Published online Jun 14, 2018. doi: 10.3748/wjg.v24.i22.2327
Peer-review started: April 7, 2018
First decision: April 27, 2018
Revised: May 3, 2018
Accepted: May 18, 2018
Article in press: May 18, 2018
Published online: June 14, 2018
Processing time: 65 Days and 9.5 Hours
Colorectal cancer (CRC) is the second leading cause of cancer worldwide. CRC is still associated with a poor prognosis among patients with advanced disease. On the contrary, due to its slow progression from detectable precancerous lesions, the prognosis for patients with early stages of CRC is encouraging. While most robust methods are invasive and costly, actual patient-friendly screening methods for CRC suffer of lack of sensitivity and specificity. Therefore, the development of sensitive, non-invasive and cost-effective methods for CRC detection and prognosis are necessary for increasing the chances of a cure. Beyond its beneficial functions for the host, increasing evidence suggests that the intestinal microbiota is a key factor associated with carcinogenesis. Many clinical studies have reported a disruption in the gut microbiota balance and an alteration in the faecal metabolome of CRC patients, suggesting the potential use of a microbial-based test as a non-invasive diagnostic and/or prognostic tool for CRC screening. This review aims to discuss the microbial signatures associated with CRC known to date, including dysbiosis and faecal metabolome alterations, and the potential use of microbial variation markers for non-invasive early diagnosis and/or prognostic assessment of CRC and advanced adenomas. We will finally discuss the possible use of these markers as predicators for treatment response and their limitations.
Core tip: Many clinical studies have reported a disruption in the gut microbiota balance and in the faecal metabolome in colorectal cancer. In this review, we describe the modifications in the microbiota composition and metabolome observed in colorectal cancer (CRC) tissue and stool samples. Then, we detail how these microbiota modifications may represent novel and promising non-invasive diagnostic and/or prognostic markers for CRC and advanced adenoma.
- Citation: Villéger R, Lopès A, Veziant J, Gagnière J, Barnich N, Billard E, Boucher D, Bonnet M. Microbial markers in colorectal cancer detection and/or prognosis. World J Gastroenterol 2018; 24(22): 2327-2347
- URL: https://www.wjgnet.com/1007-9327/full/v24/i22/2327.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i22.2327
Approximately 5% of the population in Western countries will develop colorectal cancer (CRC) during their lifetime[1]. CRC is a heterogeneous disease with a wide range of long-term outcomes and responses to treatment. Despite recent advances in the genetic and molecular characterization of tumours, CRC remains associated with a poor prognosis and very low rates of long-term survival among patients with advanced disease[2,3]. On the contrary, the 5-year survival of patients treated during the early stages of CRC, which represent only one-third of all CRC patients, is encouraging, ranging from 72% to 100%[4]. Therefore, early detection of both premalignant lesions and CRC is crucial for increasing the chances of a cure[5]. Moreover, accurate determination of the prognosis is crucial for the practitioners, in order to optimize and personalize treatment strategies. Given the poor prognosis of this cancer, it is essential to validate new diagnostic and prognostic markers. Studies on the implications of the gut microbiota in CRC are now increasing[6-11]. Identification of a CRC-associated dysbiosis and/or the targeted detection of procarcinogenic bacterial species and/or their effectors in stool or tissue samples may represent promising tools to overcome the limitations and the poor performance of current CRC screening tools and prognostic/predictive factors. This review will focus on gut microbiota-related markers, which may be new biological markers for CRC screening or prognosis determination.
Individual (opportunistic) and population-based (organized) screening strategies have been established permitting the reduction of CRC mortality either by detecting CRC or removing premalignant lesions[12]. Several CRC screening tests have been used and have different degrees of performance that vary worldwide and are primarily driven by costs, lifestyle and endoscopic resource constraints. In addition to the classic invasive endoscopic approaches, there has been great interest in developing and evaluating non-invasive methods, including radiographic examinations and stool- and blood-based tests, with the goal of creating efficient, safe, convenient and cost-effective screening tools[13-18].
Endoscopy is currently the most effective modality for detecting asymptomatic premalignant polyps and CRC and for preventing CRC development by the immediate removal of premalignant lesions[13]. However, this procedure requires prior bowel cleansing and causes postprocedural discomfort, which are both responsible for low participation rates in colonoscopy CRC screening programmes[13]. Moreover, the need for general anaesthesia and the invasiveness of the procedure exposes patients to potentially severe complications[19,20]. Computed tomography (CT) colonography, also known as “virtual colonoscopy”, is a non-invasive alternative to colonoscopy[21-23]. However, CT colonography presents some drawbacks such as its high cost, lack of standardized methods, need for bowel cleansing, poor performance for detecting small or flat lesions, and lack of ability to perform immediate biopsy[14,24]. For these reasons, other non-invasive stool- and blood-based tests are more promising tools for initial CRC screening in two-step approaches.
Non-invasive stool-based early CRC screening modalities involve either detecting occult bleeding in the stool, such as with guaiac fecal occult blood test (gFOBT) and faecal immunohistochemical testing (FIT), or detecting DNA, RNA and protein markers of neoplasia in faeces. The rationale for the use of such tools is that the mucosa of both polyps and CRC is more fragile and vascularized and thus bleeds and desquamates more easily than normal colonic mucosa. The effectiveness of gFOBT and FIT have been established for reducing both CRC incidence and mortality, thus permitting their use in many CRC screening programmes[2,19,21,25-36]. However, these techniques have been criticized for their relatively low specificity[14]. Therefore, new molecular CRC screening methods targeting abnormal proteins or mRNA expression (e.g., PKM2, β-actin expression), gene mutations (e.g., KRAS, APC, P53), microsatellite instability (MSI) or aberrantly methylated promoter regions present in stool or body fluids are a developing area of investigation to establish innovative large-scale screening programmes[13,14,37-45]. As many tissues release some of their constituents in the bloodstream, the detection of circulating tumour cells DNA (cfDNA) in the blood is a recent area of great interest in CRC screening[46-49]. Although these biomarker tests show promising sensitivities, their high costs and high test positivity rates may lead to greater demands on endoscopy departments, and the absence of large-scale validation currently remains the main limitation of their routine use in population-based CRC screening programmes[18,40,41].
Many prognostic factors including many socioeconomic and clinical factors are routinely used to optimize and personalize treatment strategies[50-53]. However, pathological factors are currently the most powerful tools that are routinely used to adapt therapeutic strategies, especially for conditioning the administration of non-operative treatments, such as chemotherapy and radiotherapy. The TNM international staging system remains the gold-standard for determining both CRC prognosis and treatment indications[4,54-56]. Unfortunately, this classification is largely imperfect, which is highlighted by the constant changes it undergoes with the advances in the field of CRC research. Furthermore, it has some limitations. In particular, it is inaccurate for some subgroups of patients whose prognosis and indications for adjuvant treatments remain unclear. Therefore, some other pathological prognostic factors are routinely used, such as the presence of lymphovenous and/or perineural invasion[57-60], tumour budding[61,62] and tumour differentiation grade[63,64].
In addition, studies have shown tumour genomics and cellular alterations can supplement clinicopathological factors. Aneuploidy[65], tumour-infiltrating lymphocytes[66,67], overexpression of the CEA in tumours[68], allelic loss in DCC, TP53, APC and MCC genes[69,70], TP53 gene mutations[71], expression of CD44 protein[72], high levels of thymidylate synthetase[73], and the detection of MSI and both RAS and BRAF genes mutations all play prominent roles in determining CRC prognosis and in personalizing treatments. They are independent strong prognostic factors[74-82] and are also predictive of the response to some widely used chemotherapy regimens, including targeted therapies and immunotherapy[74,76,83-94].
Serum prognostic factors [e.g., C-reactive protein (CRP), carcinoembryonic antigen (CEA), and cfDNA] can also be very useful tools. Both CRP and serum albumin levels are associated with a worse prognosis but are not routinely used as prognostic factors for CRC[95-101]. The CEA level is a well-recognized and routinely used independent prognostic factor for CRC[102,103]. Persistently elevated CEA levels after treatment are correlated to both a higher risk of recurrence and decreased survival. Similarly, normal CEA levels after treatment are associated with improved recurrence-free and overall survival[104]. In addition to CRC screening, investigations evaluating cfDNA have recently gained attention in the field of CRC research. The detection and the quantification of specific gene mutations (RAS and BRAF genes) or DNA methylation alterations[105,106] are correlated with CRC prognosis[107,108], independent of the tumour stage. In the future, monitoring of cfDNA levels may guide therapeutic strategies[109,110] and improve patient follow-up, allowing the earlier detection of recurrence.
As we previously mentioned, all these screening strategies and prognostic markers present many limitations. Therefore, there is still a need to develop other effective non-invasive screening and prognostic tools to replace or supplement those currently in use, with the aim of decreasing CRC mortality and optimizing treatment strategies. The study of intestinal microbiota provides new leads to identify powerful biomarkers.
The intestinal microbiota and its host have coevolved to establish a symbiotic relationship leading to digestive system homeostasis. Many metabolic, immunological, structural and protection functions that are essential to the health of the host are performed by the intestinal microbiota. Thus, one of the major functions of the gut microbiota is to ferment complex carbohydrates in order to produce a large amount of metabolites (short chain fatty acids (SCFAs), bile acids, choline, essential vitamins, etc.), some of which represent energetic and nutrient substrates for intestinal cells[111,112]. Others are involved in the regulation of cellular processes or participate in regulating hepatic lipid and glucose homeostasis[112,113]. Another function of the gut microbiota is the maintenance of epithelial homeostasis since commensal bacteria can promote epithelial integrity[114]. They can induce structural and functional maturation of the colonic epithelium with strong modulation of mucus cells[115]. The intestinal microbiota also plays a fundamental role in the development, maturation and function of the host immune system[115,116] and also confers resistance to colonization by pathogenic microorganisms by competing both for nutrients and attachment sites to the intestinal epithelium, by producing and secreting antimicrobial compounds, such as bacteriocins, and by strengthening epithelial tight junctions[117].
Approaches used to study the gut microbiome: The gut microbiota has a very large diversity of microbial populations such as bacteria, archaea, eukaryotes and viruses. Because the colonic microbiota is mostly represented by bacteria, the bacterial community is the most studied. Both tissue and faecal samples provide information about the structure of bacterial populations. The analysis of tissues is more relevant for evaluating the mechanisms of microbiota involvement in physiopathology. However, sampling of mucosa-associated bacteria by endoscopic biopsy of intestinal tissue is an invasive procedure. It is more difficult to implement than collection of a faecal sample, especially in healthy donors. Therefore, the vast majority of studies on the diversity and richness of the gut microbiota are conducted on stool samples, which are more useful for identifying diagnostic or prognostic markers for pathologies such as CRC.
The first data concerning the gut microbiota were provided by cultural approaches. However, these approaches allow only limited evaluation of this ecosystem, since less than 30% of intestinal bacteria have been cultivated to date. Since the 1990s, the advent of molecular tools targeting the bacterial 16S ribosomal RNA (rRNA) gene has revolutionized our knowledge of the gut microbiota from both faeces and tissues. Thus, genetic fingerprinting techniques (terminal restriction fragment length polymorphism (T-RFLP), denaturing gradient gel electrophoresis (DGGE), hybridization approaches (fluorescence in situ hybridization (FISH), microarrays) and clone library analysis have been applied to provide a more complete description of its structure[118-122]. Currently, quantitative PCR (Q-PCR) and 16S rRNA gene next-generation sequencing (NGS) are the most used methods for describing the composition of the intestinal bacterial community and therefore for comparing the gut microbiota of healthy individual to that of patients with diseases. Thus, modifications in the microbiota composition, also called dysbiosis, in faeces and tissues have been described in CRC[123-130]. These different approaches allow only taxonomic identification of microbial communities. However, the gut is a complex and variable ecosystem. Understanding its function and the specific role of the different bacterial populations is essential. A metagenomics approach (shotgun sequencing) makes taxa identification possible and allows exploration of the metabolic potential of the intestinal microbiota[126,131,132]. Thus, the American Human Microbiome Project (HMP) and the European “MetaHIT” (Metagenomics of the Human Intestinal Tract) consortia were formed in order to generate a catalogue of genes carried by the gut microbiota[133]. Intestinal microbial dysbiosis was recently investigated in patients with CRC[125,126,131,132]. Combining the metagenomics data available in databases with the application of other high-throughput meta-omics approaches, it is now possible to gain a better insight into the microbial function and to reveal the link between genetic potential and functionality in the gut microbiota.
Cancer-associated microbiome alterations: To date, there is no consensus in terms of the microbiota modifications observed in CRC using high-throughput sequencing of the bacterial 16S rRNA gene and metagenomics both in faecal and tissue samples compared with samples from healthy individuals. The complexity of the gut microbial ecosystem associated with other technical or biological parameters, such as the geographical location of the studied populations, the lifestyle, the diet, the sample type (faeces, mucosa, tumour), the location of the tissue sampled (i.e., right or left colon), the age of the individual, the stage of the disease, the number of patients studied, the molecular approach used (metagenomics, 16S rRNA gene NGS), the regions of the 16S rRNA targeted (V1-V2, V3, V3-V5, V4-V6 regions) for the NGS approach, and the taxonomic level considered, contribute to the lack of consensus on the definition of the composition of dysbiotic microbiota in patients with CRC.
Alpha diversity metrics, which represent the specific richness and the distribution of individuals within the species were found to be increased in adenoma and adenocarcinoma tissues compared with in normal mucosa[134-136]. However, these parameters were unchanged in faecal samples[125,134,137,138], suggesting that the increase in diversity was limited to the neoplastic lesions and did not extend to the entire mucosa.
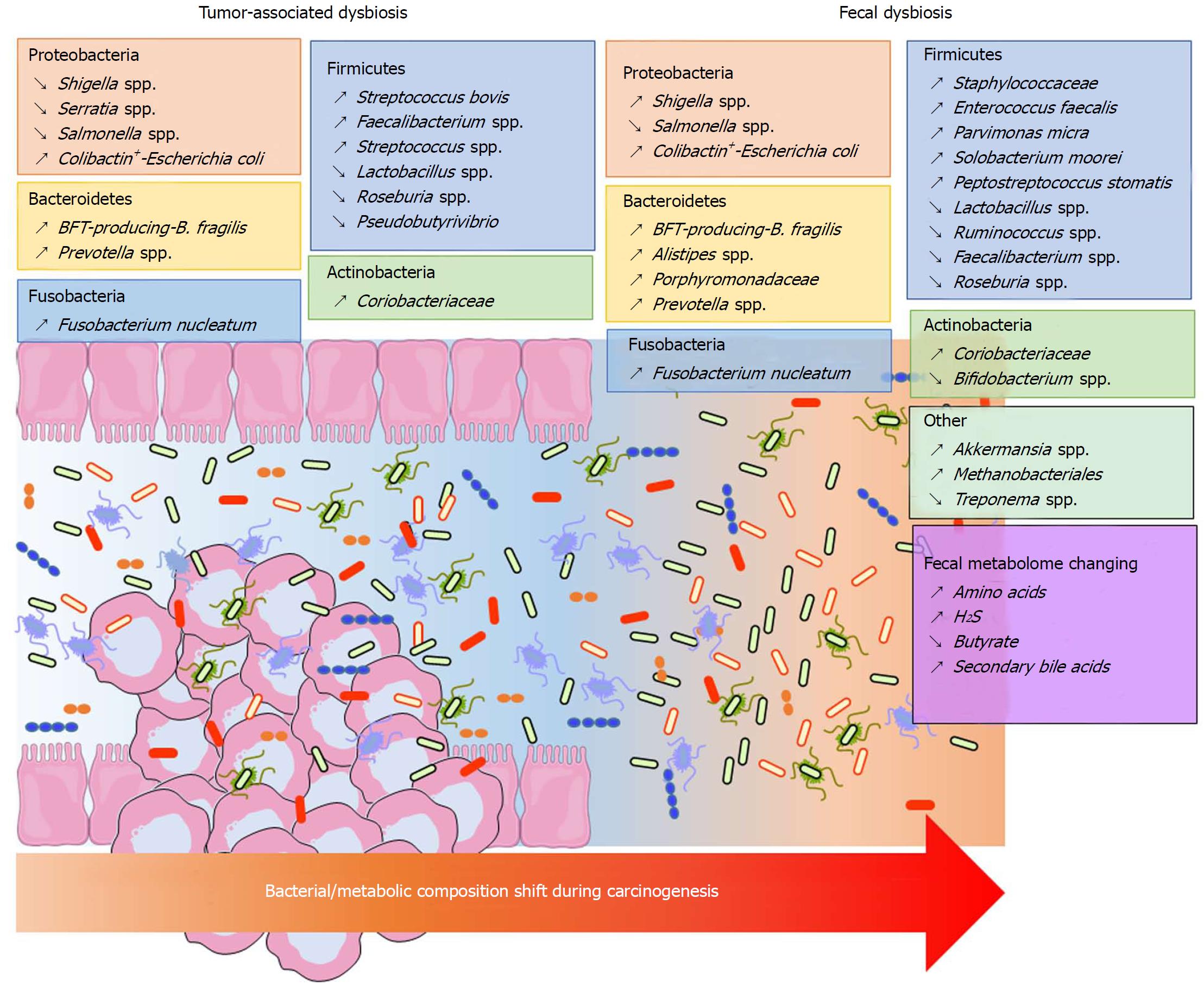
Significant shifts can be observed in the composition of the bacterial community at the phylum taxonomic level but also on a finer phylogenetic scale in CRC samples. However, these modifications vary depending on the analytic techniques used and the sample location (mucosa, adenocarcinoma, stool). It is accepted that Bacteroidetes, Fusobacteria and Proteobacteria are enriched, while Firmicutes is decreased in CRC patients[10,139] (Figure 1). A similar dysbiosis has been described in CRC animal models[140,141]. In a taxonomy-based analyses conducted in 2013, Ahn et al[142] described a lower relative abundance of Clostridia and increased Fusobacterium and Porphyromonas in case subjects compared with control subjects. Additionally, the genera Bacteroides and Prevotella, Bacteroides fragilis, Enterococcus faecalis, Streptococcus bovis, Streptococcus gallolyticus and Escherichia coli were overrepresented, while Bacteroides vulgatus, Lactobacillus and Faecalibacterium prausnitzii were decreased in the faeces or tumours of patients with CRC[123-125,131,143-146]. Other studies found that Parvimonas micra, Solobacterium moorei, Peptostreptococcus stomatis were consistently enriched in the microbiomes of the stools of CRC patients[126,131]. Many studies showed an enrichment of Fusobacterium nucleatum in stool and tissue samples from patients with colorectal carcinoma, which were confirmed by quantitative polymerase chain reaction (PCR)[126,131,147-149] (Figure 1). In addition to taxonomic studies, a metagenomics analysis showed an enrichment of virulence-associated bacterial genes in the tumour microenvironment[132]. Many studies reported a high prevalence of toxin genes expressed by some species described above. Most of enterotoxigenic B. fragilis (ETBF) strains detected in mucosal samples from patients with CRC harboured the bft gene, which encodes the bacterial toxin B. fragilis toxin (BFT)[149] (Figure 1). ETBF and BFT carrying strains were detected more often in stool samples from CRC patients compared with samples from controls[150]. Many studies reported that mucosa-associated BFT-producing B. fragilis was more prevalent in late-stage CRC[130,149]. Other toxins named cyclomodulins [cytolethal distending toxin (CDT), colibactin, cytotoxic necrotizing factor and cycle inhibiting factor], which are carried by E. coli, were more prevalent in CRC samples[145,146,151,152]. In particular, the colibactin toxin, which is synthesised from the pks island, was preferentially detected in strains isolated from CRC patients[145,146,152] (Figure 1). These toxin genes could be targets of interest for developing new prognostic or diagnostic factors.
The majority of published data were obtained in the adenocarcinoma stage. However, the intestinal mucosa might present different ecological niches according to the stage of colorectal tumour progression. Due to difficulties in sampling patients during the early stages of the disease, studies focusing on the temporal evolution of microbiota dysbiosis during colorectal carcinogenesis are scarce. Nevertheless, metacommunities can be defined from the bacterial community configurations by Nakatsu et al[153]. In early stages of CRC, bacteria belonging to Fusobacterium, Parvimonas, Gemella and Leptotrichia genera were more abundant[153]. Moreover, the occurrence of Escherichia coli, Pseudomonas veronii and Lactococcus were specific to adenomatous lesions[135,153], whereas Fusobacterium enrichment was observed in carcinomas[153]. Additionally, the abundance of Bacteroides fragilis and Granulicatella were progressively increased during the adenoma-carcinoma sequence. Lu et al[135] showed that there was a clear increase in both Proteobacteria and Bacteroidetes in tissues from patients with colorectal adenoma compared with tissues from healthy volunteers. Kostic et al[147] found that Fusobacterium spp. were enriched in human colonic adenomas relative to the surrounding tissues and in stool samples from patients with colorectal adenoma compared with samples from healthy subjects. In addition, Lactococcus and Pseudomonas were enriched in preneoplastic tissue, whereas Enterococcus, Bacillus, and Solibacillus were reduced. A high frequency of colibactin-producing E. coli and enterotoxigenic Bacteroides fragilis were also observed in adenomas from patients with Familial Adenomatous Polyposis[154]. Recently, Eklöf et al[151] showed that the prevalence of colibactin-producing E. coli were progressively increased in the adenoma-carcinoma sequence.
Finally, most of these studies do not take into account the heterogeneity of this pathology. As an example, Prevotella and Firmicutes populations were more abundant in individuals with proximal tumours, whereas Bacteroidetes populations were overrepresented in distal cancers[155]. More recently, a drastic difference in the microbial composition has been observed in the mucosa of colitis-associated CRC patients, with an increase in the Enterobacteriaceae family and the Sphingomonas genus and a decrease in Fusobacterium and Ruminococcus genera, when compared with the mucosa of sporadic CRC patients[156]. Similarly, we observed that colibactin-producing E. coli were more frequently identified in microsatellite stable (MSS) CRC[157]. In the same way, high colonization by F. nucleatum and negative-colibactin E. coli bacteria were detected in patients with MSI (microsatellite instability) compared with the MSS phenotype[149,157,158]. All these data suggest that the dysbiosis signature of CRC might be different according to the tumour phenotype and/or molecular alterations.
In conclusion, all these data show that there is an adenoma- and a CRC-associated signature in the microbiome (Figure 1). Metagenomics studies showed a redundancy in the metabolic functions of the intestinal microbiota. Therefore, studies of metabolites might reflect the CRC-associated microbiota dysbiosis with a lower heterogeneity than taxonomic determination.
Metabolite analysis procedures: The metabolome has been defined as the complete complement of all small molecule (< 1500 Da) metabolites found in a specific cell, organ or organism[159]. Global metabolomics alterations reflect changes due to environmental factors, diet, drugs, exercise, genetic variation and regulation, changes in the gut microflora, and altered kinetic activity or levels of enzymes[160]. The most widely used analytical high-throughput tools for metabolomics studies are nuclear magnetic resonance (NMR) and mass spectrometry (MS), and they both can provide complementary snapshots of the metabolome of body fluids[161]. Sample preparation is arguably the most critical step in metabolomics, but there is no universal, robust and repeatable approach defined. NMR spectroscopy benefits from being highly reproducible[162] and offers the potential to precisely quantify compounds in complex mixtures without complex sample preparation requirements[163]. Gas chromatography coupled with MS (GC-MS), which is used for the analysis of volatile low molecular weight metabolites, and liquid chromatography coupled with MS (LC-MS), which is used for the analysis of non-volatile components, have a higher sensitivity than NMR, but the sample preparation must be more fastidious. For more details regarding sample preparation in metabolomics, several authors previously described methods for NMR, GC-MS and LC-MS analysis[163].
During the last decade, using NMR and/or MS, metabolic profiles of colorectal tumour biopsies and matched colon mucosa samples revealed potential biomarkers in carbohydrate metabolism, short-chain fatty acid and other lipid metabolism, nucleotide biosynthesis, and secondary bile and steroid metabolism[164,165]. The repertoire of metabolic derangements derived from cancer cells, their microenvironment and the gut microbiota composition and activity may culminate in a distinct metabolic phenotype (also called metabotype) that characterizes the pathology of CRC. Metabolome analysis of blood and urine samples from CRC patients were performed, and several studies reported relevant biomarkers for CRC[166-168]. However, the transient localization of faeces in the colon and rectum makes it an ideal matrix for gaining information about the health and pathological state of the colon and rectum, and thus for colorectal cancer diagnosis[169]. Faecal samples consist of a diverse range of endogenous metabolites including tumour cell metabolites and gut microbiota metabolites[169].
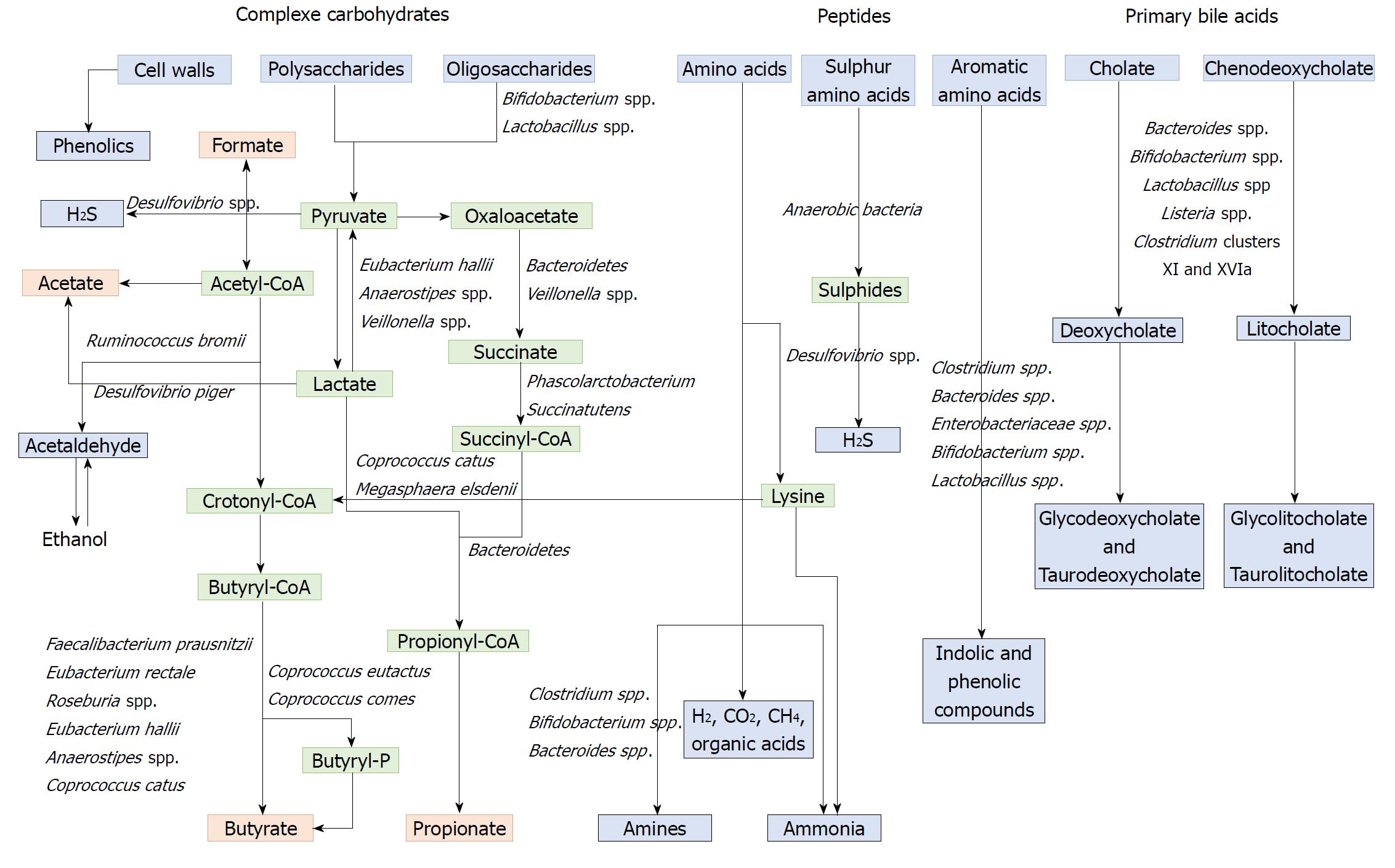
Microbial influence on the faecal metabolome: When ingested, the majority of food nutrients are absorbed in the small intestine. The residues that reach the colon are complex nutrients, generally fibres (complex carbohydrates), but also protein residues and host products such as mucin and bile acids secreted by the liver in response to fat ingestion[170]. These compounds are metabolized by the microbial populations into a diversity of metabolites that can be use by the host colonic cells[171]. The gut microbiota makes an important contribution to human metabolism by providing enzymatic functions that are not encoded by the human genome, such as the breakdown of polysaccharides and polyphenols and the synthesis of vitamins by fermentation and anaerobic degradation[172].
Polysaccharides and non-digestible carbohydrates are the primary substrates for microbial fermentation[173]. Saccharolytic fermentation of carbohydrates produces beneficial metabolites such as short-chain fatty acids[174]. However, if there is limited carbohydrate intake, bacteria use alternative energy sources, resulting in the production of other metabolites that may be more detrimental to human health[172]. The three SCFAs acetate, propionate and butyrate, which are considered beneficial metabolites, are mainly produced from polysaccharides and oligosaccharides through their metabolism into pyruvate (Figure 2). Propionate and acetate can be produced from lactate, while butyrate and acetate can be produced from acetyl CoA (Figure 2)[171]. Butyrate is an important energy source for gut epithelial cells, while propionate and acetate are mostly metabolized in the liver for cholesterol synthesis, gluconeogenesis or lipogenesis[175]. For more information, the complete synthesis and functions of SCFAs was extensively and previously described[113,175]. Microbial use of bile salts is a classic example of the relationship between the gut microbiota metabolism and the host metabolism. The primary bile acids cholic acid and chenodeoxycholic acid are produced in the liver from cholesterol and are conjugated to glycine or taurine. They are secreted by the liver following high fat intakes[176]. The intestinal microbiota can affect the biotransformation of bile acids via deconjugation, dehydrogenation, epimerization, and 7α/β-dehydroxylation of primary bile acids to generate secondary bile acids[177]. A number of gut bacteria possess bile salt hydrolase enzymes that are capable of hydrolysing the amide bond between the bile acid and its conjugated amino acid, producing the secondary bile acids deoxycholate and litocholate (Figure 2). Bile salt hydrolase genes were identified in Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, and Listeria[178]. Deconjugation of bile acids by microbial hydrolases leads to an increased level of secondary bile acids, especially deoxycholic acid, which is also known to exert carcinogenic activity[179]. Polyphenols from fruits and vegetables are poorly absorbed in the intestine, and 90-95% are metabolized by the gut microbiota[180]. The diversity of polyphenol compounds is wide and has been summarized by Marín et al[181] and Rowland et al[172]. Several gut microbial enzymes can transform plant polyphenols, such as phenolic acids, flavonoids, and lignans into simple phenolic metabolites[172,181,182]. These microbial phenolic metabolites are mostly known for their antioxidant activity and have been shown to inhibit the production of pro-inflammatory mediators[183]. Microbial protein fermentation generates potentially toxic and pro-carcinogenic metabolites involved in CRC, such as phenols, sulfides, ammonia and nitrosamines[7]. A subset of bacteria, including Bacteroidetes and Firmicutes species, produce potentially bioactive substances via the degradation of amino acids, especially nitrogenous compounds that can exert carcinogenic effects[184-186]. Moreover, sulfides produced in the gut by the bacterial reduction of dietary sulfate are enterotoxic[187]. Microbial metabolism may contribute to the toxicity of alcohol, especially in the gastrointestinal tract where aerobic and facultative anaerobic bacteria convert ethanol to acetaldehyde, which is known to be a highly toxic and pro-carcinogenic compound[188]. A specific role for bacterial biofilms in colonic metabolome was also reported[189]. With organic acids, gas is the major products of microbial fermentation in anaerobic systems. Anaerobic bacterial fermentation can lead to the synthesis of hydrogen, carbon dioxide, or methane for the majority. Some minor gases, such as NH3, NH2, volatile amines, indolic and phenolic compounds , that are mainly associated with peptide fermentation, are also produced[190,191] (Figure 2). Among them, H2S is produced by sulfate-reducing bacteria (Desulfovibrio spp.) by sulfur amino acids and taurine metabolism.
Given the key role of the gut microbiota in dietary metabolism and the growing evidence of its potential impact on host health, studies were performed to elucidate the metabolic functions of the gut microbiota. These studies mostly used metagenomic, metatranscriptomic, or metaproteomic approaches, but using metabolomics to study the gut microbiota appears to be a promising method. Indeed, metabolomics allow the measurement of both host and microbial metabolites, including metabolites in host samples derived from the microbiota and host metabolites processed by the microbial population.
Metabolome alterations in CRC: The analysis of the CRC-associated metabolome has highlighted differences in the biochemical composition of CRC patients’ stools. Few animal studies have investigated the metabolomic profile of faeces in CRC models. Significant alterations were observed in fatty acid metabolites and metabolites associated with bile acids (hypoxanthine xanthine, taurine) in faeces of Hmga1 mice as analysed by ultra-performance liquid chromatography UPLC/MS/MS and GC-MS[192]. Increased levels of short, arginine-enriched, tetra-peptide fragments were reported in the transgenic mouse faeces[192]. Similarly, CRC patients were found to have higher levels of some amino acids and alterations in the levels of some SCFA in their stools compared with controls[193]. Butyrate and several butyrate-producing bacteria were depleted in CRC patients’ stools[194]. Chen et al[143] observed that butyrate and butyrate-producing bacteria were more prevalent in healthy controls than in advanced colorectal adenoma patients, with a lower prevalence of Clostridium, Roseburia, and Eubacterium spp. and a higher prevalence of Enterococcus and Streptococcus spp. Moreover, faecal β-glucuronidase activity is increased in patients with CRC compared with healthy controls[195]. High-fat diets, which are positively correlated with the incidence of CRC, lead to an increase in bile secretion, and increased faecal bile acid concentrations were reported in patients with CRC[196]. High faecal bile concentrations were positively correlated with the incidence of CRC, with potential tumour-promoting effects of bile acids themselves[176]. Furthermore, strong antimicrobial bile acid activities lead to significant changes in the gut microbiota composition, with a relative increase in some Gammaproteobacteria and Bacteroidetes species that are associated with CRC[197]. The dietary intake of preformed N-nitroso compounds is positively correlated with CRC in European populations[185], but these compounds can also be formed endogenously via acid-driven nitrosation in the stomach and by the nitrosation of amines that are derived from the microbial fermentation of protein in the large intestine[198]. Higher levels of H2S were detected in CRC patients than those in healthy controls, but no increases in Desulfovibrio spp. were observed in faecal samples from patients with CRC[199]. Using metabolome screening, a metabolic influence of microbial biofilms on colonic tissues and the related occurrence of cancer has been reported. It was found that host cancer and specific bacterial biofilm structures contributed to the polyamine metabolite pool (N1,N12-diacetylspermine), which might have an impact on cancer development and progression[189].
All these data suggest that faecal metabolomics may not only be used for diagnostic purposes but may also serve as a prognostic tool for CRC treatment in the future.
Even if a precise microbial signature associated with CRC and a precancerous lesion have not been exactly defined, different works have shown that dysbiosis may provide new significant biomarkers for CRC diagnosis or prognostic determination.
Several studies were able to determine a correlation between faecal microbial dysbiosis and CRC prognosis/diagnosis. In 2014, Zackular et al[140] used faecal samples from 30 CRC patients, 30 colonic adenoma patients and 30 healthy controls to establish a classification model. The authors observed significant differences in the gut microbiome of healthy controls when compared with those with adenoma or CRC, and faecal microbial screening had an accuracy of 0.798 AUC for predicting CRC. By combining their gut microbiome data with known clinical risk factors (body mass index, age, race), they were able to significantly improve the ability of faecal microbial screening to discriminate between the three clinical groups when compared with risk factors alone. Zeller et al[131], with 53 CRC patients, 42 adenoma patients and 61 controls, established a 16S-based classifier for CRC detection and validated their model with an accuracy of 0.82. More recently, Ai et al[200] sequenced the 16S rRNA gene from the faeces of 141 Chinese participants and evaluated the performance of different classifiers for predicting CRC based on faecal microbiota. The authors reported a strong variation in the CRC prediction between the different models tested, with the Bayes Net algorithm displaying the best performance. Their prediction test, with 0.93 AUC, was more accurate than the FOBT, while combining both tests improved the accuracy. This list of studies that assessed the use of changes in the faecal microbiota for colorectal adenoma and CRC screening is non-exhaustive. Among these studies, while an association between CRC and microbiota is clear, there is limited agreement in the taxa reported.
For this reason, Shah et al[201] conducted a microbiome-based meta-analysis for CRC, using the results from 9 studies and including 79 colorectal adenomas patients, 195 CRC patients and 235 controls, in order to identify a common microbial marker in stool samples. In addition to the previously reported taxa, they highlighted a significant increase in Parvimonas micra ATCC 33270, Streptococcus anginosus, Parabacteroides distasonis and other members of Proteobacteria. Their microbiome-based CRC versus control classification produced an area under the receiver operating characteristic (AUROC) curve of 80.3%, which was 91.3% when combined with clinical markers from the bioinformatics pipeline. Similarly, Amitay et al[202] conducted a systematic review of 19 studies (including most of the studies cited above) examining the differences in the gut microbial community in faecal samples from people diagnosed with CRC or adenomas and from people without colorectal neoplasia to generate faecal multi-bacterial models for early detection of adenomas and CRC. Overall, they concluded that there was limited but encouraging evidence that the differences in the faecal gut microbiome between people diagnosed with CRC or adenoma and healthy controls could be used to develop new faecal tests. The authors suggested that future research should focus on developing unified documented and reproducible protocols for studying the human gut microbiome from faecal samples for more comparable results between cohorts[202].
In many studies, patients had a corresponding decrease in the Firmicutes/Bacteroidetes ratio, which might be an important marker for intestinal dysbiosis in colorectal precancerous lesions. F. nucleatum, BFT-producing B. fragilis, and colibactin-producing E. coli are known to be associated with a more aggressive TNM stage[130,145,149,158]. Their detection by PCR in tissues or faeces might allow the development of rapid tests for new prognostic factors. With a 16S rRNA gene sequencing approach, F. nucleatum, B. fragilis and Faecalibacterium prausnitzii were identified as useful prognostic biomarkers for CRC[203]. Indeed, in this study, F. nucleatum and B. fragilis were more abundant in the groups with a worse prognosis, while Faecalibacterium prausnitzii was more abundant in the survival group. Similarly, Mima et al[158] showed that the amount of F. nucleatum DNA in colorectal cancer tissue was associated with shorter survival. Another study demonstrated that the detection of F. nucleatum associated with the non-invasive screening FIT could be a promising marker for detecting neoplastic lesions[204]. Recently, Eklöf et al[151] explored the use of microbial markers for bacteria harbouring the pks island, which codes for colibactin synthesis, and F. nucleatum in stool as potential screening markers for CRC. Authors suggested that the presence of the pks island and F. nucleatum detection could predict cancer with a specificity of 63.1% and a sensitivity of 84.6%, suggesting the potential value of these microbial parameters as putative non-invasive biomarkers for CRC detection[151]. Multicentric clinical studies need to be performed to validate all these promising results in a larger cohort. Moreover, these bacterial markers might provide a CRC-associated microbiome risk profile that might aid in the early identification of individuals who are at risk and require close surveillance.
Screening the faecal metabolome is a promising non-invasive procedure for obtaining a unique metabolic fingerprint to diagnose or determine the prognosis of CRC. To our knowledge, only a few studies with different metabolomics methods have shown the diagnostic potential of faecal samples for human colorectal cancer. They are summarized in Table 1. In 2009, Bezabeh et al[205] used 1H NMR to detect colorectal neoplasia in 111 CRC human faecal samples and compared them with samples from 412 healthy controls. NMR-based metabolic profiling of faecal water extracts from patients with colorectal cancer and healthy individuals was able to identify potential diagnostic markers, such as SCFA (acetate and butyrate) and amino acids (proline and cysteine)[206]. More recently, the analysis of lyophilized stools by HPLC-GC/MS-MS resulted in the identification of 41 relevant faecal metabolites, such as xenobiotics, heme, peptides/amino acids, vitamins and co-factors, that had increased or decreased concentrations in CRC samples[207]. Volatile metabolome profiling of faecal samples using GC/TOF-MS also had diagnostic potential for detecting colorectal cancer. Phua et al[208] evaluated a small cohort and identified three specific markers (fructose, linoleic acid, and nicotinic acid) that were found at lower levels in the faecal volatile metabolome of CRC patients than in that of healthy subjects. A similar study using an electronic nose on samples from 40 CRC patients, 60 advanced adenoma patients, and 57 healthy controls was able to strongly distinguish the faecal volatile organic compounds (VOC) profile of patients with CRC and advanced adenoma from controls[209]. Given the volatility of the analysed compounds, improper sample collection and storage can drastically reduce the repeatability and robustness of the method. Batty et al[210] proposed the use of selected ion flow tube mass spectrometry (SIFT-MS) to classify 62 human faecal samples with a positive result on FOBT. Indeed, their method showed a 75% correct discrimination between CRC samples and low risk samples[210]. More recently, Sinha et al[211] intended to establish a microbe-metabolite correlation through the analysis of lyophilized faecal samples of 42 CRC patients and 89 healthy controls using HPLC-GC/MS-MS. They reported that CRC was independently associated with lower levels of Clostridia, Lachnospiraceae, p-aminobenzoate and conjugated linoleate and with higher levels of Fusobacterium, Porphyromonas, p-hydroxy-benzaldehyde, and palmitoyl-sphingomyelin. The authors identified a strong microbe-metabolite correlation in CRC patients. In conclusion, the diagnostic potential of metabolic profiling of faeces is strongly supported by several human studies, but these analytical techniques require standard procedures in order to obtain comparable and robust high-quality data.
| Matrix | Cohort | Study observations | Analytical method | Ref. |
| Aqueous dispersion of stools | 111 CRC, 412 healthy controls | Potential to detect colorectal neoplasia | One-dimensional 1H magnetic resonance spectroscopy | [205] |
| Fecal water extract | 21 CRC 11 healthy controls | Reproducible and effective method for detecting colorectal cancer markers. (↘) SCFA (acetate, butyrate) appears to be the most effective marker in CRC. | NMR | [206] |
| Lyophilized human faeces | 11 CRC 10 healthy controls | (↘) butyric acid, linoleic acid, glycerol, (↘) secondary bile acid associated with (↘) Ruminococcus spp., (↗) leucine, valine, acetic acid, valeric acid, isobutyric acid, isovaleric acid, (↗) A. muciniphila associated with (↗) proline, serine, threonine | GC-MS | [193] |
| Lyophilized human faeces | 11 CRC, 10 healthy controls | (↘) fructose, linoleic acid, and nicotinic acid in CRC stools. | GC/TOF-MS | [208] |
| Volatile organic compounds in the headspace of lyophilized stool samples | 40 CRC, 60 advanced adenomas, 57 healthy controls | Discrimination of fecal VOC profiles of patients with adenomas and CRC. | Electronic nose | [209] |
| Lyophilized human faeces | 48 CRC, 102 healthy controls | 41 metabolites significantly associated with CRC (↘) xenobiotics (↘) heme, peptides/amino acids, vitamins, co-factors, other CRC associated molecules. | HPLC-GC/MS-MS | [207] |
| Human faeces | 31 CRC, 31 controls with positive FOBT | Discrimination of CRC samples with better specificity and sensitivity than FOBT. (↘) ammonia, sulfides, acetaldehyde | SIFT-MS | [210] |
| Lyophilized human faeces | 42 CRC, 89 healthy controls | Microbe-metabolite correlation in CRC: (↗) Clostridia, Lachnospiraceae, p-aminobenzoate and conjugated linoleate. (↗) Fusobacterium, Porphyromonas, p-hydroxy-benzaldehyde, and palmitoyl-sphingomyelin. | HPLC-GC/MS-MS | [211] |
| Human faeces | 13 CRC | Discrimination of CRC samples with 7 metabolites: alphahydroxyisovalerate, isovalerate, N1-methyl-2-pyridine-5-carboxamide, 7-ketodeoxycholate, deoxycholate, valerate, and tryptophylglycine | UPLC-MS/MS; GC/MS | [165] |
It has long been recognized that the gut microbiota can modify the pharmacokinetics of various drugs including anticancer therapies thus influencing therapeutic outcomes[212-217] and/or side effects[218-220]. Irinotecan, which is a first-line chemotherapeutic agent for metastatic CRC, causes adverse and dose-limiting effects that are largely influenced by bacterial β-glucuronidase[214,219]. Metagenomic and metabolomic profiling of patients’ gut microbiota could thus be informative before choosing this drug to predict side effects. Moreover, specific members of the gut microbiota might also drive chemoresistance to treatment. Indeed, Fusobacterium nucleatum, causes resistance to oxaliplatin and 5-FU through autophagy induction in colorectal cancer cell lines in vitro and in vivo in mouse models of colorectal xenografts[221]. Conversely, the sensitivity of cancer cells to many chemotherapeutic agents has been shown to be modulated by intra-tumoural microbial agents[212,215,216,221,222]. Moreover, the gut microbiota also has a long-term impact on both the efficacy and toxicity of anticancer therapies. In particular, it can modulate the concomitant anti-tumour immune response[223-227]. Indeed, various pre-clinical mouse models of subcutaneous tumours (melanoma, lung cancer, sarcoma) have suggested that specific crosstalk between bacteria and immune cells may affect responses to anticancer chemotherapies[223-227]. In terms of CRC in particular, the efficacy of oxaliplatin, which is a drug routinely used for CRC treatment, has been shown to be strongly reinforced by gut microbiota in mice that were transplanted with MC38 colorectal syngeneic tumours, due to myeloid cells with increased anti-tumour functions[224]. In addition, it has been described that the gut microbiota contributes to anti-tumour functions of adoptively transferred T cells in several models, including colorectal tumours implanted in mice[225,228,229]. This suggests that the patients’ microbiota should also be taken into account in future clinical studies involving infusion of autologous anticancer T cells.
Immune checkpoint therapies are often used in association with chemotherapies and can also be positively or negatively impacted by the gut microbiota in terms of toxicity[230-233] and therapeutic effect, as shown for anti-CTLA-4[231,233], anti-PD-L1[234] and anti-PD-1[235-237] antibodies. Interestingly, three clinical investigations demonstrated that the gut microbiota can be used to predict responder and non-responder patients to PD-1/PDL-1 immunotherapies in several solid epithelial tumours[235-237]. In terms of CRC, many studies have shown the capacity of the gut microbiota to modulate tumour-infiltrating myeloid cell[238-240] and T cell responses[241-244], while improvement in immunotherapy efficacy by the microbiota is mediated precisely by these immune pathways in solid human epithelial tumours (e.g., melanoma)[235-237]. Moreover, in a subcutaneous CRC mouse model, a positive impact of some microbial species on anti-CTLA-4 immunotherapy was observed[233]. Given the severe colitis observed in some patients receiving immunotherapies (e.g., antibodies to CTLA4 and PD-L1) and the role of gut microbes in colitis, it is possible that the gut microbiota influences this toxicity. Investigations are thus needed to elucidate the potential role of the microbiota on immunotherapy efficacy and toxicity in preclinical models and patients. Gut microbiota profiling of CRC patients might be useful in the future to predict treatment responses and/or side effects of cancer chemotherapies and immunotherapies. All these data could lead to “personalized medicine 2.0” that is being developed in an ongoing study investigating the impact of the intestinal microbiome on treatment responses and the toxicity of capecitabine or TAS-102 chemotherapies in patients with metastatic and/or irresectable CRC[245].
Perioperative antibiotic prophylaxis strategies targeting the gut microbiota are widely used in colorectal surgery. The aim of such approaches is to prevent both surgical site and wound infections, protecting against potential contamination by the microorganisms that are spread from surgically induced alterations of the intestinal epithelial barrier. However, these antibiotic prophylaxis strategies may also impair the beneficial impact of the gut microbiota on intestinal immune function and healing (for review[246]). Few studies have investigated the impact that bowel preparation prior to colorectal surgery may have on the mucosa-associated and luminal colonic microbiota[247]. After the suppression of beneficial bacteria, the host may lose its colonization resistance to pathogens and may have decreased local stimulation of systemic immunity and the healing process[248,249]. After surgery, if the commensal microbiota does not repopulate the tract, the situation can lead to the development of pathogenic bacteria such as C. difficile[250]. Moreover, data from animal and human studies have suggested that some of the bacteria present at the anastomotic site may respond to surgical stress by activating virulence pathways, which results in alterations in the healing process. An investigation of the faecal microbiota of patients before and after colorectal surgery for CRC revealed that Enterococcus faecalis and Pseudomonas aeruginosa were the dominant pathogens present postoperatively in the stools, with a several log-fold increase during the postoperative recovery period[251]. Postoperative sepsis (e.g., anastomotic leakage, perianastomotic abscess) is the most feared complication after colorectal surgery and is responsible for potentially deadly complications and significant alterations in the patient’s quality of life. The critical role of the intestinal microbiome in sepsis has been illustrated by studies on germ-free animals, which demonstrated improved survival in these animals compared with conventional animals[252]. Therefore, the restoration and/or maintenance of a microbiota favouring intestinal healing and preventing surgical site infections after colorectal surgery could be a promising approach for the development of new therapeutic strategies, thus targeting the gut microbiota to improve surgical outcomes. Moreover, the parallel development of tools, such as the “personalized microbiota composition analysis”, to be performed pre- and postoperatively to evaluate the clinical relevance of gut microbiota modulation to positively influence the clinical outcomes and to optimize the perioperative strategies appears promising.
The complexity of the microbiome turns the need for microbial marker-based diagnosis techniques into a real challenge. Numerous studies have reported associations between microbial markers, such as F. nucleatum, or colibactin-producing E. coli, and CRC, but to date, there is no universal microbial marker defined for CRC detection. Several limitations should be taken into consideration for the future development of new tests. First, the very high variability of the microbiota composition between individuals due to sex, age, diet, lifestyle, genetic background, medication use, ethnicity, or geographical location make finding a universal microbial marker almost impossible. Antibiotic therapy greatly influences the expression of microbial markers and is a critical limitation to microbial marker use. Moreover, standardization in terms sample collection and storage, RNA or metabolite extraction, sample analysis, and data processing is essential to compare studies. If studies that screen for microbiota composition use the same method of 16S rRNA sequencing, the use of metabolomics technologies will require exhaustive standardization between studies. Stary et al[253] shared a very interesting and detailed list of recommendations for using microbial markers for CRC screening. (1) Any studies should be prepared carefully, taking into account the recommendations and limitations of techniques published previously. (2) Validation of CRC screening markers on specific populations should be encouraged because differences in the gut microbiome are observed in different geographical locations or in different racial/ethnic groups. (3) Whenever possible, conventional culture should be used to confirm the findings from sequencing studies. Particularly, the candidate marker status of species or genes revealed by molecular techniques should be confirmed or refuted by culture and vice versa. Systematic high-throughput culturomics should be developed because cultivation represents an approach that is economical. (4) Screening techniques for CRC risk should evaluate all the known candidate markers, combining particular species, genotoxin production and possibly further strain characteristics whenever relevant. And (5) the potential for practical detection should always be considered. For example, tumour screening, which requires colonoscopy, is costly, uncomfortable for patient and is the gold standard tool for CRC screening. Faecal samples are better non-invasive specimens for developing these microbial-associated-markers. However, faeces are likely to contain a large number of microbial species unrelated to the disease site, which may introduce noise in the detection of potential biomarkers of the disease. Because some bacteria that are associated with CRC, such as F. nucleatum, are indigenous to the human oral cavity, analysing the oral microbiome may be an alternative screening method for CRC. Saliva is a biological fluid that may be suitable for biomarker detection. The oral microbial compositions may theoretically reflect the oral and general health status. Flemer et al[254] found that several oral taxa (such as Streptococcus and Prevotella spp.) were differentially abundant in CRC patients versus controls. Moreover, they developed a classification model based on the oral swab microbiota that distinguished individuals with CRC or polyps from controls (sensitivity: 53% CRC, 67% polyps; specificity: 96%). Importantly, when data from both the faecal and oral swab microbiota were considered in this model, the sensitivity increased to 76% for CRC and 88% for polyps. In addition to Stary’s recommendations, we showed that microbial markers might be different depending on the tumour CRC phenotype[157]. Moreover, a deeper understanding of the gut microbiota structure and function may help to identify several bacteria that when combined may provide a real CRC-associated microbial signature.
Finally, the therapeutic efficacy of anticancer drugs could also be improved by active modulation of the gut microbiota through the use of probiotics, prebiotics or specific inhibitors. This perspective is supported by the fact that immunotherapy resistance observed in germ-free mice, in antibiotic-treated mice, and in those that have previously received faecal microbiota transplantation (FMT) from non-responder patients, can be reversed by FMT from responder patients[233,235-237]. Moreover, it has been highlighted that specific bacterial species, such as Lactobacillus johnsonii[226], Enterococcus hirae[223,226] Barnesiella intestinihominis[223], Akkermansia muciniphila[237], Bacteroides[233] and Bifidobacterium[234,236] species, are beneficial in this context, which may lead the way to innovative “oncobiotics” strategies that combine anticancer and microbiota-targeting agents.
In this review, we discuss both predictive and prognostic microbial-associated markers identified in CRC. The faecal-associated microbiota may be dynamically linked to colon cancer, which, in turn, may offer evidence for microflora-associated diagnostic, preventive, and prognostic approaches for CRC. However, it is clear that additional clinical studies are necessary to validate these parameters to improve the diagnosis and therapeutic management of CRC.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: France
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Lorenzo-Zúñiga V, Mayol J, Sunakawa Y S- Editor: Gong ZM L- Editor: A E- Editor: Wang C
| 1. | Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1848] [Cited by in RCA: 2073] [Article Influence: 188.5] [Reference Citation Analysis (0)] |
| 2. | Rees CJ, Bevan R. The National Health Service Bowel Cancer Screening Program: the early years. Expert Rev Gastroenterol Hepatol. 2013;7:421-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Jones RP, Jackson R, Dunne DF, Malik HZ, Fenwick SW, Poston GJ, Ghaneh P. Systematic review and meta-analysis of follow-up after hepatectomy for colorectal liver metastases. Br J Surg. 2012;99:477-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 4. | O’Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420-1425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1063] [Cited by in RCA: 1162] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 5. | Winawer SJ. The history of colorectal cancer screening: a personal perspective. Dig Dis Sci. 2015;60:596-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 6. | Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 607] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 7. | Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1054] [Cited by in RCA: 1232] [Article Influence: 102.7] [Reference Citation Analysis (2)] |
| 8. | Gao R, Gao Z, Huang L, Qin H. Gut microbiota and colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017;36:757-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 9. | Yu YN, Fang JY. Gut Microbiota and Colorectal Cancer. Gastrointest Tumors. 2015;2:26-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 10. | Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 613] [Cited by in RCA: 556] [Article Influence: 61.8] [Reference Citation Analysis (5)] |
| 11. | Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 946] [Cited by in RCA: 1226] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 12. | Zhang J, Cheng Z, Ma Y, He C, Lu Y, Zhao Y, Chang X, Zhang Y, Bai Y, Cheng N. Effectiveness of Screening Modalities in Colorectal Cancer: A Network Meta-Analysis. Clin Colorectal Cancer. 2017;16:252-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 13. | Bevan R, Rutter MD. Colorectal Cancer Screening-Who, How, and When? Clin Endosc. 2018;51:37-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 14. | Bray C, Bell LN, Liang H, Collins D, Yale SH. Colorectal Cancer Screening. WMJ. 2017;116:27-33. [PubMed] |
| 15. | Hadjipetrou A, Anyfantakis D, Galanakis CG, Kastanakis M, Kastanakis S. Colorectal cancer, screening and primary care: A mini literature review. World J Gastroenterol. 2017;23:6049-6058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 79] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 16. | Hamzehzadeh L, Yousefi M, Ghaffari SH. Colorectal Cancer Screening: A Comprehensive Review to Recent Non-Invasive Methods. Int J Hematol Oncol Stem Cell Res. 2017;11:250-261. [PubMed] |
| 17. | Issa IA, Noureddine M. Colorectal cancer screening: An updated review of the available options. World J Gastroenterol. 2017;23:5086-5096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 434] [Cited by in RCA: 391] [Article Influence: 48.9] [Reference Citation Analysis (11)] |
| 18. | Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, Maida C, Cammarota G, Gasbarrini A, Scarpulla G. Screening of colorectal cancer: present and future. Expert Rev Anticancer Ther. 2017;17:1131-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 125] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 19. | Leuraud K, Jezewski-Serra D, Viguier J, Salines E. Colorectal cancer screening by guaiac faecal occult blood test in France: Evaluation of the programme two years after launching. Cancer Epidemiol. 2013;37:959-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 20. | Rutter MD, Nickerson C, Rees CJ, Patnick J, Blanks RG. Risk factors for adverse events related to polypectomy in the English Bowel Cancer Screening Programme. Endoscopy. 2014;46:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 144] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 21. | Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM; American College of Gastroenterology. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected]. Am J Gastroenterol. 2009;104:739-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 981] [Cited by in RCA: 1059] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 22. | Pox CP. Controversies in colorectal cancer screening. Digestion. 2014;89:274-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 23. | Sali L, Mascalchi M, Falchini M, Ventura L, Carozzi F, Castiglione G, Delsanto S, Mallardi B, Mantellini P, Milani S. Reduced and Full-Preparation CT Colonography, Fecal Immunochemical Test, and Colonoscopy for Population Screening of Colorectal Cancer: A Randomized Trial. J Natl Cancer Inst. 2015;108:pii: djv319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 24. | Pooler BD, Kim DH, Pickhardt PJ. Indeterminate but Likely Unimportant Extracolonic Findings at Screening CT Colonography (C-RADS Category E3): Incidence and Outcomes Data From a Clinical Screening Program. AJR Am J Roentgenol. 2016;207:996-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ. 2014;348:g2467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 663] [Cited by in RCA: 610] [Article Influence: 55.5] [Reference Citation Analysis (2)] |
| 26. | Vleugels JL, van Lanschot MC, Dekker E. Colorectal cancer screening by colonoscopy: putting it into perspective. Dig Endosc. 2016;28:250-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, Ederer F. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2174] [Article Influence: 67.9] [Reference Citation Analysis (1)] |
| 28. | Shaukat A, Mongin SJ, Geisser MS, Lederle FA, Bond JH, Mandel JS, Church TR. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369:1106-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 646] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 29. | Holme Ø, Bretthauer M, Fretheim A, Odgaard-Jensen J, Hoff G. Flexible sigmoidoscopy versus faecal occult blood testing for colorectal cancer screening in asymptomatic individuals. Cochrane Database Syst Rev. 2013;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 30. | Heresbach D, Manfredi S, D’halluin PN, Bretagne JF, Branger B. Review in depth and meta-analysis of controlled trials on colorectal cancer screening by faecal occult blood test. Eur J Gastroenterol Hepatol. 2006;18:427-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Pitkäniemi J, Seppä K, Hakama M, Malminiemi O, Palva T, Vuoristo MS, Järvinen H, Paimela H, Pikkarainen P, Anttila A. Effectiveness of screening for colorectal cancer with a faecal occult-blood test, in Finland. BMJ Open Gastroenterol. 2015;2:e000034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Katičić M, Antoljak N, Kujundžić M, Stamenić V, Skoko Poljak D, Kramarić D, Stimac D, Strnad Pešikan M, Samija M, Ebling Z. Results of National Colorectal Cancer Screening Program in Croatia (2007-2011). World J Gastroenterol. 2012;18:4300-4307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Steele RJ, McClements PL, Libby G, Black R, Morton C, Birrell J, Mowat NA, Wilson JA, Kenicer M, Carey FA. Results from the first three rounds of the Scottish demonstration pilot of FOBT screening for colorectal cancer. Gut. 2009;58:530-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 132] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 34. | Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, Levin TR, Lieberman D, Robertson DJ. Colorectal Cancer Screening: Recommendations for Physicians and Patients From the U.S. Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2017;153:307-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 513] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 35. | Navarro M, Nicolas A, Ferrandez A, Lanas A. Colorectal cancer population screening programs worldwide in 2016: An update. World J Gastroenterol. 2017;23:3632-3642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 374] [Cited by in RCA: 409] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 36. | Hewitson P, Glasziou P, Irwig L, Towler B, Watson E. Screening for colorectal cancer using the faecal occult blood test, Hemoccult. Cochrane Database Syst Rev. 2007;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 279] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 37. | Li R, Liu J, Xue H, Huang G. Diagnostic value of fecal tumor M2-pyruvate kinase for CRC screening: a systematic review and meta-analysis. Int J Cancer. 2012;131:1837-1845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Haug U, Rothenbacher D, Wente MN, Seiler CM, Stegmaier C, Brenner H. Tumour M2-PK as a stool marker for colorectal cancer: comparative analysis in a large sample of unselected older adults vs colorectal cancer patients. Br J Cancer. 2007;96:1329-1334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Tonus C, Sellinger M, Koss K, Neupert G. Faecal pyruvate kinase isoenzyme type M2 for colorectal cancer screening: a meta-analysis. World J Gastroenterol. 2012;18:4004-4011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 39] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME; Colorectal Cancer Study Group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704-2714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 525] [Article Influence: 25.0] [Reference Citation Analysis (3)] |
| 41. | Schreuders EH, Grobbee EJ, Spaander MC, Kuipers EJ. Advances in Fecal Tests for Colorectal Cancer Screening. Curr Treat Options Gastroenterol. 2016;14:152-162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 42. | Ahlquist DA. Multi-target stool DNA test: a new high bar for noninvasive screening. Dig Dis Sci. 2015;60:623-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 43. | Dhaliwal A, Vlachostergios PJ, Oikonomou KG, Moshenyat Y. Fecal DNA testing for colorectal cancer screening: Molecular targets and perspectives. World J Gastrointest Oncol. 2015;7:178-183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 44. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1239] [Article Influence: 112.6] [Reference Citation Analysis (1)] |
| 45. | Knudsen AB, Zauber AG, Rutter CM, Naber SK, Doria-Rose VP, Pabiniak C, Johanson C, Fischer SE, Lansdorp-Vogelaar I, Kuntz KM. Estimation of Benefits, Burden, and Harms of Colorectal Cancer Screening Strategies: Modeling Study for the US Preventive Services Task Force. JAMA. 2016;315:2595-2609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 377] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 46. | Hauptman N, Glavač D. Colorectal Cancer Blood-Based Biomarkers. Gastroenterol Res Pract. 2017;2017:2195361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 47. | Hench IB, Hench J, Tolnay M. Liquid Biopsy in Clinical Management of Breast, Lung, and Colorectal Cancer. Front Med (Lausanne). 2018;5:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 48. | Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017;8:55632-55645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 49. | Nordgård O, Tjensvoll K, Gilje B, Søreide K. Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Br J Surg. 2018;105:e110-e120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 50. | Law WL, Choi HK, Lee YM, Ho JW. The impact of postoperative complications on long-term outcomes following curative resection for colorectal cancer. Ann Surg Oncol. 2007;14:2559-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 186] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 51. | McArdle CS, McMillan DC, Hole DJ. Impact of anastomotic leakage on long-term survival of patients undergoing curative resection for colorectal cancer. Br J Surg. 2005;92:1150-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 352] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 52. | Nespoli A, Gianotti L, Totis M, Bovo G, Nespoli L, Chiodini P, Brivio F. Correlation between postoperative infections and long-term survival after colorectal resection for cancer. Tumori. 2004;90:485-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 53. | Mynster T, Christensen IJ, Moesgaard F, Nielsen HJ. Effects of the combination of blood transfusion and postoperative infectious complications on prognosis after surgery for colorectal cancer. Danish RANX05 Colorectal Cancer Study Group. Br J Surg. 2000;87:1553-1562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 130] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 54. | Adloff M, Arnaud JP, Ollier JC, Schloegel M. [Can the prognosis of patients treated surgically in cancer of the rectum or colon be improved by follow-up? Prospective study of 909 patients]. Chirurgie. 1989;115:228-36; discussion 236-7. [PubMed] |
| 55. | Moertel CG, Fleming TR, Macdonald JS, Haller DG, Laurie JA, Goodman PJ, Ungerleider JS, Emerson WA, Tormey DC, Glick JH. Levamisole and fluorouracil for adjuvant therapy of resected colon carcinoma. N Engl J Med. 1990;322:352-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1678] [Cited by in RCA: 1625] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 56. | Greene FL, Stewart AK, Norton HJ. A new TNM staging strategy for node-positive (stage III) colon cancer: an analysis of 50,042 patients. Ann Surg. 2002;236:416-21; discussion 421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 163] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 57. | Compton CC, Fielding LP, Burgart LJ, Conley B, Cooper HS, Hamilton SR, Hammond ME, Henson DE, Hutter RV, Nagle RB. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:979-994. [PubMed] |
| 58. | Compton CC. Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol. 2003;16:376-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 278] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 59. | Huh JW, Kim HR, Kim YJ. Lymphovascular or perineural invasion may predict lymph node metastasis in patients with T1 and T2 colorectal cancer. J Gastrointest Surg. 2010;14:1074-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 60. | Liebig C, Ayala G, Wilks J, Verstovsek G, Liu H, Agarwal N, Berger DH, Albo D. Perineural invasion is an independent predictor of outcome in colorectal cancer. J Clin Oncol. 2009;27:5131-5137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 369] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 61. | Nakamura T, Mitomi H, Kikuchi S, Ohtani Y, Sato K. Evaluation of the usefulness of tumor budding on the prediction of metastasis to the lung and liver after curative excision of colorectal cancer. Hepatogastroenterology. 2005;52:1432-1435. [PubMed] |
| 62. | Zlobec I, Lugli A, Baker K, Roth S, Minoo P, Hayashi S, Terracciano L, Jass JR. Role of APAF-1, E-cadherin and peritumoral lymphocytic infiltration in tumour budding in colorectal cancer. J Pathol. 2007;212:260-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 107] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 63. | Purdie CA, Piris J. Histopathological grade, mucinous differentiation and DNA ploidy in relation to prognosis in colorectal carcinoma. Histopathology. 2000;36:121-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 64. | Cho YB, Chun HK, Yun HR, Kim HC, Yun SH, Lee WY. Histological grade predicts survival time associated with recurrence after resection for colorectal cancer. Hepatogastroenterology. 2009;56:1335-1340. [PubMed] |
| 65. | Zarbo RJ, Nakhleh RE, Brown RD, Kubus JJ, Ma CK, Mackowiak P. Prognostic significance of DNA ploidy and proliferation in 309 colorectal carcinomas as determined by two-color multiparametric DNA flow cytometry. Cancer. 1997;79:2073-2086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 66. | Ohtani H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PubMed] |
| 67. | Baker K, Zlobec I, Tornillo L, Terracciano L, Jass JR, Lugli A. Differential significance of tumour infiltrating lymphocytes in sporadic mismatch repair deficient versus proficient colorectal cancers: a potential role for dysregulation of the transforming growth factor-beta pathway. Eur J Cancer. 2007;43:624-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 68. | Jantscheff P, Terracciano L, Lowy A, Glatz-Krieger K, Grunert F, Micheel B, Brümmer J, Laffer U, Metzger U, Herrmann R. Expression of CEACAM6 in resectable colorectal cancer: a factor of independent prognostic significance. J Clin Oncol. 2003;21:3638-3646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 132] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 69. | Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 2005;41:2060-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 123] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 70. | Shibata D, Reale MA, Lavin P, Silverman M, Fearon ER, Steele G Jr, Jessup JM, Loda M, Summerhayes IC. The DCC protein and prognosis in colorectal cancer. N Engl J Med. 1996;335:1727-1732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 198] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 71. | Schwandner O, Schiedeck TH, Bruch HP, Duchrow M, Windhoevel U, Broll R. p53 and Bcl-2 as significant predictors of recurrence and survival in rectal cancer. Eur J Cancer. 2000;36:348-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 72. | Mulder JW, Kruyt PM, Sewnath M, Oosting J, Seldenrijk CA, Weidema WF, Offerhaus GJ, Pals ST. Colorectal cancer prognosis and expression of exon-v6-containing CD44 proteins. Lancet. 1994;344:1470-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 239] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 73. | Koopman M, Venderbosch S, Nagtegaal ID, van Krieken JH, Punt CJ. A review on the use of molecular markers of cytotoxic therapy for colorectal cancer, what have we learned? Eur J Cancer. 2009;45:1935-1949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 74. | Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 978] [Cited by in RCA: 988] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 75. | Frankel TL, Vakiani E, Nathan H, DeMatteo RP, Kingham TP, Allen PJ, Jarnagin WR, Kemeny NE, Solit DB, D’Angelica MI. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer. 2017;123:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 76. | Karagkounis G, Torbenson MS, Daniel HD, Azad NS, Diaz LA Jr, Donehower RC, Hirose K, Ahuja N, Pawlik TM, Choti MA. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer. 2013;119:4137-4144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 77. | Stremitzer S, Stift J, Gruenberger B, Tamandl D, Aschacher T, Wolf B, Wrba F, Gruenberger T. KRAS status and outcome of liver resection after neoadjuvant chemotherapy including bevacizumab. Br J Surg. 2012;99:1575-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 78. | Vauthey JN, Zimmitti G, Kopetz SE, Shindoh J, Chen SS, Andreou A, Curley SA, Aloia TA, Maru DM. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg. 2013;258:619-26; discussion 626-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 286] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 79. | Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931-5937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 464] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 80. | Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, Agarwal A, Maru DM, Sieber O, Desai J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623-4632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 597] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 81. | Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011-2019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1314] [Cited by in RCA: 1452] [Article Influence: 103.7] [Reference Citation Analysis (0)] |
| 82. | Gafà R, Maestri I, Matteuzzi M, Santini A, Ferretti S, Cavazzini L, Lanza G. Sporadic colorectal adenocarcinomas with high-frequency microsatellite instability. Cancer. 2000;89:2025-2037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 83. | Benatti P, Gafà R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res. 2005;11:8332-8340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 282] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 84. | Bendardaf R, Lamlum H, Ristamäki R, Korkeila E, Syrjänen K, Pyrhönen S. Mismatch repair status is a predictive factor of tumour response to 5-fluorouracil and irinotecan chemotherapy in patients with advanced colorectal cancer. Tumour Biol. 2007;28:212-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 85. | Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 340] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 86. | Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, Piñol V, Xicola RM, Bujanda L, Reñé JM. Mismatch repair status in the prediction of benefit from adjuvant fluorouracil chemotherapy in colorectal cancer. Gut. 2006;55:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 171] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 87. | Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1585] [Cited by in RCA: 1634] [Article Influence: 74.3] [Reference Citation Analysis (0)] |
| 88. | Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 1161] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 89. | Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2504] [Cited by in RCA: 2403] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 90. | Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1610] [Cited by in RCA: 1731] [Article Influence: 144.3] [Reference Citation Analysis (0)] |
| 91. | Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2901] [Cited by in RCA: 3122] [Article Influence: 195.1] [Reference Citation Analysis (1)] |
| 92. | Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, Cabiddu M, Iacovelli R, Bossi I, Lonati V. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015;51:587-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 373] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 93. | Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, Sorich MJ. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112:1888-1894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 237] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 94. | van Brummelen EMJ, de Boer A, Beijnen JH, Schellens JHM. BRAF Mutations as Predictive Biomarker for Response to Anti-EGFR Monoclonal Antibodies. Oncologist. 2017;22:864-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 95. | McMillan DC, Wotherspoon HA, Fearon KC, Sturgeon C, Cooke TG, McArdle CS. A prospective study of tumor recurrence and the acute-phase response after apparently curative colorectal cancer surgery. Am J Surg. 1995;170:319-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 115] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 96. | Canna K, McMillan DC, McKee RF, McNicol AM, Horgan PG, McArdle CS. Evaluation of a cumulative prognostic score based on the systemic inflammatory response in patients undergoing potentially curative surgery for colorectal cancer. Br J Cancer. 2004;90:1707-1709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 97. | Crozier JE, McKee RF, McArdle CS, Angerson WJ, Anderson JH, Horgan PG, McMillan DC. The presence of a systemic inflammatory response predicts poorer survival in patients receiving adjuvant 5-FU chemotherapy following potentially curative resection for colorectal cancer. Br J Cancer. 2006;94:1833-1836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 98. | Al-Shaiba R, McMillan DC, Angerson WJ, Leen E, McArdle CS, Horgan P. The relationship between hypoalbuminaemia, tumour volume and the systemic inflammatory response in patients with colorectal liver metastases. Br J Cancer. 2004;91:205-207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 86] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 99. | McMillan DC, Watson WS, O’Gorman P, Preston T, Scott HR, McArdle CS. Albumin concentrations are primarily determined by the body cell mass and the systemic inflammatory response in cancer patients with weight loss. Nutr Cancer. 2001;39:210-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 349] [Article Influence: 15.2] [Reference Citation Analysis (1)] |
| 100. | Cengiz O, Kocer B, Sürmeli S, Santicky MJ, Soran A. Are pretreatment serum albumin and cholesterol levels prognostic tools in patients with colorectal carcinoma? Med Sci Monit. 2006;12:CR240-CR247. [PubMed] |
| 101. | McMillan DC, Canna K, McArdle CS. Systemic inflammatory response predicts survival following curative resection of colorectal cancer. Br J Surg. 2003;90:215-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 360] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 102. | Takada S, Namiki M, Takahara S, Matsumiya K, Kondoh N, Kokado Y, Matsumoto K, Nakamura T, Okuyama A. Serum HGF levels in acute renal rejection after living related renal transplantation. Transpl Int. 1996;9:151-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 103. | Harrison LE, Guillem JG, Paty P, Cohen AM. Preoperative carcinoembryonic antigen predicts outcomes in node-negative colon cancer patients: a multivariate analysis of 572 patients. J Am Coll Surg. 1997;185:55-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 81] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 104. | Hohenberger P, Schlag PM, Gerneth T, Herfarth C. Pre- and postoperative carcinoembryonic antigen determinations in hepatic resection for colorectal metastases. Predictive value and implications for adjuvant treatment based on multivariate analysis. Ann Surg. 1994;219:135-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 78] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 105. | Malapelle U, Mayo de-Las-Casas C, Rocco D, Garzon M, Pisapia P, Jordana-Ariza N, Russo M, Sgariglia R, De Luca C, Pepe F. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer. 2017;116:802-810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 106. | Ng JM, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci. 2015;16:2472-2496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 107. | Basnet S, Zhang ZY, Liao WQ, Li SH, Li PS, Ge HY. The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis. J Cancer. 2016;7:1105-1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 108. | El Messaoudi S, Mouliere F, Du Manoir S, Bascoul-Mollevi C, Gillet B, Nouaille M, Fiess C, Crapez E, Bibeau F, Theillet C. Circulating DNA as a Strong Multimarker Prognostic Tool for Metastatic Colorectal Cancer Patient Management Care. Clin Cancer Res. 2016;22:3067-3077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 131] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 109. | Kitahara M, Hazama S, Tsunedomi R, Takenouchi H, Kanekiyo S, Inoue Y, Nakajima M, Tomochika S, Tokuhisa Y, Iida M. Prediction of the efficacy of immunotherapy by measuring the integrity of cell-free DNA in plasma in colorectal cancer. Cancer Sci. 2016;107:1825-1829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 110. | Toledo RA, Cubillo A, Vega E, Garralda E, Alvarez R, de la Varga LU, Pascual JR, Sánchez G, Sarno F, Prieto SH. Clinical validation of prospective liquid biopsy monitoring in patients with wild-type RAS metastatic colorectal cancer treated with FOLFIRI-cetuximab. Oncotarget. 2017;8:35289-35300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 111. | LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 790] [Cited by in RCA: 959] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 112. | Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5:e73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 596] [Cited by in RCA: 885] [Article Influence: 98.3] [Reference Citation Analysis (0)] |
| 113. | Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189-200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1566] [Cited by in RCA: 2366] [Article Influence: 262.9] [Reference Citation Analysis (0)] |
| 114. | Natividad JM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res. 2013;69:42-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 330] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 115. | Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2011] [Cited by in RCA: 2402] [Article Influence: 200.2] [Reference Citation Analysis (0)] |
| 116. | Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974-977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1311] [Cited by in RCA: 1209] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 117. | Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1048] [Cited by in RCA: 944] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 118. | Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 303] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 119. | Matsumoto M, Sakamoto M, Hayashi H, Benno Y. Novel phylogenetic assignment database for terminal-restriction fragment length polymorphism analysis of human colonic microbiota. J Microbiol Methods. 2005;61:305-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 120. | Dehingia M, Devi KT, Talukdar NC, Talukdar R, Reddy N, Mande SS, Deka M, Khan MR. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci Rep. 2015;5:18563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 121. | Sagheddu V, Patrone V, Miragoli F, Morelli L. Abundance and Diversity of Hydrogenotrophic Microorganisms in the Infant Gut before the Weaning Period Assessed by Denaturing Gradient Gel Electrophoresis and Quantitative PCR. Front Nutr. 2017;4:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 122. | Swidsinski A, Dörffel Y, Loening-Baucke V, Gille C, Reißhauer A, Göktas O, Krüger M, Neuhaus J, Schrödl W. Impact of humic acids on the colonic microbiome in healthy volunteers. World J Gastroenterol. 2017;23:885-890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 123. | Balamurugan R, Janardhan HP, George S, Chittaranjan SP, Ramakrishna BS. Bacterial succession in the colon during childhood and adolescence: molecular studies in a southern Indian village. Am J Clin Nutr. 2008;88:1643-1647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 124. | Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 630] [Cited by in RCA: 620] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 125. | Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 967] [Article Influence: 96.7] [Reference Citation Analysis (0)] |
| 126. | Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 781] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 127. | Amitay EL, Werner S, Vital M, Pieper DH, Höfler D, Gierse IJ, Butt J, Balavarca Y, Cuk K, Brenner H. Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. 2017;38:781-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 128. | Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 558] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 129. | Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010;9:249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 171] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 130. | Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 450] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 131. | Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 651] [Cited by in RCA: 855] [Article Influence: 77.7] [Reference Citation Analysis (0)] |
| 132. | Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015;7:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 175] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 133. | Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM. Enterotypes of the human gut microbiome. Nature. 2011;473:174-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5822] [Cited by in RCA: 5032] [Article Influence: 359.4] [Reference Citation Analysis (2)] |
| 134. | Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, Moris F, Rodrigo L, Mira A, Collado MC. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 196] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 135. | Lu Y, Chen J, Zheng J, Hu G, Wang J, Huang C, Lou L, Wang X, Zeng Y. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep. 2016;6:26337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 136. | Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, Wettergren Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017;4:e000145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 255] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 137. | Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, Randall TA, Galanko J, Benson A, Sandler RS. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012;6:1858-1868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 169] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 138. | Goedert JJ, Gong Y, Hua X, Zhong H, He Y, Peng P, Yu G, Wang W, Ravel J, Shi J. Fecal Microbiota Characteristics of Patients with Colorectal Adenoma Detected by Screening: A Population-based Study. EBioMedicine. 2015;2:597-603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 139. | Lucas C, Barnich N, Nguyen HTT. Microbiota, Inflammation and Colorectal Cancer. Int J Mol Sci. 2017;18:pii: E1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 232] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 140. | Zackular JP, Rogers MA, Ruffin MT 4th, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7:1112-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 395] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 141. | Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, Fodor AA, Jobin C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5:4724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 287] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 142. | Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 710] [Article Influence: 59.2] [Reference Citation Analysis (1)] |
| 143. | Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 788] [Cited by in RCA: 735] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 144. | Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 343] [Article Influence: 28.6] [Reference Citation Analysis (1)] |
| 145. | Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Déchelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20:859-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 330] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 146. | Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1374] [Cited by in RCA: 1664] [Article Influence: 128.0] [Reference Citation Analysis (1)] |
| 147. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1498] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 148. | Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 442] [Cited by in RCA: 429] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 149. | Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 2015;10:e0119462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 195] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 150. | Keenan JI, Aitchison A, Purcell RV, Greenlees R, Pearson JF, Frizelle FA. Screening for enterotoxigenic Bacteroides fragilis in stool samples. Anaerobe. 2016;40:50-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 151. | Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, Alexeyev O, Rutegård J, Wikberg ML, Palmqvist R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141:2528-2536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 152. | Prorok-Hamon M, Friswell MK, Alswied A, Roberts CL, Song F, Flanagan PK, Knight P, Codling C, Marchesi JR, Winstanley C. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63:761-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 153. | Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 558] [Cited by in RCA: 512] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 154. | Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 761] [Cited by in RCA: 788] [Article Influence: 112.6] [Reference Citation Analysis (0)] |
| 155. | Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, O’Riordain M, Shanahan F, O’Toole PW. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633-643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 427] [Cited by in RCA: 564] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 156. | Richard ML, Liguori G, Lamas B, Brandi G, da Costa G, Hoffmann TW, Pierluigi Di Simone M, Calabrese C, Poggioli G, Langella P. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes. 2017; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 157. | Gagnière J, Bonnin V, Jarrousse AS, Cardamone E, Agus A, Uhrhammer N, Sauvanet P, Déchelotte P, Barnich N, Bonnet R. Interactions between microsatellite instability and human gut colonization by Escherichia coli in colorectal cancer. Clin Sci (Lond). 2017;131:471-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 158. | Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973-1980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 754] [Article Influence: 83.8] [Reference Citation Analysis (0)] |
| 159. | Wishart DS. Current progress in computational metabolomics. Brief Bioinform. 2007;8:279-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 160. | Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 595] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 161. | Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 897] [Cited by in RCA: 794] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 162. | Dumas ME, Maibaum EC, Teague C, Ueshima H, Zhou B, Lindon JC, Nicholson JK, Stamler J, Elliott P, Chan Q. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP Study. Anal Chem. 2006;78:2199-2208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 252] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 163. | Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J Pharm Biomed Anal. 2014;87:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 249] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 164. | Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J Proteome Res. 2009;8:352-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 350] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 165. | Brown DG, Rao S, Weir TL, O’Malia J, Bazan M, Brown RJ, Ryan EP. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016;4:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 193] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 166. | Farshidfar F, Weljie AM, Kopciuk KA, Hilsden R, McGregor SE, Buie WD, MacLean A, Vogel HJ, Bathe OF. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer. 2016;115:848-857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 167. | Farshidfar F, Kopciuk KA, Hilsden R, McGregor SE, Mazurak VC, Buie WD, MacLean A, Vogel HJ, Bathe OF. A quantitative multimodal metabolomic assay for colorectal cancer. BMC Cancer. 2018;18:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 168. | Uchiyama K, Yagi N, Mizushima K, Higashimura Y, Hirai Y, Okayama T, Yoshida N, Katada K, Kamada K, Handa O. Serum metabolomics analysis for early detection of colorectal cancer. J Gastroenterol. 2017;52:677-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 169. | Mal M. Noninvasive metabolic profiling for painless diagnosis of human diseases and disorders. Future Sci OA. 2016;2:FSO106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 170. | O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 774] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 171. | Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1207] [Cited by in RCA: 1406] [Article Influence: 108.2] [Reference Citation Analysis (0)] |
| 172. | Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57:1-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 993] [Cited by in RCA: 1606] [Article Influence: 200.8] [Reference Citation Analysis (0)] |
| 173. | Ouwehand AC, Derrien M, de Vos W, Tiihonen K, Rautonen N. Prebiotics and other microbial substrates for gut functionality. Curr Opin Biotechnol. 2005;16:212-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 106] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 174. | Miller TL, Wolin MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol. 1996;62:1589-1592. [PubMed] |
| 175. | van der Beek CM, Dejong CHC, Troost FJ, Masclee AAM, Lenaerts K. Role of short-chain fatty acids in colonic inflammation, carcinogenesis, and mucosal protection and healing. Nutr Rev. 2017;75:286-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 247] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 176. | Barrasa JI, Olmo N, Lizarbe MA, Turnay J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol In Vitro. 2013;27:964-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 137] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 177. | Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1644] [Cited by in RCA: 2037] [Article Influence: 101.9] [Reference Citation Analysis (0)] |
| 178. | Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580-13585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 773] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 179. | Ocvirk S, O’Keefe SJ. Influence of Bile Acids on Colorectal Cancer Risk: Potential Mechanisms Mediated by Diet - Gut Microbiota Interactions. Curr Nutr Rep. 2017;6:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 180. | Manach C, Scalbert A, Morand C, Rémésy C, Jiménez L. Polyphenols: food sources and bioavailability. Am J Clin Nutr. 2004;79:727-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5019] [Cited by in RCA: 4518] [Article Influence: 215.1] [Reference Citation Analysis (0)] |
| 181. | Marín L, Miguélez EM, Villar CJ, Lombó F. Bioavailability of dietary polyphenols and gut microbiota metabolism: antimicrobial properties. Biomed Res Int. 2015;2015:905215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 445] [Cited by in RCA: 499] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 182. | Russell W, Duthie G. Plant secondary metabolites and gut health: the case for phenolic acids. Proc Nutr Soc. 2011;70:389-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 183. | Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutr Biochem. 2013;24:1415-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 1044] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 184. | Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, Flint HJ. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57:523-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 300] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 185. | Loh YH, Jakszyn P, Luben RN, Mulligan AA, Mitrou PN, Khaw KT. N-Nitroso compounds and cancer incidence: the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am J Clin Nutr. 2011;93:1053-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 143] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 186. | Gill CI, Rowland IR. Diet and cancer: assessing the risk. Br J Nutr. 2002;88 Suppl 1:S73-S87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 125] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 187. | Roediger WE, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci. 1997;42:1571-1579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 198] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 188. | Homann N. Alcohol and upper gastrointestinal tract cancer: the role of local acetaldehyde production. Addict Biol. 2001;6:309-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 52] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 189. | Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF, Felding BH, Ivanisevic J, Cho K, Wick EC, Hechenbleikner EM. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 272] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 190. | Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70:443-459. [PubMed] |
| 191. | Suarez F, Furne J, Springfield J, Levitt M. Insights into human colonic physiology obtained from the study of flatus composition. Am J Physiol. 1997;272:G1028-G1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 192. | Williams MD, Xian L, Huso T, Park JJ, Huso D, Cope LM, Gang DR, Siems WF, Resar L, Reeves R. Fecal Metabolome in Hmga1 Transgenic Mice with Polyposis: Evidence for Potential Screen for Early Detection of Precursor Lesions in Colorectal Cancer. J Proteome Res. 2016;15:4176-4187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 193. | Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8:e70803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 414] [Cited by in RCA: 491] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 194. | Weaver GA, Krause JA, Miller TL, Wolin MJ. Short chain fatty acid distributions of enema samples from a sigmoidoscopy population: an association of high acetate and low butyrate ratios with adenomatous polyps and colon cancer. Gut. 1988;29:1539-1543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 100] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 195. | Kim DH, Jin YH. Intestinal bacterial beta-glucuronidase activity of patients with colon cancer. Arch Pharm Res. 2001;24:564-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 135] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 196. | Ou J, DeLany JP, Zhang M, Sharma S, O’Keefe SJ. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr Cancer. 2012;64:34-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 197. | Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1318] [Cited by in RCA: 1649] [Article Influence: 137.4] [Reference Citation Analysis (0)] |
| 198. | Roisin Hughes IR. Metabolic Activities of the Gut Microflora in Relation to Cancer. Microb Ecol Health Dis. 2000;12:179-185. [RCA] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 199. | Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroenterol Hepatol. 2012;9:504-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 200. | Ai L, Tian H, Chen Z, Chen H, Xu J, Fang JY. Systematic evaluation of supervised classifiers for fecal microbiota-based prediction of colorectal cancer. Oncotarget. 2017;8:9546-9556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 201. | Shah MS, DeSantis TZ, Weinmaier T, McMurdie PJ, Cope JL, Altrichter A, Yamal JM, Hollister EB. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67:882-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 202. | Amitay EL, Krilaviciute A, Brenner H. Systematic review: Gut microbiota in fecal samples and detection of colorectal neoplasms. Gut Microbes. 2018;1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 203. | Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, Li J, Zhang D, Zhou Y. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget. 2016;7:46158-46172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 162] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 204. | Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441-1448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 205. | Bezabeh T, Somorjai R, Dolenko B, Bryskina N, Levin B, Bernstein CN, Jeyarajah E, Steinhart AH, Rubin DT, Smith IC. Detecting colorectal cancer by 1H magnetic resonance spectroscopy of fecal extracts. NMR Biomed. 2009;22:593-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 206. | Monleón D, Morales JM, Barrasa A, López JA, Vázquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009;22:342-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 146] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 207. | Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014;35:2089-2096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 118] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 208. | Phua LC, Chue XP, Koh PK, Cheah PY, Ho HK, Chan EC. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol Ther. 2014;15:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 209. | de Meij TG, Larbi IB, van der Schee MP, Lentferink YE, Paff T, Terhaar Sive Droste JS, Mulder CJ, van Bodegraven AA, de Boer NK. Electronic nose can discriminate colorectal carcinoma and advanced adenomas by fecal volatile biomarker analysis: proof of principle study. Int J Cancer. 2014;134:1132-1138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 210. | Batty CA, Cauchi M, Lourenço C, Hunter JO, Turner C. Use of the Analysis of the Volatile Faecal Metabolome in Screening for Colorectal Cancer. PLoS One. 2015;10:e0130301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 211. | Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016;11:e0152126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 168] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 212. | Bronckaers A, Balzarini J, Liekens S. The cytostatic activity of pyrimidine nucleosides is strongly modulated by Mycoplasma hyorhinis infection: Implications for cancer therapy. Biochem Pharmacol. 2008;76:188-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 213. | Espín JC, González-Sarrías A, Tomás-Barberán FA. The gut microbiota: A key factor in the therapeutic effects of (poly)phenols. Biochem Pharmacol. 2017;139:82-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 410] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 214. | Guthrie L, Gupta S, Daily J, Kelly L. Human microbiome signatures of differential colorectal cancer drug metabolism. NPJ Biofilms Microbiomes. 2017;3:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 215. | Vande Voorde J, Sabuncuoğlu S, Noppen S, Hofer A, Ranjbarian F, Fieuws S, Balzarini J, Liekens S. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J Biol Chem. 2014;289:13054-13065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 121] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 216. | Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 1199] [Article Influence: 149.9] [Reference Citation Analysis (0)] |
| 217. | Geller LT, Straussman R. Intratumoral bacteria may elicit chemoresistance by metabolizing anticancer agents. Mol Cell Oncol. 2017;5:e1405139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 218. | Takasuna K, Hagiwara T, Hirohashi M, Kato M, Nomura M, Nagai E, Yokoi T, Kamataki T. Involvement of beta-glucuronidase in intestinal microflora in the intestinal toxicity of the antitumor camptothecin derivative irinotecan hydrochloride (CPT-11) in rats. Cancer Res. 1996;56:3752-3757. [PubMed] |
| 219. | Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831-835. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 863] [Cited by in RCA: 800] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 220. | Nakayama H, Kinouchi T, Kataoka K, Akimoto S, Matsuda Y, Ohnishi Y. Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl)uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics. 1997;7:35-43. [PubMed] |
| 221. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-563.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 1473] [Article Influence: 184.1] [Reference Citation Analysis (0)] |
| 222. | Lehouritis P, Cummins J, Stanton M, Murphy CT, McCarthy FO, Reid G, Urbaniak C, Byrne WL, Tangney M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci Rep. 2015;5:14554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 216] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 223. | Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity. 2016;45:931-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 630] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 224. | Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1253] [Cited by in RCA: 1669] [Article Influence: 139.1] [Reference Citation Analysis (0)] |
| 225. | Kuczma MP, Ding ZC, Li T, Habtetsion T, Chen T, Hao Z, Bryan L, Singh N, Kochenderfer JN, Zhou G. The impact of antibiotic usage on the efficacy of chemoimmunotherapy is contingent on the source of tumor-reactive T cells. Oncotarget. 2017;8:111931-111942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 226. | Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1147] [Cited by in RCA: 1542] [Article Influence: 128.5] [Reference Citation Analysis (0)] |
| 227. | Xu X, Zhang X. Effects of cyclophosphamide on immune system and gut microbiota in mice. Microbiol Res. 2015;171:97-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 228. | Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, Palmer DC, Boni A, Muranski P, Yu Z. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117:2197-2204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 475] [Cited by in RCA: 448] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 229. | Uribe-Herranz M, Bittinger K, Rafail S, Guedan S, Pierini S, Tanes C, Ganetsky A, Morgan MA, Gill S, Tanyi JL. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight. 2018;3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 230. | Carbonnel F, Soularue E, Coutzac C, Chaput N, Mateus C, Lepage P, Robert C. Inflammatory bowel disease and cancer response due to anti-CTLA-4: is it in the flora? Semin Immunopathol. 2017;39:327-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 231. | Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 585] [Cited by in RCA: 920] [Article Influence: 131.4] [Reference Citation Analysis (0)] |
| 232. | Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, No D, Gobourne A, Littmann E, Huttenhower C. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 560] [Cited by in RCA: 769] [Article Influence: 85.4] [Reference Citation Analysis (0)] |
| 233. | Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1834] [Cited by in RCA: 2540] [Article Influence: 254.0] [Reference Citation Analysis (0)] |
| 234. | Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1979] [Cited by in RCA: 2830] [Article Influence: 283.0] [Reference Citation Analysis (1)] |
| 235. | Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2999] [Cited by in RCA: 3297] [Article Influence: 471.0] [Reference Citation Analysis (0)] |
| 236. | Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1390] [Cited by in RCA: 2126] [Article Influence: 303.7] [Reference Citation Analysis (1)] |
| 237. | Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2493] [Cited by in RCA: 3791] [Article Influence: 473.9] [Reference Citation Analysis (0)] |
| 238. | Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1659] [Cited by in RCA: 1912] [Article Influence: 159.3] [Reference Citation Analysis (0)] |
| 239. | Park HE, Kim JH, Cho NY, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch. 2017;471:329-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 240. | Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, Chung L, Finard BB, Wu X, Fathi P. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10:421-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 241. | Bhattacharya N, Yuan R, Prestwood TR, Penny HL, DiMaio MA, Reticker-Flynn NE, Krois CR, Kenkel JA, Pham TD, Carmi Y. Normalizing Microbiota-Induced Retinoic Acid Deficiency Stimulates Protective CD8(+) T Cell-Mediated Immunity in Colorectal Cancer. Immunity. 2016;45:641-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 130] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 242. | Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, Trella E, Galati-Fournier V, Oertli D, Däster SR. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut. 2018; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 243. | Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 500] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 244. | Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1113] [Cited by in RCA: 1304] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 245. | Aarnoutse R, de Vos-Geelen JMPGM, Penders J, Boerma EG, Warmerdam FARM, Goorts B, Olde Damink SWM, Soons Z, Rensen SSM, Smidt ML. Study protocol on the role of intestinal microbiota in colorectal cancer treatment: a pathway to personalized medicine 2.0. Int J Colorectal Dis. 2017;32:1077-1084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 246. | Guyton K, Alverdy JC. The gut microbiota and gastrointestinal surgery. Nat Rev Gastroenterol Hepatol. 2017;14:43-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 247. | Bachmann R, Leonard D, Delzenne N, Kartheuser A, Cani PD. Novel insight into the role of microbiota in colorectal surgery. Gut. 2017;66:738-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 248. | Sassone-Corsi M, Raffatellu M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol. 2015;194:4081-4087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 224] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 249. | Karrasch T, Jobin C. Wound healing responses at the gastrointestinal epithelium: a close look at novel regulatory factors and investigative approaches. Z Gastroenterol. 2009;47:1221-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 250. | Lee L, Liberman S, Charlebois P, Stein B, Kaneva P, Carli F, Feldman LS. The impact of complications after elective colorectal resection within an enhanced recovery pathway. Tech Coloproctol. 2018;22:191-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 251. | Ohigashi S, Sudo K, Kobayashi D, Takahashi T, Nomoto K, Onodera H. Significant changes in the intestinal environment after surgery in patients with colorectal cancer. J Gastrointest Surg. 2013;17:1657-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 252. | Souza DG, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. J Immunol. 2004;173:4137-4146. [PubMed] |
| 253. | Stary L, Mezerova K, Skalicky P, Zboril P, Raclavsky V. Are we any closer to screening for colorectal cancer using microbial markers?A critical review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2017;161:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 254. | Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, Hurley E, O’Riordain M, Shanahan F, O’Toole PW. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2017; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 441] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 255. | Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1596] [Cited by in RCA: 1963] [Article Influence: 178.5] [Reference Citation Analysis (0)] |