile range (IQR). We expressed categorical variables asa percentage.
The results of the 13C-urea breath test served as the reference standard for assessing the diagnostic accuracy of the THD fecal test in detecting H. pylori infection. We calculated sensitivity, specificity, negative and positive predictive values, and diagnostic accuracy for the THD fecal test together with 95% confidence intervals (CI), according to standard definitions. Since the prevalence of the condition in the enrolled population influences sensitivity, specificity, and predictive values, we also calculated the positive and negative likelihood ratios. To identify the impact of our findings on clinical decision-making, we calculated the post-test probability after positive and negative results onthe THD fecal test for populations with different pre-test probabilities of H. pylori infection based on likelihood ratios.
We pre-planned to handle uninterpretable and missing index test or reference standard findings with both the “complete case” approach and “best-worst case” imputation, to avoid overestimation of diagnostic accuracy parameters[28]. In detail, these results were removed for the “complete case” analysis, while they were considered as false-negatives and false-positivesfor the “worst case” analysis or as true-negatives and true-positives for the “best-case” analysis.
We estimatedthe point prevalence of H. pylori resistance to clarithromycin and levofloxacinas the number of participant with the specific antibiotic resistance divided by the total number of participants diagnosed with H. pylori infection at THD fecal test.
We assumed a 94% sensitivity and 97% specificity for the THD fecal test, based on monoclonal stool antigen test accuracy parameters for the lack of high-quality evidence on molecular fecal tests[8]. Assuming a H. pylori infection prevalence of 34.3%[1], a marginal error of 0.05, and a drop-out of 10%, we calculated a sample size of at least 280 participants, to provide 80% power with an α of 0.05 to detect the pre-specified sensitivity and specificity values for the THD fecal test[29].
We used Statistical Analysis Software (SAS Institute Inc., Cary, NC, United States) 9.4 for all the analyses.
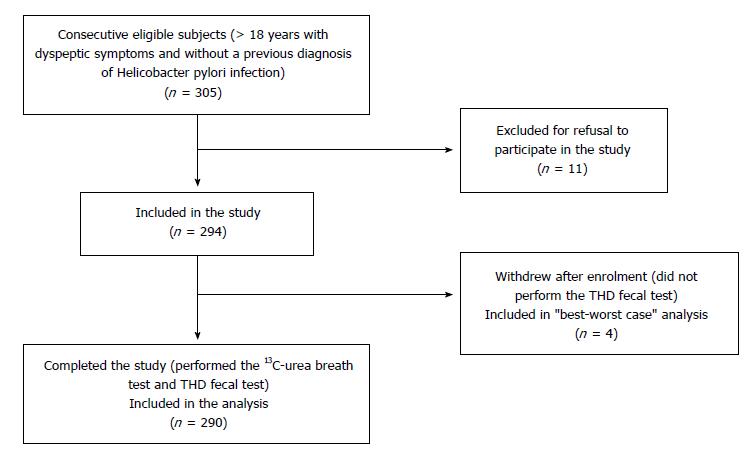
During the enrollment period, 305 consecutive participants were eligible for inclusion in the study. Of these, 11 were excluded for preference not to participate.Four out of the 294 participants withdrew from the study after the enrollment visit did not undergo THD fecal testing (Figure 1).
Table 1 describes the characteristics of the 294 participants included in the study. The median age was 37.0 years (IQR: 29.0 to 46.0 years). There were 177 (60.2%) women. The 13C-urea breath test (reference standard test) was positive in 95 (32.3%) participants. Forty (13.6%) participants were older than 50 years and were scheduled to undergo upper endoscopy with histology. Of these, 23 (7.8%) participants, 7 with and 16 without H. pylori infection, agreed to undergo this examination. There was full concordance between the 13C-urea breath test and histology in detecting H. pylori infection in these participants. With regard to dyspeptic symptoms reported at enrollment visit, 115 (39.1%) participants experienced post-prandial fullness, 50 (17.0%) early satiation, 130 (44.2%) epigastric pain, and 170 (57.8%) epigastric burning. There were no reported adverse events during performance of the index or reference standard test.
| Characteristic | Value (n= 294) |
| Age, median (IQR), yr | 37.0 (29.0-46.0) |
| Female sex, number | 177 (60.2) |
| Helicobacter pylori infection, number | 95 (32.3)1 |
| Upper endoscopy with histology, number | 23 (7.8)2 |
| Dyspeptic symptoms, number | |
| Post-prandial fullness | 115 (39.1) |
| Early satiation | 50 (17.0) |
| Epigastric pain | 130 (44.2) |
| Epigastric burning | 170 (57.8) |
| Concomitant diseases, number | |
| Cardiovascular disease | 8 (2.7) |
| Chronic kidney disease/dialysis | 0 (0) |
| Chronic liver disease/cirrhosis | 3 (1.0) |
| Other chronic diseases3 | 10 (3.4) |
The amplification curves of H. pylori DNA sequences from the real time polymerase chain reaction are shown in Appendix 3.
Direct comparisons between the 13C-urea breath test and THD fecal test results for detecting H. pylori infection are reported in Table 2.
The diagnostic accuracy parameters of the THD fecal test for detecting H. pylori infection, using 13C-urea breath test as the reference standard, are shown in Table 3. In the “complete-case” analysis, including the 290 participants who completed the study, the THD fecal test had a sensitivity of 90.2% (CI: 84.2% to 96.3%), specificity of 98.5% (CI: 96.8% to 100%), positive predictive value of 96.5% (CI: 92.6% to 100%), and negative predictive value of 95.6% (CI: 92.8% to 98.4%). The accuracy of the index test in correctly identifying participants with and without H. pylori infection was 95.9% (CI: 93.6% to 98.2%). The THD fecal test positive likelihood ratio was 59.5 (CI: 19.3 to 183.4), and the negative likelihood ratio was 0.10 (CI: 0.05 to 0.18). In the “best-worst case” analysis, including all 294 participants enrolled in the study, there were small changes in THD fecal test diagnostic accuracy parameters (Table 3).
| Parameter | Complete-case analysis | Best-case analysis | Worst-case analysis |
| (n = 290) | (n = 294)1 | (n = 294)1 | |
| Sensitivity, % (95%CI) | 90.2 (84.2 to 96.3) | 90.5 (84.6 to 96.4) | 87.4 (80.7 to 94.1) |
| Specificity, % (95%CI) | 98.5 (96.8 to 100) | 98.5 (96.8 to 100) | 98.0 (96.0 to 99.9) |
| PPV, % (95%CI) | 96.5 (92.6 to 100) | 96.6 (92.9 to 100) | 95.4 (91.0 to 99.8) |
| NPV, % (95%CI) | 95.6 (92.8 to 98.4) | 95.6 (92.8 to 98.4) | 94.2 (91.0 to 97.4) |
| Accuracy, % (95%CI) | 95.9 (93.6 to 98.2) | 95.9 (93.7 to 98.2) | 94.6 (92.0 to 97.2) |
| Positive LR, estimate (95%CI) | 59.5 (19.3 to 183.4) | 60.0 (19.5 to 185.0) | 43.5 (16.4 to 115.0) |
| Negative LR, estimate (95%CI) | 0.10 (0.05 to 0.18) | 0.10 (0.05 to 0.18) | 0.13 (0.08 to 0.22) |
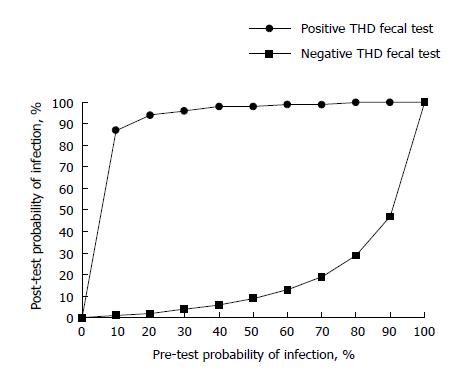
Figure 2 displays the estimation of the probability of H. pylori infection after positive and negative THD fecal test results for populations with different prevalence of disease, based on likelihood ratios calculated from our data. As shown, the probability of infection after a positive index test finding is higher than 90% in populations with a disease prevalence ≥ 20%. The probability of infection after a negative index test finding is lower than 10% and 20% in populations with disease prevalence ≤ 50% and ≤ 70%, respectively.
Table 4 summarizes the H. pylori resistance rates to clarithromycin and levofloxacin in the 83 H. pylori infected participants identified with the THD fecal test. Thirty-four (41.0%) participants had bacterial genotypic changes consistent with antibiotic-resistant H. pylori infection. Of these, 27 (32.5%) had bacterial strains resistant to clarithromycin, 3 (3.6%) to levofloxacin, and 4 (4.8%) to both antibiotics. The overall resistance rates were 37.3% (31 participants) to clarithromycin and 8.4% (7 participants) to levofloxacin.
| Clarithromycin | Total | ||
| Susceptible | Resistant | ||
| Levofloxacin | |||
| Susceptible | 49 (59.1) | 27 (32.5) | 76 (91.6) |
| Resistant | 3 (3.6) | 4 (4.8) | 7 (8.4) |
| Total | 52 (62.7) | 31 (37.3) | 83 (100) |
In this diagnostic test accuracy study involving unselected consecutive participants and blinded outcome assessment, we observed that the non-invasive THD fecal test has high performance for the diagnosis of H. pylori infection among patients with dyspeptic symptoms. This test showed a 90.2% sensitivity and 98.5% specificity, with a diagnostic accuracy of 95.9%. Considering the worldwide prevalence of H. pylori infection, ranging from 70% in Africa to 24% in Oceania[1], there was > 90% post-test probability of bacterial infection after a positive THD fecal test result and < 20% probability after a negative finding. In Western Europe, the prevalence of infection is around 34%[1], leading to post-test probabilities > 95% and < 5% after a positive and negative test, respectively. The THD fecal test identified H. pylori resistance rates of 32.5% to clarithromycin, 3.6% to levofloxacin, and 4.8% to both antibiotics.
A recent meta-analysis found that the 23S rRNA subunit gene is the most accurate marker for detecting H. pylori in stools using molecular analyses based on real time polymerase chain reaction. The pooled analysis of six diagnostic accuracy studies showed estimated sensitivity of 82% (95%CI: 77% to 86%) and specificity of 99% (95%CI: 98% to 100%) for the bacterial 23S rRNA subunit gene[13]. Comparing our results with findings from existing primary studies[14-19] included in this meta-analysis is difficult, mainly due to limitations in methodological reporting in these studies and differences in the selection of population and the reference standard used. Four studies[14,15,17,19] included children (age < 18 years), one[18] did not specify the participants’ age, and one[16] included adults (range 20 to 81 years), with an estimated disease prevalence ranging between 21% and 81% among studies. The reference standard was monoclonal stool antigen test in three studies[16-18], a combination of histology, rapid urease test, and culture in two studies[14,19], and a combination of histology, culture, 13C-urea breath test and monoclonal stool antigen test in one study[15]. In two studies[14,19], consecutive participants were enrolled, while in the remaining four[15-18], a convenience sample was selected. No authors provided the sample size estimation. There was no reported blinding of the assessment of the index and reference standard test findings, except for pathologists analyzing gastric biopsy samples in one study[15]. Thus, the use of convenience samples and lack of blinding may have led to an overestimation of the diagnostic accuracy of these molecular tests for the non-invasive diagnosis of H. pylori infection. Despite the selection of consecutive participants and the blinded assessment of the index and reference standard results in our study, we found greater test sensitivity and similar high specificity compared to the pooled estimate of earlier studies. This may be due to the high performance of the THD device for obtaining adequate stool sample-derived product to extract H. pylori DNA. Our sensitivity estimate is concordant with that reported in a recent publication[30], although this last study is burdened by the same methodological limitations and risk of bias as previous researches.
Our H. pylori resistance rates to clarithromycin and levofloxacinwere consistent with those reported in previous epidemiologic studies from the same geographic area[31]. Molecular analyses performed on stool samples showed bacterial resistance rates of 36%-41% to clarithromycin[16,30], in agreement with our estimate of 37.3%. There is limited evidence on the use of molecular fecal tests to detect H. pylori resistance to levofloxacin. Recently, the molecular analysis “Genotype HelicoDR assay” (Hain Lifescience GmbH, Nehren, Germany) has been applied to fecal samples for detecting clarithromycin and fluoroquinolone resistance using molecular analysis on gastric biopsy, with proven high accuracy, as the reference standard[32]. This test identified a resistance rate to fluoroquinolone of 13% in stools, which was somewhat higher than the present study (8.4%). However, there was low agreement between stool and biopsy findings for resistance to both clarithromycin (53%) and fluoroquinolone (35%), indicating a poor performance of this test for the non-invasive assessment of H. pylori resistances to antibiotics. By contrast, the THD fecal test has shown high concordance with real time polymerase chain reaction-based molecular analysis on gastric tissue for detecting bacterial resistance to clarithromycin[20].
This study investigated the diagnostic accuracy parameters of a novel molecular tool (THD fecal test) for the non-invasive diagnosis of H. pylori infection in consecutive participants with dyspepsia. The strengths of the study include an a priori sample size, prospectively enrolled consecutive participants, and blinded assessment of the index and reference standard results to increase the certainty of the findings. Our study also assessed the feasibility of detecting bacterial resistance to clarithromycin and levofloxacin, using a non-invasive approach.
Our study has some limitations that should be considered when interpreting the results. First, we diagnosed H. pylori infection using a single non-invasive test, while a second confirmation test (histology) was performed only in the subgroup of participants older than 50 years. However, the 13C-urea breath test is recommended as the gold-standard non-invasive diagnostic approach for detecting this bacterial infection, with 96% sensitivity and 93% specificity[4,7]. We also found complete agreement between 13C-urea breath test and histology in participants older than 50 years. Moreover, current international guidelines[4,5,9] recommend a test-and-treat strategy for H. pylori infection in young (≤ 50 years in Italy) dyspeptic people without alarm symptoms to avoid the costs, inconvenience and discomfort of endoscopy. Thus, our approach reflects the current clinical practice and the most appropriate diagnostic strategy for this population. Second, we did not perform molecular analysis of bacterium resistance to clarithromycin and levofloxacin on gastric biopsy samples to confirm the results obtained on stool samples. Thus, we did not calculate diagnostic accuracy parameters of the THD fecal test for detecting the bacterial resistance to these antibiotics. However, we have previously demonstrated full concordance between THD fecal test and molecular analysis on gastric tissue findings for detecting H. pylori resistance to clarithromycin[20].
In conclusion, our results indicate that the THD fecal test has high diagnostic performance for non-invasive detection of H. pylori infection in patients with dyspeptic symptoms while enabling identification of bacterium resistance to clarithromycin and levofloxacin. The THD fecal test may assist in the conduct of randomized trials to evaluate the benefits and harms of tailored eradication strategies in first-line. On these bases, the THD fecal test may inform clinical decision-making and guide individualized treatments for H. pylori infection.
Diagnostic approaches for Helicobacter pylori (H. pylori) infection include invasive and non-invasive testing. The non-invasive 13C-urea breath test and stool monoclonal antigen test have high accuracy for diagnosing the infection, although the stool test has low acceptability in some contexts and needs local validation. The need for upper endoscopy is a limitation to the use of invasive tests. Molecular tests are promising approaches for diagnosing H. pylori infection, due to the added advantage of identifying bacterial DNA mutations associated with antibiotic resistance.
The application of molecular diagnostic tests on gastric biopsy samples is limited by the need for invasive endoscopic procedure. Thus, the non-invasive application of these tests on fecal samples is gaining increasing interest. An accurate non-invasive molecular test may guide first-line eradicating treatments with the potential advantages of increasing bacterial eradication rates and reducing the development of H. pylori resistance to antibiotics. However, existing studies on molecular tests for H. pylori detection in stools show suboptimal quality.
We aimed to assess the accuracy of a new non-invasive molecular test, the THD fecal test, for the diagnosis of H. pylori infection, using 13C-urea breath test as the reference standard. Additionally, we estimated the point prevalence of H. pylori DNA mutations conferring resistance to clarithromycin and levofloxacin.
We conducted a prospective two-center diagnostic test accuracy study. We enrolled consecutive people ≥ 18 years old without previous diagnosis of H. pylori infection, referred for dyspepsia between February and October 2017. At enrollment, all participants underwent 13C-urea breath test. Participants aged over 50 years were scheduled to undergo upper endoscopy with histology. Participants collected stool samples 1-3 d after enrollment for the THD fecal test. The detection of bacterial 23S rRNA subunit gene indicated H. pylori infection. We also used the index diagnostic test to examine mutations conferring resistance to clarithromycin and levofloxacin. Independent investigators analyzed the index test and reference standard test results blinded to the other test findings, participants’ information and histology results. We estimated diagnostic accuracy parameters, together with their 95% confidence intervals. The novelty of our research methods included an a priori sample size, a prospective enrollment of consecutive participants, and the blindingof outcome assessors. This approach increased the certainty of our findings.
Out of 294 participants, 95 (32.3%) had a positive 13C-urea breath test. Four (1.4%) participants withdrew from the study after the enrollment visit. In the 290 participants who completed the study, the THD fecal test sensitivity was 90.2% (CI: 84.2%-96.3%), specificity 98.5% (CI: 96.8%-100%), positive predictive value 96.5% (CI: 92.6%-100%), negative predictive value 95.6% (CI: 92.8%-98.4%), accuracy 95.9% (CI: 93.6%-98.2%), positive likelihood ratio 59.5 (CI: 19.3-183.4), negative likelihood ratio 0.10 (CI: 0.05-0.18). Out of 83 H. pylori infected participants identified with the THD fecal test, 27 (32.5%) had bacterial strains resistant to clarithromycin, 3 (3.6%) to levofloxacin, and 4 (4.8%) to both antibiotics.
Our results indicate that the THD fecal test has high diagnostic accuracy for the non-invasive diagnosis of H. pylori infection in patients with dyspeptic symptoms, while enabling identification of bacterium resistance to clarithromycin and levofloxacin. The certainty of our findings is based on the rigorous methodological approach used in the assessment of the THD fecal test diagnostic performance. THD fecal testing may inform clinical decision-making and guide individualized therapies to eradicate H. pylori infection.
The spread of H. pylori resistance to antibiotics has prompted the investigation of the efficacy of antibiotic susceptibility-guided therapies. THD fecal testing may assist in the conduct of randomized trials to evaluate the benefits and harms of tailored eradication strategies in first-line.
THD Spa, Correggio (Italy), for providing free of charge THD fecal test for all participants included in the study.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report classification:
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Buzas G, Day AS, Ulaşoğlu C S- Editor: Wang XJ L- Editor: Filipodia E- Editor: Huang Y
| 1. | Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153:420-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1361] [Cited by in RCA: 2048] [Article Influence: 256.0] [Reference Citation Analysis (0)] |
| 2. | Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1848] [Cited by in RCA: 1907] [Article Influence: 82.9] [Reference Citation Analysis (3)] |
| 3. | Graham DY, Lew GM, Klein PD, Evans DG, Evans DJ Jr, Saeed ZA, Malaty HM. Effect of treatment of Helicobacter pylori infection on the long-term recurrence of gastric or duodenal ulcer. A randomized, controlled study. Ann Intern Med. 1992;116:705-708. [PubMed] |
| 4. | Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2220] [Cited by in RCA: 1983] [Article Influence: 247.9] [Reference Citation Analysis (1)] |
| 5. | Zagari RM, Romano M, Ojetti V, Stockbrugger R, Gullini S, Annibale B, Farinati F, Ierardi E, Maconi G, Rugge M. Guidelines for the management of Helicobacter pylori infection in Italy: The III Working Group Consensus Report 2015. Dig Liver Dis. 2015;47:903-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 6. | Fallone CA, Chiba N, van Zanten SV, Fischbach L, Gisbert JP, Hunt RH, Jones NL, Render C, Leontiadis GI, Moayyedi P. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology. 2016;151:51-69.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 695] [Cited by in RCA: 634] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 7. | Ferwana M, Abdulmajeed I, Alhajiahmed A, Madani W, Firwana B, Hasan R, Altayar O, Limburg PJ, Murad MH, Knawy B. Accuracy of urea breath test in Helicobacter pylori infection: meta-analysis. World J Gastroenterol. 2015;21:1305-1314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 124] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (4)] |
| 8. | Gisbert JP, de la Morena F, Abraira V. Accuracy of monoclonal stool antigen test for the diagnosis of H. pylori infection: a systematic review and meta-analysis. Am J Gastroenterol. 2006;101:1921-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 188] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 9. | Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am J Gastroenterol. 2017;112:212-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 744] [Cited by in RCA: 1018] [Article Influence: 127.3] [Reference Citation Analysis (1)] |
| 10. | Ierardi E, Giorgio F, Iannone A, Losurdo G, Principi M, Barone M, Pisani A, Di Leo A. Noninvasive molecular analysis of Helicobacter pylori: Is it time for tailored first-line therapy? World J Gastroenterol. 2017;23:2453-2458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Kjøller M, Fischer A, Justesen T. Transport conditions and number of biopsies necessary for culture of Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1991;10:166-167. [PubMed] |
| 12. | De Francesco V, Margiotta M, Zullo A, Hassan C, Troiani L, Burattini O, Stella F, Di Leo A, Russo F, Marangi S. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. Ann Intern Med. 2006;144:94-100. [PubMed] |
| 13. | Khadangi F, Yassi M, Kerachian MA. Review: Diagnostic accuracy of PCR-based detection tests for Helicobacter Pylori in stool samples. Helicobacter. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 14. | Vécsei A, Innerhofer A, Binder C, Gizci H, Hammer K, Bruckdorfer A, Riedl S, Gadner H, Hirschl AM, Makristathis A. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin Gastroenterol Hepatol. 2010;8:309-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 15. | Lottspeich C, Schwarzer A, Panthel K, Koletzko S, Rüssmann H. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J Clin Microbiol. 2007;45:1718-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Noguchi N, Rimbara E, Kato A, Tanaka A, Tokunaga K, Kawai T, Takahashi S, Sasatsu M. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J Med Microbiol. 2007;56:1174-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Booka M, Okuda M, Shin K, Miyashiro E, Hayashi H, Yamauchi K, Tamura Y, Yoshikawa N. Polymerase chain reaction--restriction fragment length polymorphism analysis of clarithromycin-resistant Helicobacter pylori infection in children using stool sample. Helicobacter. 2005;10:205-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Rimbara E, Noguchi N, Yamaguchi T, Narui K, Kawai T, Sasatsu M. Development of a highly sensitive method for detection of clarithromycin-resistant Helicobacter pylori from human feces. Curr Microbiol. 2005;51:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Scaletsky IC, Aranda KR, Garcia GT, Gonçalves ME, Cardoso SR, Iriya K, Silva NP. Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter. 2011;16:311-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Giorgio F, Ierardi E, Sorrentino C, Principi M, Barone M, Losurdo G, Iannone A, Giangaspero A, Monno R, Di Leo A. Helicobacter pylori DNA isolation in the stool: an essential pre-requisite for bacterial noninvasive molecular analysis. Scand J Gastroenterol. 2016;51:1429-1432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Lijmer JG, Moher D, Rennie D, de Vet HC. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1702] [Cited by in RCA: 2109] [Article Influence: 210.9] [Reference Citation Analysis (0)] |
| 22. | Stanghellini V, Chan FK, Hasler WL, Malagelada JR, Suzuki H, Tack J, Talley NJ. Gastroduodenal Disorders. Gastroenterology. 2016;150:1380-1392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 818] [Cited by in RCA: 973] [Article Influence: 108.1] [Reference Citation Analysis (0)] |
| 23. | Gatta L, Vakil N, Ricci C, Osborn JF, Tampieri A, Perna F, Miglioli M, Vaira D. Effect of proton pump inhibitors and antacid therapy on 13C urea breath tests and stool test for Helicobacter pylori infection. Am J Gastroenterol. 2004;99:823-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 95] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Graham DY, Opekun AR, Hammoud F, Yamaoka Y, Reddy R, Osato MS, El-Zimaity HM. Studies regarding the mechanism of false negative urea breath tests with proton pump inhibitors. Am J Gastroenterol. 2003;98:1005-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 105] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 25. | Malfertheiner P. Diagnostic methods for H. pylori infection: Choices, opportunities and pitfalls. United European Gastroenterol J. 2015;3:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Gisbert JP, Pajares JM. Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection -- a critical review. Aliment Pharmacol Ther. 2004;20:1001-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 257] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 27. | Li ZX, Huang LL, Liu C, Formichella L, Zhang Y, Wang YM, Zhang L, Ma JL, Liu WD, Ulm K. Cut-off optimization for 13 C-urea breath test in a community-based trial by mathematic, histology and serology approach. Sci Rep. 2017;7:2072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, Irwig L, Levine D, Reitsma JB, de Vet HC. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6:e012799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1355] [Cited by in RCA: 1571] [Article Influence: 174.6] [Reference Citation Analysis (0)] |
| 29. | Buderer NM. Statistical methodology: I. Incorporating the prevalence of disease into the sample size calculation for sensitivity and specificity. Acad Emerg Med. 1996;3:895-900. [PubMed] |
| 30. | Beckman E, Saracino I, Fiorini G, Clark C, Slepnev V, Patel D, Gomez C, Ponaka R, Elagin V, Vaira D. A Novel Stool PCR Test for Helicobacter pylori May Predict Clarithromycin Resistance and Eradication of Infection at a High Rate. J Clin Microbiol. 2017;55:2400-2405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 31. | Ierardi E, Giorgio F, Losurdo G, Di Leo A, Principi M. How antibiotic resistances could change Helicobacter pylori treatment: A matter of geography? World J Gastroenterol. 2013;19:8168-8180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 80] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 32. | Brennan DE, Omorogbe J, Hussey M, Tighe D, Holleran G, O’Morain C, Smith SM, McNamara D. Molecular detection of Helicobacter pylori antibiotic resistance in stool vs biopsy samples. World J Gastroenterol. 2016;22:9214-9221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (2)] |