Published online Jun 21, 2018. doi: 10.3748/wjg.v24.i23.2482
Peer-review started: March 23, 2018
First decision: April 10, 2018
Revised: April 17, 2018
Accepted: May 18, 2018
Article in press: May 18, 2018
Published online: June 21, 2018
Processing time: 84 Days and 19.2 Hours
To evaluate the association of 12 tag single nucleotide polymorphisms (tagSNPs) in three onco-long non-coding RNA (lncRNA) genes (HOTTIP, CCAT2, MALAT1) with the risk and prognosis of hepatocellular cancer (HCC).
Twelve tagSNPs covering the three onco-lncRNAs were genotyped by the KASP method in a total of 1338 samples, including 521 HCC patients and frequency-matched 817 controls. The samples were obtained from an unrelated Chinese population at the First Hospital of China Medical University from 2012-2015. The expression quantitative trait loci (eQTL) analyses were conducted to explore further the potential function of the promising SNPs.
Three SNPs in HOTTIP, one promoter SNP in MALAT1, and one haplotype of HOTTIP were associated with HCC risk. The HOTTIP rs17501292, rs2067087, and rs17427960 SNPs were increased to 1.55-, 1.20-, and 1.18-fold HCC risk under allelic models (P = 0.012, 0.017 and 0.049, respectively). MALAT1 rs4102217 SNP was increased to a 1.32-fold HCC risk under dominant models (P = 0.028). In addition, the two-way interaction of HOTTIP rs17501292-MALAT1 rs619586 polymorphisms showed a decreased effect on HCC risk (Pinteraction = 0.028, OR = 0.30) and epistasis with each other. HOTTIP rs3807598 variant genotype showed significantly longer survival time in HBV negative subgroup (P = 0.049, HR = 0.12), and MALAT1 rs591291 showed significantly better prognosis in female and HBV negative subgroups (P = 0.022, HR = 0.37; P = 0.042, HR = 0.25, respectively). In the study, no significant effect was observed in eQTL analysis.
Specific lncRNA (HOTTIP and MALAT1) SNPs have potential to be biomarkers for HCC risk and prognosis.
Core tip: We aim to evaluate the association of twelve tag single nucleotide polymorphisms (tagSNPs) in three onco-lncRNA genes (HOTTIP, CCAT2, MALAT1) with the risk and prognosis of hepatocellular cancer (HCC). Twelve tagSNPs covering the three onco-lncRNAs were genotyped by the KASP method in a total of 1338 samples. We found three SNPs in HOTTIP, one promoter SNP in MALAT1 and one haplotype of HOTTIP gene were associated with HCC risk. In addition, HOTTIP rs3807598 variant genotype showed significantly longer survival time in hepatitis B virus (HBV) negative subgroup, and MALAT1 rs591291 showed significantly better prognosis in female and HBV negative subgroups.
- Citation: Wang BG, Xu Q, Lv Z, Fang XX, Ding HX, Wen J, Yuan Y. Association of twelve polymorphisms in three onco-lncRNA genes with hepatocellular cancer risk and prognosis: A case-control study. World J Gastroenterol 2018; 24(23): 2482-2490
- URL: https://www.wjgnet.com/1007-9327/full/v24/i23/2482.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i23.2482
Hepatocellular cancer (HCC) is a common malignant tumor with high incidence and mortality, and it is the main histologic type of primary liver cancer[1,2]. Similar to most other solid tumors, HCC patients are considered to be incurable due to the extensive heterogeneity in the clinical manifestations and biological characteristics[3]. HCC heterogeneity can be manifested via diverse genetic, epigenetic, and histogenic features, as well as ethnic differences in patients[4]. Knowledge of genetic and epigenetic variations could aid the early detection and personalized management of HCC. To date, the research hotspots regarding genetic and epigenetic variations are not only coding genes but also noncoding RNAs.
Long non-coding RNAs (lncRNAs) are a type of noncoding RNA with a length of 200 bp, which can function as miRNA sponges to compete with mRNAs by acting as so-called competing endogenous RNAs (ceRNAs)[5]. Genetic variations such as single-nucleotide polymorphisms (SNPs) can alter the expression of coding genes and lncRNAs[6]. To date, several lncRNAs have been reported to be involved in carcinogenesis, such as H19, HOTHAIR, HOTTIP, CCAT2, and MALAT1. Regarding studies of genetic variation, only H19 and HOTAIR SNPs have been well investigated. For example, several meta-analyses showed that H19 and HOTHAIR SNPs were associated with cancer risk[7-9], and the HOTAIR rs920778 SNP was found to be associated with ovarian cancer prognosis[10]. Only four studies have focused on the polymorphisms of the onco-lncRNAs HOTTIP, CCAT2, and MALAT1 (Gene ID: 100316868, 101805488, 378938, respectively)[11-14]. Among them, Gong et al[11] found that these SNPs were significantly associated with lung cancer susceptibility or platinum-based chemotherapy response. However, no SNPs in the above-mentioned lncRNAs have been reported to be associated with HCC risk, and few comprehensive and systematic analyses have been performed on polymorphisms in these three onco-lncRNAs. It thus remains unclear whether the promising SNPs in these lncRNAs have potential to be used as biomarkers for HCC risk and prognosis.
In the present study, we adopted a candidate gene association study strategy with the selected 12 potentially functional tagSNPs covering the three onco-lncRNAs HOTTIP, CCAT2, and MALAT1 to determine whether these SNPs are associated with HCC risk and prognosis and whether promising SNPs could affect the expression of corresponding lncRNAs. We aimed to identify predictive biomarkers for HCC risk and prognosis, establish experimental basis for the comprehension of the HCC etiology, and improve our understanding of the pathogenesis and disease progression of HCC.
This research project was approved by the Ethical Committee of the First Hospital of China Medical University and written informed consent was obtained from each subject at the time of recruitment. The study was designed to be composed of two parts: risk and prognosis. In the risk study, a total of 1338 participants were recruited, including 521 patients who underwent surgical operation for HCC at the First Hospital of China Medical University between 2012 and 2015. The inclusion and exclusion criteria were as follows: (1) The participants who underwent surgical operation were diagnosed with HCC by pathological confirmation, in accordance with the World Health Organization (WHO) classification; and (2) removal of other pathological types of liver cancer (gallbladder cell carcinoma, mix-type liver cancer. and hepatosarcoma). A total of 817 frequency-matched controls were also recruited, some of whom were from a health screening program from the Zhuanghe area, Liaoning Province, China, performed between 2002 and 2012, while others were from a health screening program at the First Hospital of China Medical University performed between 2012 and 2015.
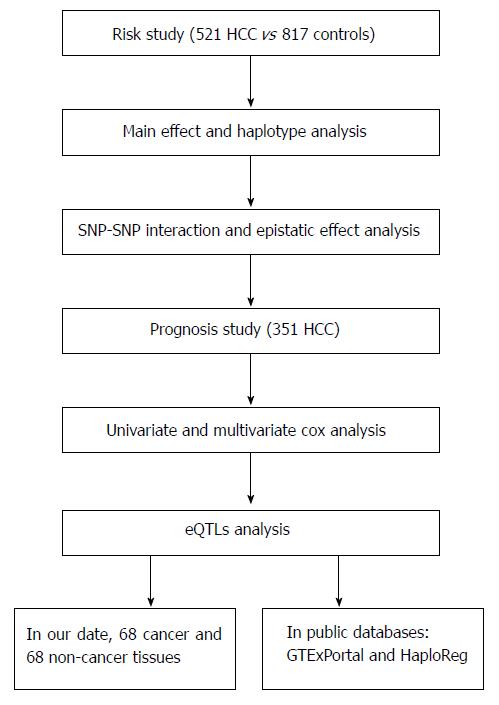
To investigate further the association of these lncRNA polymorphisms with clinicopathological parameters and overall survival of HCC patients, we used data of 351 HCC cases for which information on death or survival was available. Patients with (1) distant metastasis found preoperatively or (2) incomplete pathological data entries were excluded from the survival analysis. Follow-up was completed by July 1st, 2017. For the promising SNPs, lncRNA expression was investigated to explore the possible mechanism by which they exerted their effects, according to our experimental and bioinformatic data. The study design is shown in Figure 1. This research project was approved by the Ethical Committee of the First Hospital of China Medical University and written informed consent was obtained when the patients and controls were recruited.
We selected polymorphisms using 1000G data (http://www.internationalgenome.org/home), as reported previously[15-17]. The tagSNPs were selected separately using the following criteria: (1) Haploview with the Tagger function was used; (2) the population of the HapMap selected Chinese Han Beijing (CHB) population; (3) those for which pairwise tagging had r2 of ≥ 0.8; and (4) those with a minor allele frequency of ≥ 5%. The selection area was enlarged by 10 kb both upstream and downstream for these three lncRNA genes. FastSNP and fSNP searches were used to predict the potential SNP function (http://compbio.cs.queensu.ca/F-SNP/)[18,19]. A total of 12 SNPs from the three lncRNA genes were selected by integrating these two publicly available tools. Locations and characteristics of the selected SNPs are shown in Table 1.
| Gene | Chr. Pos. | SNP1 | Loc. | Genotype | Controls (%) | Cases (%) | P2 value | OR (95%CI) | PHWE value |
| HOTTIP | 7p15.2 | rs17501292 | Exon | TT | 732 (91.2) | 453 (87.1) | 1 (Ref.) | 0.190 | |
| TG | 71 (8.8) | 66 (12.7) | 0.021 | 1.52 (1.06-2.17) | |||||
| GG | 0 (0) | 1 (0.2) | NA | NA | |||||
| TG + GG vs TT | 0.017 | 1.54 (1.08-2.20) | |||||||
| G vs T | 0.012 | 1.55 (1.10-2.18) | |||||||
| rs2067087 | Exon | GG | 174 (21.7) | 88 (16.9) | 1 (Ref.) | 0.674 | |||
| GC | 405 (50.6) | 263 (50.7) | 0.236 | 1.16 (0.90-1.49) | |||||
| CC | 222 (27.7) | 168 (32.4) | 0.015 | 1.49 (1.08-2.08) | |||||
| CC vs GG + GC | 0.035 | 1.35 (1.02-1.82) | |||||||
| C vs G | 0.017 | 1.20 (1.03-1.41) | |||||||
| rs17427960 | Intron | CC | 172 (21.7) | 85 (16.7) | 1 (Ref.) | 0.613 | |||
| CA | 387 (48.8) | 259 (50.9) | 0.707 | 1.05 (0.81-1.35) | |||||
| AA | 234 (29.5) | 165 (32.4) | 0.032 | 1.78 (1.03-2.00) | |||||
| AA vs CC + CA | 0.028 | 1.39 (1.03-1.85) | |||||||
| A vs C | 0.049 | 1.18 (1.00-1.37) | |||||||
| MALAT1 | 22q13.2 | rs4102217 | Promoter | GG | 608 (75.1) | 362 (69.6) | 1 (Ref.) | 0.055 | |
| GC | 180 (22.2) | 148 (28.5) | 0.011 | 1.39 (1.08-1.79) | |||||
| CC | 22 (2.7) | 10 (1.9) | 0.481 | 0.76 (0.36-1.63) | |||||
| GC + CC vs GG | 0.028 | 1.32 (1.03-1.69) | |||||||
| C vs G | 0.097 | 1.20 (0.97-1.50) |
Genomic DNA was extracted using a previously reported method[20] and diluted to a working concentration of 20 ng/µL for genotyping. The genotyping assay was performed by Gene Company (Shanghai, China), using allele-specific PCR with KASPar (KASP) reagents (LGC Genomics, Hoddesdon, United Kingdom). For quality control, we repeatedly genotyped 10% of the total samples at one time. The concordance rate of these repeated samples reached 100%, which demonstrated that the genotyping results were reliable.
The extraction of total RNA from 68 HCC specimens and corresponding samples from nearby noncancerous regions was performed as described previously[15], and a total of 2.0 μg of isolated RNA was converted into cDNA using Quantscript RT Kit (Tiangen Biotech, Beijing, China). The RNA expression levels of the promising lncRNA genes (HOTTIP and MALAT1) and an internal-control gene (GAPDH) were measured using SYBR Premix Ex Taq II (TaKaRa Biotech, Dalian, China) in an Eppendorf Mastercycler Gradient System (Eppendorf AG, Hamburg, Germany). Each reaction was performed in duplicate and controls without a template were also tested every time. The primers are summarized in Supplementary Table 1.
To perform functional candidate polymorphism and expression quantitative trait locus (eQTL) analyses on the promising genes, we mined the data from the following databases: GTExPortal (https://www.gtexportal.org/home/) and Haploreg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php)[21].
Between-group differences in sex variability, as well as accordance with Hardy-Weinberg equilibrium, were compared by the χ2 test and by analysis of variance for age variability. Multivariate logistic regression with adjustments for age and sex was used to show the association between selected lncRNA polymorphisms and HCC risk. The haplotypes of each gene were analyzed using SHEsis software[22]. The two-way pairwise interactions of lncRNA SNP-SNP were calculated using multivariate logistic regression. Univariate and multivariate survival analyses were carried out by the log-rank test and the Cox proportional hazards model. The differences of relative lncRNA levels between two groups were tested by Student’s t-test. P value < 0.05 was considered to be significant.
The demographic characteristics of HCC cases and control subjects are shown in Supplementary Table 2. All polymorphism genotype distributions in cases and controls are shown in Table 1, including 12 SNPs in three lncRNA genes (HOTTIP: rs3807598, rs17501292, rs2067087, rs17427960, rs78248039; CCAT2: rs3843549, rs138947056, rs6983267; MALAT1: rs4102217, rs591291, rs11227209, and rs619586). Among them, most SNPs accorded with Hardy-Weinberg equilibrium, except for CCAT2 rs6983267 (PHWE = 0.029). This SNP was thus excluded from further association analysis.
Among these remaining 11 SNPs, four in the HOTTIP and MALAT1 genes were associated with HCC risk, and they all increased HCC risk (HOTTIP: rs17501292, rs2067087, rs17427960; MALAT1: rs4102217; Table 1, Supplementary Table 3). Among them, two SNPs (HOTTIP: rs17501292; MALAT1: rs4102217) showed significant in a dominant model and the other two showed in a recessive model. HOTTIP rs17501292 and MALAT1 rs4102217 were associated with an increased risk of HCC (P = 0.017 and 0.028, OR = 1.54 and 1.32, respectively) in a dominant model. In addition, HOTTIP rs2067087 and rs17427960 variant genotypes also showed associations with an increased risk of HCC (P = 0.035 and 0.028, OR = 1.35 and 1.39, respectively) in a recessive model. Stratified analysis based on gender, age, smoking, and drinking was performed to analyze the association between each SNP and HCC risk. The results are shown in Supplementary Table 4 and suggest these variables have potential predictive value for specific subgroup populations in HCC risk.
We chose to exclude haplotypes with a frequency of less than 0.03 from the analysis. We found only one haplotype in the HOTTIP gene that was associated with HCC risk. Compared with other haplotypes, patients with the C-G-T-A haplotype of HOTTIP rs3807598-rs17501292-rs2067087-rs17427960 showed a 1.91-fold increased risk of HCC (P = 0.006, 95%CI: 1.20-3.05; Table 2).
| Haplotype | Control (%) | Case (%) | OR (95%CI) | P value |
| HOTTIP1 | ||||
| CGCA | 33.54 (2.4) | 407.00 (4.5) | 1.91 (1.20-3.05) | 0.006 |
| CTGC | 663.39 (47.3) | 389.97 (43.4) | 0.85 (0.71-1.01) | 0.066 |
| GTCA | 604.75 (43.1) | 401.77 (44.7) | 1.08 (0.91-1.28) | 0.406 |
| CCAT22 | ||||
| AAG | 623.60 (39.8) | 434.27 (42.7) | 1.13 (0.96-1.33) | 0.136 |
| AAT | 937.40 (59.8) | 577.73 (56.9) | 0.89 (0.75-1.04) | 0.136 |
| MALAT13 | ||||
| CTCA | 217.99 (13.8) | 167.00 (16.3) | 1.22 (0.08-1.52) | 0.080 |
| GCCA | 934.81 (59.3) | 599.74 (58.6) | 0.97 (0.82-1.14) | 0.718 |
| GTCA | 296.19 (18.8) | 1720 (16.6) | 0.86 (0.70-1.06) | 0.160 |
| GTGG | 85.00 (5.4) | 58.94 (5.8) | 1.07 (0.76-1.51) | 0.689 |
For the data mining of two-way SNP-SNP interactions, we analyzed all possible pair combinations between all of these 11 SNPs and found that the pairwise interaction of HOTTIP rs17501292-MALAT1 rs619586 was significant (Pinteraction = 0.028, OR = 0.30, 95%CI: 0.10-0.88; Table 3).
We further analyzed the epistatic effect of HOTTIP rs17501292 and MALAT1 rs619586 and found in the subset with MALAT1 rs619586 AA wild type and HOTTIP rs17501292 SNP an increased risk of HCC under a dominant model (P = 0.002, OR = 1.85); however, in the subset with the HOTTIP rs17501292 TG+GG genotype, the MALAT1 rs619586 SNP decreased the risk of HCC under a dominant model (P = 0.050, OR = 0.36; Supplementary Table 5).
| Variables | MALAT1 rs619586 | |
| AA | AG + GG | |
| HCC vs CON (n = 335 vs 572) | ||
| HOTTIP rs17501292 | ||
| TT | ||
| Case/control | 372/617 | 78/107 |
| OR (95%CI) | 1 | 1.21(0.88-1.66) |
| TG + GG | ||
| Case/control | 61/55 | 6/15 |
| OR (95%CI) | 1.84 (1.25-2.71) | 0.66 (0.26-1.73) |
| Pinteraction = 0.028, OR (95%CI) = 0.30 (0.10-0.88) | ||
We analyzed the association of all 11 SNPs with the overall survival of HCC patients but found no significant association in either univariate or multivariate Cox proportional hazard analysis (Supplementary Table 6). In the stratified analysis, those with the HOTTIP rs3807598 variant genotype were shown to have a significantly longer survival time in the HBV-negative subgroup (P = 0.049, HR = 0.12, 95%CI = 0.02-0.99), and MALAT1 rs591291 showed an association with a significantly better prognosis in the female and HBV-negative subgroups (P = 0.022, HR = 0.37, 95%CI = 0.16-0.87; P = 0.042, HR = 0.25, 95%CI = 0.07-0.95, respectively; Table 4, Supplementary Table 7).
| Gene | SNP | Stratified | Stratified factors | Genotype | HCC | Death | MST (M) | P value | Hazard ratio (95%CI) |
| HOTTIP | rs3807598 | n = 136 | |||||||
| HBV | Positive | CC | 30 (22.06) | 12 | 90.0 | 1 (reference) | |||
| CG | 73 (53.68) | 23 | 90.1b | 0.391 | 0.74 (0.37-1.48) | ||||
| GG | 33 (24.26) | 11 | 64.1b | 0.680 | 0.84 (0.37-1.91) | ||||
| n = 31 | |||||||||
| Negative | CC | 10 (32.26) | 7 | 21.0 | 1 (reference) | ||||
| CG | 13 (41.94) | 4 | 41.8b | 0.374 | 0.60 (0.16-1.97) | ||||
| GG | 8 (25.80) | 1 | 70.3b | 0.049 | 0.12 (0.02-0.99) | ||||
| n = 285 | |||||||||
| MALAT1 | rs591291 | n = 286 | |||||||
| Gender | Male | CC | 98 (34.27) | 39 | 48.0 | 1 (reference) | |||
| TC | 146 (51.05) | 56 | 56.0 | 0.865 | 0.97 (0.64-1.45) | ||||
| TT | 42 (14.68) | 19 | 47.0 | 0.678 | 1.12 (0.65-1.94) | ||||
| n = 64 | |||||||||
| Female | CC | 22 (34.38) | 13 | 32.0 | 1 (reference) | ||||
| TC | 37 (57.81) | 9 | 56.0 | 0.022 | 0.37 (0.16-0.87) | ||||
| TT | 5 (7.81) | 0 | NA | 0.286 | 0.04 (0.00-16.09) | ||||
| n = 139 | |||||||||
| HBV | Positive | CC | 54 (38.85) | 18 | 69.0 | 1 (reference) | |||
| TC | 69 (49.64) | 21 | 92.1b | 0.816 | 0.93 (0.49-1.74) | ||||
| TT | 16 (11.51) | 7 | 90.0 | 0.965 | 0.98 (0.41-2.35) | ||||
| n = 31 | |||||||||
| Negative | CC | 8 (25.81) | 5 | 6.0 | 1 (reference) | ||||
| TC | 16 (51.61) | 4 | 27.0 | 0.042 | 0.25 (0.07-0.95) | ||||
| TT | 7 (22.58) | 3 | 21.0 | 0.215 | 0.40 (0.09-1.71) |
We used eQTL analysis to investigate the effect of the SNPs identified to be associated with HCC risk on lncRNA expression. In neither the cancerous group nor the noncancerous group was a significant difference observed for the effect of the positive SNPs on lncRNA expression levels (Table 5). Among them, only the heterozygote genotype of intronic rs17427960 of the HOTTIP gene was associated with higher lncRNA-HOTTIP expression, with borderline significance (CA vs CC: P = 0.063; Table 5). Next, we searched public databases for the SNPs for which positive results were obtained in the eQTL analysis (rs17501292, rs2067087, rs17427960, rs4102217). The results from the GTExPortal showed that rs4102217 is a functional SNP in 34 different tissues, such as pancreas and stomach (Supplementary Table 8), and that rs17501292 is a functional SNP in tibial artery tissue; however, no eQTL data for rs2067087 and rs17427960 were found in the public databases. In addition, in Haploreg, it was shown that these SNPs are associated with several kinds of regulatory motifs if the SNP bases altered (Supplementary Figure 1).
| Variable | Non-cancer tissues | Cancer tissues | ||||||
| n | ΔCt (Mean ± SD) | Normalized 2-ΔΔCt | P1 value | n | ΔCt (Mean ± SD) | Normalized 2-ΔΔCt | P1 value | |
| The effect of HOTTIP rs17501292 genotypes to HOTTIP mRNA expression | ||||||||
| TT | 10 | 12.32 ± 4.06 | 1 (0.60, 16.68) | Ref. | 25 | 11.84 ± 3.87 | 1.39 (0.10, 20.39) | Ref. |
| TG | 1 | NA | NA | NA | 2 | 15.30 ± 1.65 | NA | 0.735 |
| GG | 0 | NA | NA | NA | 0 | NA | NA | NA |
| The effect of HOTTIP rs2067087 genotypes to HOTTIP mRNA expression | ||||||||
| GG | 2 | 8.64 ± 2.06 | 1 (0.24, 4.17) | Ref. | 5 | 11.66 ± 1.43 | 1 (0.37, 2.69) | Ref. |
| GC | 5 | 12.25 ± 4.17 | 0.13 (0.01, 1.23) | 0.253 | 13 | 11.85 ± 3.53 | 0.88 (0.08, 10.13) | 0.101 |
| CC | 4 | 13.70 ± 3.86 | 0.03 (0.00, 0.86) | 0.453 | 8 | 12.23 ± 5.42 | 0.67 (0.02, 28.84) | 0.444 |
| The effect of HOTTIP rs17427960 genotypes to HOTTIP mRNA expression | ||||||||
| CC | 2 | 8.64 ± 2.06 | 1 (0.24, 4.17) | Ref. | 6 | 12.47 ± 2.34 | 1 (0.20, 5.06) | Ref. |
| CA | 5 | 11.55 ± 3.21 | 0.13 (0.01, 1.15) | 0.254 | 13 | 11.88 ± 3.72 | 2.13 (0.04, 125.37) | 0.063 |
| AA | 3 | 13.56 ± 4.71 | 0.03 (0.00, 0.86) | 0.465 | 6 | 11.38 ± 5.88 | 1.51 (0.11, 19.84) | 0.348 |
| The effect of MALAT1 rs4102217 genotypes to MALAT1 mRNA expression | ||||||||
| GG | 52 | -2.25 ± 4.40 | 1 (0.05, 21.11) | Ref. | 51 | -0.98 ± 4.88 | 1 (0.03, 29.45) | Ref. |
| GC | 16 | -3.76 ± 3.60 | 2.85 (0.23, 34.54) | 0.404 | 16 | -2.79 ± 4.55 | 3.51 (0.15, 82.14) | 0.742 |
| CC | NA | NA | NA | NA | 1 | NA | NA | NA |
In this study, we preliminarily screened all of the tagSNPs covering three onco-lncRNAs, HOTTIP, CCAT2, and MALAT1, for associations with HCC risk and prognosis. We identified four promising risk-associated SNPs, one haplotype, and a two-way pairwise interaction combination associated with HCC risk. We also found that patients carrying the HOTTIP rs3807598 and MALAT1 rs591291 variant genotypes had a longer survival time in the HBV-negative subgroup. Further molecular experiments were also conducted to investigate whether the tagSNPs could affect the expression of the corresponding lncRNAs. Our study provides an experimental basis for seeking predictive biomarkers for the risk and prognosis of HCC.
LncRNAs function as ceRNAs to compete with mRNAs for access to miRNAs, which could regulate the expression of coding genes[5]. Most studies on lncRNA expression have focused on H19 and HOTAIR as well as other lncRNAs such as PRNCR1, HOTTIP, CCAT1, CCAT2, and MALAT1. HOTTIP, CCAT2, and MALAT1 are all onco-lncRNAs, which have similar biological functions in promoting cell proliferation and invasion[23-29]. They can also promote HCC metastasis and epithelial-mesenchymal transition[30-33]. The HOTTIP gene is located in 7p15.2 and has three exons, the CCAT2 gene is located in 8q24.21 and has one exon, and the MALAT1 gene is located in 11q13.1 and has two exons. The most common SNPs reported for these genes are HOTTIP rs3807598, CCAT2 rs6983267, and MALAT1 rs619586. The first of these was found to be predictive of hematological toxicity in a three-way interaction pattern[34], and the latter two were indicated to be associated with platinum-based chemotherapy response in lung cancer[11]. In this study, we found that SNPs in two exons (rs17501292 and rs2067087) and one intron (rs17427960) of the HOTTIP gene as well as an SNP in the promoter (rs4102217) of the MALAT1 gene were associated with HCC risk; these variant alleles increased HCC risk in the range from 1.18- to 1.55-fold. These four SNPs are reported here for the first time to be associated with cancer risk. Concerning the commonly studied HOTTIP rs3807598 and MALAT1 rs619586 SNPs, no significant associations with HCC risk were found in this study, which is consistent with the findings in a report by Liu[35]. In addition, we found that none of the CCAT2 SNPs was associated with HCC risk. Following the identification of the possible significant SNPs, we further analyzed the relationship between the HOTTIP C-G-C-A haplotype of rs3807598-rs17501292-rs2067087-rs17427960 and HCC risk. The results showed an increase in HCC risk of 1.91-fold in those with this haplotype, and the OR value was greater than that for each SNP alone. Taking these findings together, it is newly indicated that the HOTTIP SNPs rs17501292, rs2067087, and rs17427960 and the MALAT1 SNP rs4102217 have potential to be biomarkers for HCC risk.
Combined interaction analysis for multiple SNPs from different genes is more sensitive and powerful than one-dimensional SNP analysis[36]. For individual SNPs at single loci that were previously shown to have no or a weak effect on disease risk, an epistatic effect may appear when they are analyzed in combination[37]. One of the most significant findings in this study was the SNP-SNP interaction identified for the HOTTIP rs17501292-MALAT1 rs619586 polymorphisms, which was confirmed by the epistatic effect analysis. In the main-effect analysis, HOTTIP rs17501292 had a weak effect, and MALAT1 rs619586 had no effect on the risk of HCC. However, the pairwise analysis of these two in combination showed that they had an interactive effect on HCC risk. Subsequently, we analyzed the epistatic effect of these two SNPs and found that MALAT1 rs619586 was associated with a decreased risk of HCC only in the presence of the HOTTIP rs17501292 TG+GG genotype. A similar epistatic effect between coding genes was also found in our previous study[38]. Further investigations are needed to verify our findings and the mechanism involved in the epistatic phenomenon.
In the prognostic analysis, we found no significant association of the studied SNPs with the overall survival of HCC patients. However, in the stratified analysis based on gender, we found that MALAT1 rs591291 was associated with significantly better prognosis in the female subgroup. When stratified by HBV infection status, we found that patients carrying HOTTIP rs3807598 and MALAT1 rs591291 variant genotypes had longer survival times. As some biomarkers are specific for certain subgroups and have potential to be used for the diagnosis or individualized therapy of specific subgroups[39], the above-mentioned polymorphisms could have value in predicting HCC prognosis for certain subgroups.
eQTL is an analysis in which the combination of mRNA expression and genotype data is applied to determine which variants are correlated with the transcription levels of genes[40]. We analyzed the SNPs potentially associated with HCC risk in our own data and then reanalyzed them in two public databases for the eQTL analysis. HOTTIP rs17501292 and rs2067087 are both located in exon 2 of this gene, while rs17427960 is in intron 2. In contrast, rs4102217 is located at -1255 bp of the MALAT1 gene, within the promoter region. Among these four SNPs, we found that only the heterozygous genotype of intronic rs17427960 of the HOTTIP gene was associated with a higher lncRNA-HOTTIP expression level, with borderline significance. The public databases offered some supportive evidence for this from findings in other tissues, suggesting that rs4102217 in the MALAT1 promoter is a functional SNP in 34 different tissues, such as pancreas and stomach, and that exonic rs17501292 of HOTTIP is a functional SNP in tibial artery tissue. In addition, some regulatory motifs, which were predicted by the bioinformatical software listed in Supplementary Figure 1, are transcription factors like PAX-4 and AP1. Thus, it is reasonable to assume that these SNPs could regulate certain motifs, leading to higher expression of oncogenic lncRNA and thus an elevation of HCC risk. However, further functional research is required to confirm this.
In summary, we found that the SNPs rs17501292, rs2067087, and rs17427960 in the HOTTIP gene, rs4102217 in the MALAT1 gene, and a haplotype of HOTTIP increased the risk of HCC. In addition the SNPs HOTTIP rs3807598 and MALAT1 rs591291 were associated with longer survival time in the HBV-negative subgroup.
Genetic polymorphisms could be biomarkers for cancer risk and prognosis. Recent years, it was found that coding genes and non-coding genes had single nucleotide polymorphisms (SNPs). LncRNAs had important roles in the tumor incidence, progression, and prognosis. Thus, lncRNA polymorphisms had potential to be biomarkers for cancer precaution and prognostic prediction.
The aim of this study is to screen out the effective biomarkers for the hepatocellular cancer (HCC) risk and prognosis. The selected polymorphisms would have potential for the prediction of cancer risk and prognosis.
Five hundred and twenty-one patients of HCC and frequency matched 817 controls were studied for the cancer risk study. Among them, 351 patients for which the information was all available were recruited for the prognosis study. Then, 68 HCC specimens and corresponding samples from the noncancerous region were detected for the expression level study.
For the risk and prognosis study, the samples were detected by the genomic DNA extracted and allele-specific PCR with KASPar reagents. The single nucleotide polymorphisms were selected by the Haploview software. The expression level study isolated the RNA isolated and then converted it to cDNA. The SYBR based real-time PCR were adopted for the lncRNA expression.
We found the HOTTIP rs17501292, rs2067087, and rs17427960 SNPs increased HCC risk by 1.55-, 1.20-, and 1.18-fold under an allelic model, and the MALAT1 rs4102217 SNP increased HCC risk by 1.32-fold under a dominant model. In addition, the two-way interaction of HOTTIP rs17501292 and MALAT1 rs619586 polymorphisms decreased HCC risk and exhibited epistatic effects. In the survival analysis, the HOTTIP rs3807598 variant genotype showed significantly longer survival time in the hepatitis B virus (HBV)-negative subgroup, and MALAT1 rs591291 showed an association with significantly better prognosis in the female and HBV-negative group. In this study, no significant effect in eQTL analysis was observed.
Some specific HOTTIP and MALAT1 SNPs have the potential to be biomarkers for HCC risk and prognosis.
Screening SNPs could be valuable for the identification of biomarkers for HCC risk and prognosis. It could also be used for patient care, as there would be a cohort of patients who would benefit from screening using these positive SNPs.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Gingras MC, Lepetsos P, Ratnasari N S- Editor: Wang XJ L- Editor: Filipodia E- Editor: Huang Y
| 1. | Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753-770. [PubMed] [DOI] [Full Text] |
| 2. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [PubMed] [DOI] [Full Text] |
| 3. | Roessler S, Budhu A, Wang XW. Deciphering cancer heterogeneity: the biological space. Front Cell Dev Biol. 2014;2:12. [PubMed] [DOI] [Full Text] |
| 4. | Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38-55. [PubMed] [DOI] [Full Text] |
| 5. | Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366-5383. [PubMed] [DOI] [Full Text] |
| 6. | Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600-2608. [PubMed] [DOI] [Full Text] |
| 7. | Li XF, Yin XH, Cai JW, Wang MJ, Zeng YQ, Li M, Niu YM, Shen M. Significant association between lncRNA H19 polymorphisms and cancer susceptibility: a meta-analysis. Oncotarget. 2017;8:45143-45153. [PubMed] [DOI] [Full Text] |
| 8. | Lv Z, Xu Q, Yuan Y. A systematic review and meta-analysis of the association between long non-coding RNA polymorphisms and cancer risk. Mutat Res. 2017;771:1-14. [PubMed] [DOI] [Full Text] |
| 9. | Chu H, Chen Y, Yuan Q, Hua Q, Zhang X, Wang M, Tong N, Zhang W, Chen J, Zhang Z. The HOTAIR, PRNCR1 and POLR2E polymorphisms are associated with cancer risk: a meta-analysis. Oncotarget. 2017;8:43271-43283. [PubMed] [DOI] [Full Text] |
| 10. | Qiu H, Wang X, Guo R, Liu Q, Wang Y, Yuan Z, Li J, Shi H. HOTAIR rs920778 polymorphism is associated with ovarian cancer susceptibility and poor prognosis in a Chinese population. Future Oncol. 2017;13:347-355. [PubMed] [DOI] [Full Text] |
| 11. | Gong WJ, Yin JY, Li XP, Fang C, Xiao D, Zhang W, Zhou HH, Li X, Liu ZQ. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37:8349-8358. [PubMed] [DOI] [Full Text] |
| 12. | Wang JZ, Xiang JJ, Wu LG, Bai YS, Chen ZW, Yin XQ, Wang Q, Guo WH, Peng Y, Guo H. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer. 2017;17:167. [PubMed] [DOI] [Full Text] |
| 13. | Kasagi Y, Oki E, Ando K, Ito S, Iguchi T, Sugiyama M, Nakashima Y, Ohgaki K, Saeki H, Mimori K. The Expression of CCAT2, a Novel Long Noncoding RNA Transcript, and rs6983267 Single-Nucleotide Polymorphism Genotypes in Colorectal Cancers. Oncology. 2017;92:48-54. [PubMed] [DOI] [Full Text] |
| 14. | Hu P, Qiao O, Wang J, Li J, Jin H, Li Z, Jin Y. rs1859168 A > C polymorphism regulates HOTTIP expression and reduces risk of pancreatic cancer in a Chinese population. World J Surg Oncol. 2017;15:155. [PubMed] [DOI] [Full Text] |
| 15. | Xu Q, Chen MY, He CY, Sun LP, Yuan Y. Promoter polymorphisms in trefoil factor 2 and trefoil factor 3 genes and susceptibility to gastric cancer and atrophic gastritis among Chinese population. Gene. 2013;529:104-112. [PubMed] [DOI] [Full Text] |
| 16. | Gong Y, He C, Duan Z, Sun L, Xu Q, Xing C, Yuan Y. Association of two ERCC4 tagSNPs with susceptibility to atrophic gastritis and gastric cancer in Chinese. Gene. 2013;519:335-342. [PubMed] [DOI] [Full Text] |
| 17. | Vineis P, Manuguerra M, Kavvoura FK, Guarrera S, Allione A, Rosa F, Di Gregorio A, Polidoro S, Saletta F, Ioannidis JP. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101:24-36. [PubMed] [DOI] [Full Text] |
| 18. | Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391-397. [PubMed] [DOI] [Full Text] |
| 19. | Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, Wang HH, Yao A, Chen YT, Hsu CN. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635-W641. [PubMed] [DOI] [Full Text] |
| 20. | Xu Q, Yuan Y, Sun LP, Gong YH, Xu Y, Yu XW, Dong NN, Lin GD, Smith PN, Li RW. Risk of gastric cancer is associated with the MUC1 568 A/G polymorphism. Int J Oncol. 2009;35:1313-1320. [PubMed] |
| 21. | Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238-1243. [PubMed] [DOI] [Full Text] |
| 22. | Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, He L, Shi Y. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009;19:519-523. [PubMed] [DOI] [Full Text] |
| 23. | Deng X, Zhao Y, Wu X, Song G. Upregulation of CCAT2 promotes cell proliferation by repressing the P15 in breast cancer. Biomed Pharmacother. 2017;91:1160-1166. [PubMed] [DOI] [Full Text] |
| 24. | Wu ZJ, Li Y, Wu YZ, Wang Y, Nian WQ, Wang LL, Li LC, Luo HL, Wang DL. Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-β signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:706-714. [PubMed] |
| 25. | Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840-10852. [PubMed] [DOI] [Full Text] |
| 26. | Zhang S, Wang W, Liu G, Xie S, Li Q, Li Y, Lin Z. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed Pharmacother. 2017;95:711-720. [PubMed] [DOI] [Full Text] |
| 27. | Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, Zeng X, Zhang W. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417-59434. [PubMed] [DOI] [Full Text] |
| 28. | Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother. 2017;95:922-928. [PubMed] [DOI] [Full Text] |
| 29. | Shi B, Wang Y, Yin F. MALAT1/miR-124/Capn4 axis regulates proliferation, invasion and EMT in nasopharyngeal carcinoma cells. Cancer Biol Ther. 2017;18:792-800. [PubMed] [DOI] [Full Text] |
| 30. | Xu Y, Wang B, Zhang F, Wang A, Du X, Hu P, Zhu Y, Fang Z. Long non-coding RNA CCAT2 is associated with poor prognosis in hepatocellular carcinoma and promotes tumor metastasis by regulating Snail2-mediated epithelial-mesenchymal transition. Onco Targets Ther. 2017;10:1191-1198. [PubMed] [DOI] [Full Text] |
| 31. | Zhou N, Si Z, Li T, Chen G, Zhang Z, Qi H. Long non-coding RNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration and apoptosis. Oncol Lett. 2016;12:132-138. [PubMed] [DOI] [Full Text] |
| 32. | Chen F, Bai G, Li Y, Feng Y, Wang L. A positive feedback loop of long noncoding RNA CCAT2 and FOXM1 promotes hepatocellular carcinoma growth. Am J Cancer Res. 2017;7:1423-1434. [PubMed] |
| 33. | Chen L, Yao H, Wang K, Liu X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem. 2017;118:4836-4843. [PubMed] [DOI] [Full Text] |
| 34. | Gong WJ, Peng JB, Yin JY, Li XP, Zheng W, Xiao L, Tan LM, Xiao D, Chen YX, Li X. Association between well-characterized lung cancer lncRNA polymorphisms and platinum-based chemotherapy toxicity in Chinese patients with lung cancer. Acta Pharmacol Sin. 2017;38:581-590. [PubMed] [DOI] [Full Text] |
| 35. | Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, Zhang Y, Chen J, Shen H, Hu Z. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7:e35145. [PubMed] [DOI] [Full Text] |
| 36. | Yin J, Vogel U, Ma Y, Qi R, Wang H. HapMap-based study of the DNA repair gene ERCC2 and lung cancer susceptibility in a Chinese population. Carcinogenesis. 2009;30:1181-1185. [PubMed] [DOI] [Full Text] |
| 37. | Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5:618-625. [PubMed] [DOI] [Full Text] |
| 38. | He C, Tu H, Sun L, Xu Q, Gong Y, Jing J, Dong N, Yuan Y. SNP interactions of Helicobacter pylori-related host genes PGC, PTPN11, IL1B, and TLR4 in susceptibility to gastric carcinogenesis. Oncotarget. 2015;6:19017-19026. [PubMed] [DOI] [Full Text] |
| 39. | Baker SG, Kramer BS, Sargent DJ, Bonetti M. Biomarkers, subgroup evaluation, and clinical trial design. Discov Med. 2012;13:187-192. [PubMed] |
| 40. | Gupta RM, Musunuru K. Mapping Novel Pathways in Cardiovascular Disease Using eQTL Data: The Past, Present, and Future of Gene Expression Analysis. Front Genet. 2013;3:232. [PubMed] [DOI] [Full Text] |