Published online Mar 7, 2017. doi: 10.3748/wjg.v23.i9.1552
Peer-review started: November 15, 2016
First decision: January 10, 2017
Revised: January 21, 2017
Accepted: February 16, 2017
Article in press: February 17, 2017
Published online: March 7, 2017
Processing time: 111 Days and 9.5 Hours
Clostridium difficile (C. difficile) is a spore-forming, toxin-producing, gram-positive anaerobic bacterium that is the principal etiologic agent of antibiotic-associated diarrhea. Infection with C. difficile (CDI) is characterized by diarrhea in clinical syndromes that vary from self-limited to mild or severe. Since its initial recognition as the causative agent of pseudomembranous colitis, C. difficile has spread around the world. CDI is one of the most common healthcare-associated infections and a significant cause of morbidity and mortality among older adult hospitalized patients. Due to extensive antibiotic usage, the number of CDIs has increased. Diagnosis of CDI is often difficult and has a substantial impact on the management of patients with the disease, mainly with regards to antibiotic management. The diagnosis of CDI is primarily based on the clinical signs and symptoms and is only confirmed by laboratory testing. Despite the high burden of CDI and the increasing interest in the disease, episodes of CDI are often misdiagnosed. The reasons for misdiagnosis are the lack of clinical suspicion or the use of inappropriate tests. The proper diagnosis of CDI reduces transmission, prevents inadequate or unnecessary treatments, and assures best antibiotic treatment. We review the options for the laboratory diagnosis of CDI within the settings of the most accepted guidelines for CDI diagnosis, treatment, and prevention of CDI.
Core tip: This work is a review of the strategies that may be used for laboratory diagnosis of infection with Clostridium difficile. First, we provide general recommendations for testing of samples taking in account the guidelines of the Society for Healthcare Epidemiology of America/Infectious Diseases Society of America and the American College of Gastroenterology. We reviewed diverse methods of diagnosis including, culture, toxigenic culture, cell cytotoxic neutralization assay and the use of enzyme immuno assays. Finally, we present an overview of singleplex and multiplex nucleic acid amplification tests.
- Citation: Martínez-Meléndez A, Camacho-Ortiz A, Morfin-Otero R, Maldonado-Garza HJ, Villarreal-Treviño L, Garza-González E. Current knowledge on the laboratory diagnosis of Clostridium difficile infection. World J Gastroenterol 2017; 23(9): 1552-1567
- URL: https://www.wjgnet.com/1007-9327/full/v23/i9/1552.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i9.1552
Clostridium difficile (C. difficile) is a Gram-positive and strictly anaerobic bacterium that may exist in a vegetative form that is very sensitive to oxygen. During stress, C. difficile produces spores that enable the microbe to survive harsh conditions for prolonged periods of time and facilitate its dissemination in the environment[1]. Upon ingestion, the spores resist the low pH of the stomach and reach the anaerobic environment of the gut. When the intestinal microbiote is altered because of antibiotic treatment, especially broad-spectrum antibiotics, the spores germinate. Next, C. difficile develops into its vegetative form, proliferates, and colonizes the gut[2]. C. difficile infection (CDI) is the principal cause of antibiotic-associated diarrhea. Diarrhea because of CDI may be self-limited, mild, or severe, and is one of the symptoms of a variety of clinical syndromes due to CDI. Complications of CDI are pseudomembranous colitis, fulminant colitis, and toxic megacolon[3].
CDI is an intestinal disease mediated by potent cytotoxic enzymes that damage the intestinal mucosa[4,5]. These cytotoxic enzymes, toxin A (TcdA) and toxin B (TcdB)[6,7], alter cytoskeletal actin, which leads to diminished transepithelial resistance, fluid accumulation, and destruction of the intestinal epithelium[5,8]. C. difficile toxins also induce the release of proinflammatory cytokines from enterocytes, mast cells, and macrophages[9]. The genome of toxigenic strains of C. difficle has a pathogenicity locus (PaLoc) of 19.6 kb that contains the tcdA and tcdB genes. Other PaLoc genes are tcdR and tcdC; the former encodes a positive regulator and the latter a negative regulator of the expression of the A and B toxins. Yet another PaLoc gene is tcdE, which encodes a holin-like protein that may be involved in the secretion of toxins[10]. Besides, some strains produce the C. difficile binary toxin (CDT), which is composed of an enzymatic component, CdtA, and a binding component, CdtB. These components are codified by cdtA and cdtB genes, which are located in the CDT locus (CdtLoc). CDT may potentiate the toxicity of TcdA and TcdB and lead to a more serious illness and could, therefore, be considered another virulence factor[11,12].
The vast majority of diarrhea cases are not related to a particular pathogen. From all the stool samples submitted to the laboratory for testing of C. difficile toxins, only 10% to 25% are positive[13]; most commonly in cases of antibiotic-associated diarrhea. Other pathogens that may cause antibiotic-associated diarrhea are: Staphylococcus aureus, Clostridium perfringens, Salmonella species, and Klebsiella oxytoca[14]. The clinical suspicion of CDI is the presentation of diarrhea after administration of antibiotics shortly after the beginning of treatment and up to 8 wk after treatment initiation[13]. In mild to moderate disease, diarrhea is the main symptom and passing of watery stools with foul odor is characteristic. The presence of hematochezia is rare. Moderate to severe disease is usually accompanied by systemic symptoms such as abdominal cramps, fever (up to 40 °C), leukocytosis (up to 50000 cells/mm3), and hypoalbuminemia (< 2.5 mg/dL)[13,15]. Colitis is characterized by fever, cramps, leukocytosis, and the presence of leukocytes in feces. Furthermore, a thickened colon wall is observed with computed tomography and in half of the cases pseudomembranes can be seen with endoscopy[13]. When pseudomembranes are found, a diagnostic of CDI can be made, as antibiotic-associated diarrhea due to other pathogens tends to have normal endoscopy findings[16]. In severe cases, CDI may progress to toxic megacolon with the risk of colon wall perforation[15]. The diagnosis of toxic megacolon is accomplished through radiological evidence of colonic dilatation, commonly involving the ascending or transverse colon[17]. According to the most commonly used criteria for the diagnosis of toxic megacolon[18], three of the following four criteria should be present: fever, tachycardia, leukocytosis, and anemia. Additionally, dehydration, electrolyte disturbance, and hypotension or changes in mental status (any of the criteria must be present)[17-19].
Ever since C. difficile was recognized as the causative agent of pseudomembranous colitis, the number of CDI cases has increased worldwide. Recently, Lessa et al[20] estimated that the number of CDI in the United States was 453000, from which 29000 died within 30 d after diagnosis. The healthcare costs related to CDI were estimated to be $ 4.8 billion for acute care facilities alone[20]. Since the year 2000, both the number and severity of CDI have increased due to the emergence of a more virulent strain with a higher antimicrobial resistance[21]. After strain typing by pulsed-field gel electrophoresis (PFGE), restriction endonuclease analysis, and ribotyping, this strain was denominated BI/North American PFGE type 1 (NAP1)/027[21]. So far, all BI/NAP1/027 isolates are positive for binary toxin CDT and have an 18-base pair deletion in tcdC that is associated with an increased production of toxins A and B; a single base pair deletion at position 117 of tcdC has also been related to higher toxin expression[3]. Concerning antimicrobial resistance, BI/NAP1/027 are resistant to fluoroquinolones, which provides a selective advantage to this strain[21]. This strain is disseminated worldwide; the above is evidenced by reports from America, Europe, Asia, and Oceania[22].
One of the key points when treating CDI is to discontinue unnecessary therapy with antibiotics; thus allowing the gut microbiota to recover. The Clinical Practice Guidelines for C. difficile Infection in Adults of the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA), the Guidelines for Diagnosis, Treatment, and Prevention of C. difficile Infections of the American College of Gastroenterology (ACG) and the Guidance Document for Clostridium difficile of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) recommend the use of metronidazole when treating an initial episode of mild to moderate CDI. The dosage is 500 mg orally, 3 times per day for 10 to 14 d. In case of severe initial CDI, vancomycin (125 mg orally, 4 times per day for 10 to 14 d) should be administered[23-25]. A combination of oral vancomycin (500 mg 4 times per day) and intravenous metronidazole (500 mg every 8 h) are indicated for the treatment of severe, complicated CDI according to the SHEA/IDSA and ESCMID guidelines[23,25]. On the other hand, the ACG recommends a dosage of 125 mg of vancomycin. When treating a first episode of recurrence, the regimen should be the same as an initial case, according to the severity of the infection. If there is a second episode of recurrence, a pulsed regimen of vancomycin is recommended. Fidaxomicin is narrow spectrum macrocyclic antibiotic, approved by the American Food and Drug Administration (FDA), that selectively eradicates C. difficile with a minimum effect on the intestinal microbiota. The relapse rate of fidaxomicin is lower than the one of vancomycin[26-28].
Despite the high burden of CDI and the increased interest in the disease, episodes of CDI are often misdiagnosed. A study from Spain revealed that two out of three CDI cases were either undiagnosed or misdiagnosed[29]. The main explanation may be the lack of clinical suspicion, particularly in community cases with patients who do not meet the risk criteria (age > 65 years or previous hospitalization). Besides, an inadequate test may yield false-negative results[30]. Furthermore, the interpretation of laboratory data is complicated, as the presence of C. difficile in stool does not mean CDI; the other way round, the absence of C. difficile toxins does not rule out the possibility of CDI. To interpret laboratory results, the techniques that were applied must be considered. A correct diagnosis of CDI is important because it has a substantial impact on case managment, mainly with regards to antibiotic regimens. The diagnosis of CDI is primarily based on the clinical signs and symptoms and is only confirmed by laboratory testing[31]. Misdiagnosis has two main consequences: first, patients may be undertreated or overtreated; second, the delay of proper infection control allows for further dissemination[32].
The following is a review of the strategies that may be used for laboratory diagnosis of CDI. To date, the Clinical Practice Guidelines of the SHEA/IDSA provide one of the most widely acceptable guidelines for the diagnosis and clinical management of CDI cases[23]. Also, the Guidelines of the ACG focus on the recommendations for the diagnosis and management of patients with CDI as well as for the prevention and control of outbreaks[24]. The above documents provide useful recommendations on which this review is based.
Both the Clinical Practice Guidelines of the SHEA/IDSA and the Guidelines of the ACG state that C. difficile testing is recommended only for stool samples from patients with diarrhea (Table 1), which is defined as the evacuation of loose stools, three or more times in 24 h or less[23,24]. The ESCMID recommends testing only stools of Bristol score 5 to 7[25]. The Bristol scale is a graded visual scale, composed of seven grades which range from stools with a form of separate hard lumps (score 1) to watery stools (score 7)[33]. To correlate the Bristol scale with C. difficile detection, stool samples with Bristol scale ≥ 5 were tested for C. difficile with an enzyme immunoassay (EIA) for glutamate dehydrogenase (GDH) and toxins A/B followed by a molecular assay for indeterminate results[34]. Detection of C. difficile was more frequent in semiformed stools (Bristol 5 or 6) than in watery stools (Bristol 7). Bristol 5 stool specimens accounted for the highest rate of positive testing. Therefore, Bristol 5 stool specimens should not be discarded. Further study is required to verify whether specimen with lower Bristol scores should be tested. There was no association between the Bristol score and the rates of hospital-onset CDI, severe CDI, and complications of CDI[34].
| Recommendation | Ref. |
| C. difficile testing is recommended only for stool samples from patients with diarrhea | [23,24] |
| The testing of asymptomatic patients is not recommended | [23] |
| Perirectal swabs are not accepted for C. difficile testing unless the patient has developed ileus | [23,24] |
| Repeated testing of C. difficile does not improve detection and does not change the result | [23,24] |
| Retesting, as a proof of cure, remains controversial | [23,24] |
Commonly, perirectal swabs are not accepted for C. difficile testing, except for selected cases, such as patients with ileus[23,24]. Ileus is characterized by a lack of bowel movements that causes a blockage of the intestines. For patients with ileus, an accurate sampling technique is the perirectal swab[35]. Some patients with CDI develop fever and abdominal pain but not diarrhea. These patients may develop severe complications, like fulminant colitis and intestinal perforations[36].
The Clinical Practice Guidelines of the SHEA/IDSA recommend no testing of asymptomatic patients[23] (Table 1) because asymptomatic colonization with a toxigenic strain of C. difficile is common. But even samples from patients with persistent diarrhea may yield false-positive CDI results; for example, alternate etiology was reported to be the cause of the symptoms in 25% of a cohort of 117 cases that had been diagnosed with recurrent CDI[37]. Particularly, highly sensitive molecular assays may yield false-positive results. Positive CDI tests for asymptomatic patients are common. The carriage rates of toxigenic C. difficile strains are similar between open populations (6.6%)[38] and hospitalized ones (8%)[39]. Hospitalized populations tend to be at an increased risk of developing CDI due to antibiotic treatment and prolonged exposure because of long stays at healthcare facilities[40]. The environment and skin of asymptomatic carriers have higher percentages of spores and represent a permanent source of contamination and spreading of spores to other patients and setting surfaces. The continual washing of hands of both personnel and patients is a universal preventing measure[41].
The carriage rate of C. difficile is high among infants (0 to 3 years of age). Among 85 healthy infants at day nurseries, the carriage of C. difficile was 45%, and the frequency of toxigenic strains was 13%[42]. One-year follow-up studies among newborns revealed that 74% to 100% had CDI-positive stool, often in the neonatal period[42] In a study that followed 10 infants, 81/111 samples (73%) were positive for C. difficile and 21 (26%) had toxigenic strains[42]. Another study that followed 42 infants found that 106/288 stool samples (37%) were CDI positive[43]. Most strains (71%) were toxin producers. Interestingly, six infants evacuated loose stools during the study, but only three of them could be linked to C. difficile. Furthermore, carriage of C. difficile was similar in infants suffering from loose stools than children with normal stools.
A point to consider when testing specimens from infants is the difficulty to differentiate a diarrheal stool from a normal stool, since infant stool may not be fully formed. When comparing infant cases (age ≤ 12 mo) with CDI-positive diarrheal stool with cases with CDI-negative diarrheal stool no differences in clinical symptoms were found. However, in both groups, alternative causes of diarrhea were found[44]. Rather than looking for C. difficile, the authors suggested looking for other causes, especially viral ones, of diarrhea in infants.
Neither the Clinical Practice Guidelines of the SHEA/IDSA nor the Guidelines of the ACG recommend retesting (Table 1). This recommendation is especially valid for molecular methods. Several studies have demonstrated that repeated testing for C. difficile does not improve detection nor change the result. Repeated testing, particularly with molecular tests, only increases healthcare costs and the probability of false-positive results. After having implemented a new policy that alerted physicians about the consequences and disadvantages of a C. difficile PCR test within 7 d of an initial test, a healthcare institute saw the requests for CDI retesting reduced by 91%[45]. Among the 135 retests that were performed, 122 were repeaters after a negative initial test, and only 4 of them turned positive upon repeating the test. Even lower positive conversion rates (0.05%-1%) have been reported by others after repeating PCR assays on 4213 samples with negative results in a previous test[46]. Thus, an initial test with a negative test result does not justify retesting unless there is a founded suspicion of a false-negative result.
Retesting to monitor response to treatment also remains controversial. Although a negative conversion rate of 67.6% 14 d after a positive Cepheid Xpert C. difficile test has been reported[47], cases that were clinically cured remained positive when testing for toxins[48].
A stool culture is essential to prepare isolates for molecular typing, which is required for epidemiologic studies. The SHEA/IDSA guidelines recommend toxigenic culture (TC) as the standard to which other methods should be compared. The first step is to recover C. difficile spores. Stool samples are either heated to 80 °C or mixed with an equal volume of absolute ethanol and incubated at room temperature. This way vegetative cells and contaminating microbes are eliminated while spores are recovered[49]. Next, the sample is inoculated into a differential and selective medium that allows the spores to germinate. A well-known medium to recover C. difficile from stool specimens is cycloserine-cefoxitin-fructose-agar (CCFA)[50]. Cycloserine and cefoxitin are present at concentrations that inhibit the growth of most Gram-negative and Gram-positive bacteria, without affecting the growth of C. difficile. Fructose is an important nutrient and neutral red is added as a pH indicator. A 48-h solid culture of C. difficile in CCFA presents flat, grayish, and shiny colonies with spreading edges and a typical horse manure smell. Under ultraviolet light, the colonies appear yellow-green fluorescent. Under the microscope, cells of C. difficile are Gram-positive and possess subterminal to terminal spores. Identification of C. difficile colonies may be based on colony morphology, Gram staining, and odor; confirmation of species may be assessed trough biochemical systems for the identification of anaerobes.
To improve both the recovery and identification of C. difficile cultures, the original CCFA formulation has been modified. The addition of biliary salts, particularly sodium taurocholate, promotes germination[51] and the inclusion of egg yolk allows to verify lecithinase and lipase activity of isolates[50,52]. Furthermore, enrichment broths have been formulated to recover small amounts of spores and allow them to germinate. When comparing two broths, a cycloserine-cefoxitin fructose broth proved to be more sensitive than a cycloserine-cefoxitin mannitol broth that had been supplemented with taurocholate and lysozyme, (CCMB-TAL), but CCMB-TAL yielded better recovery rates when cultures were semi-quantified[49].
Furthermore, chromogenic media have been developed. For example, C. difficile grown on ChromID C. difficile agar (bioMérieux, France) yields black colonies that often can be observed after 24 h of incubation[53]. However, lengthening the incubation from 24 to 48 h increased the sensitivity significantly from 53% to 100% (P < 0.001)[54]. ChromID C. difficile agar yields a higher 24-h recovery (sensitivity, 92%) than CCFA (sensitivity, 22%)[55]. Likewise, ChromID C. difficile agar, which had a sensitivity of 100% and a recovery of 94%, outperformed CCFA supplemented with sodium taurocholate, which had a sensitivity of 87% and a recovery of 82%[56]. Yet another study confirmed that ChromID C. difficile agar performs best when compared to CCFA, cycloserine-cefoxitin-egg-yolk agar and tryptone soy agar with sheep blood[49].
TC is a two-step reference method for the diagnosis of CDI. In step one, C. difficile strains are isolated and grown on a selective medium, and in step two, colonies are tested for toxin production on a variety of cell lines. The grown isolates are re-cultured in broth, and the supernatant is filtered and added to a cell line culture. The cytopathic effect (CPE) is evaluated and neutralized by antitoxin. This procedure may take a few days to accomplish, which makes it an impractical option for routine diagnosis[57]. Alternatively, testing of toxin production may be performed using an EIA[58,59].
The main concern about TC is the possibility of recovering non-toxigenic strains (strains that are not capable of produce toxins A and/or B). Another possibility is, though recovering a toxigenic strain, it may not be producing toxins, thus not causing clinical symptoms. When evaluating the clinical significance of TC and the citotoxicity assay on 169 samples that met CDI criteria, it was found that cases positive for both assays were more severe than cases that were positive for TC only. On the other hand, if only the cytotoxicity assay had been performed, one-third of the cases would have been missed[60]. The latter is an argument in favor of TC for CDI diagnosis.
The Cell Cytotoxic Neutralization Assay (CCNA) has been considered the gold standard for the diagnosis of CDI. In this assay, the filtrate of a recently obtained stool sample is inoculated onto various sensitive cell lines to evaluate the CPE of C. difficile toxins, particularly TcdB[57]. CPE is observed as cell rounding; some strains may induce protrusions in the cell lines, a phenomenon known as "sordellii-like" CPE[61,62]. If the CPE can be reversed by an antitoxin, the test is positive for the C. difficile toxin[57]. The assay must be performed in fresh stools, since sample freezing or a delay in its processing may result in loss of activity of toxins and false negative test results[63].
Diverse cell lines have been used for the detection of toxins: African green monkey kidney, McCoy, MRC-5, primary rhesus monkey kidney, and Vero cells. Among these cell lines, Vero cells and McCoy cells were a 100% concordant with respect to the detection of toxins[64]. When maintaining a cell line is impossible or non-desirable, there are cost-effective commercial assays available. A CCNA executed with Hs27 HFF ReadyCells is not only easy-to-use but also outperforms the EIA and TC in both sensitivity and specificity; 90.8% vs 78.6% in sensitivity and 98.3% vs 97.8% in specificity, respectively[65].
C. difficile produces and secretes GDH; this enzyme allows the bacterium to manage oxidative stress derived from the immune response by inactivating hydrogen peroxide through the production of α-ketoglutarate[66]. Although GDH screening of stool specimens for the diagnosis of CDI diagnosis is common[67], its value is limited to being a preliminary test, since both toxigenic and non-toxigenic strains produce GDH[68]. GDH is highly conserved among C. difficile strains; no differences in reactivity were found among 168 C. difficile isolates belonging to 77 different ribotypes using three different assays carried out by two different groups[68]. As the GDH assay may be positive for C. difficile strains that do not produce toxins, a positive GDH assay needs a confirmatory test (CCNA, EIA, a molecular test, or TC)[23,35,67].
GDH has been incorporated, as an initial test, into multistep algorithms (ACG guidelines). Recently, there has been a tendency towards a two-step algorithm, where in the absence of detection of toxin by EIA, clinical evaluation should also be applied to determine true CDI or colonization. This has recently been recommended by ESCMID[69].
The C. Diff Quik Chek Complete assay is a rapid membrane EIA that combines the detection of both GDH and the toxins A and B[70]. With TC as a reference, the C. Diff Quik Chek Complete had a sensitivity of 63.6% and a specificity of 98%[70]. Both assays proved to be highly sensitive (range: 97.6% to 100%) and accurate (88% true positive or true negative). Discrepant results were resolved with the Xpert C. diff assay[71]. The algorithm allowed to rule out C. difficile without additional tests when GDH is negative and to confirm CDI when both GDH and toxin A/B results are positive. Another GDH test, the automated Vidas C. difficile GDH assay (bioMérieux, Marcy l'Etoile, France), has a 95% agreement with the C. Diff Quik Chek assay (Quik Chek-60, Techlab, United States)[72]. In case of discrepant results, molecular tests are recommended by various authors, because CCNA and TC are expensive and time-consuming[35,73]. A two-step algorithm (step 1, GDH and toxin detection by EIA; step 2, loop-mediated isothermal amplification) yielded a sensitivity of 81%, a specificity and positive predictive value (PPV) of 100%, and a negative predictive value (NPV) of 96%[74]. Even though the sensitivity was lower than in other studies, the PPVs and NPVs supported the practice of reporting the specimen as positive without further testing. A more complex algorithm increased the CDI detection rate from 8% to 19%; initial positive GDH testing (VIDAS C. difficile; bioMérieux) was confirmed by toxin testing (VIDAS C. difficile Toxin A&B; bioMérieux). In case a toxin EIA did not confirm a positive GDH, additional tests [nucleic acid amplification test (NAAT), CCNA, or TC] were executed on positive GDH samples. A positive confirmatory test is considered CDI-positive sample[75].
Apart from providing improved diagnostic performance, multistep algorithms are the most cost effective for various reasons: (1) because they avoid unnecessary or inadequate treatment and its consequences[76]; and (2) an initial screening with a cheap GDH test allows rapid identification of negative samples, limiting the use of more expensive NAAT tests to only those samples that were positive for GDH[77].
One of the first strategies for the diagnosis of CDI was the detection of toxins with a specific EIA. Several EIA-based kits are commercially available in different formats, such as: lateral flow immunoassay, also known as immunochromatography or strip tests; and solid-phase assays, for example micro-wells[15]. EIA-based confirmation of CDI is practical, fast, and cheap, but it is also one of the least consistent methods (sensitivity range: 63% to 94%, specificity range: 75% to 100%)[23]. According to the SHEA and IDSA guidelines, toxin detection by EIA is less sensitive than detection by CCNA; thus, EIA should not be used alone for the diagnosis of CDI to avoid a false-negative result[23]. A two-step or three-step algorithm that combines GDH screening with EIA improves CDI diagnosis[23,24].
The inconsistent sensitivity of EIAs may be due to several factors, such as: antigenic variation among the toxins of different circulating strains, inadequate storage and transportation of samples, freeze-thaw cycles, and inter-laboratory technical variance, among others[15]. Some of the early developed EIAs accounted only for the detection of toxin A; however, there are reports of strains that do not produce toxin A[78-80]. Also, some assays detect both toxins A and B plus the detection of GDH[81].
Using TC and CCNA as reference, six commercially available EIAs and three lateral-flow assays for the detection of C. difficile toxins A and B have been compared. The sensitivities ranged from 60% to 81.6%, whereas the specificities ranged from 91.4% to 99.4%. PPVs and NPVs were diverse and depended on whether the samples originated from a low-prevalence environment (community) or a high-prevalence environment (hospital setting). Though PPVs were low for both settings, the PPV was higher in the high-prevalence setting, independent from the gold standard chosen as a reference. NPVs were high for both settings (above 95%), regardless of the reference chosen[82]. In a two-step algorithm, the initial screening with GDH detection (C.Diff Chek-60, TechLab/Wampole) yielded a sensitivity of 93.4% and a specificity of 96.6% (reference assay: TC). Next, only positive specimens were confirmed with a rapid toxin A/B assay (Tox A/B Quik Chek, TechLab/Wampole, Blacksburg, VA), which yielded a specificity and a PPV of 97.1% and 96.5%, respectively. Compared to TC, the sensitivity of the EIA-based toxin assay was low (52.9%)[83].
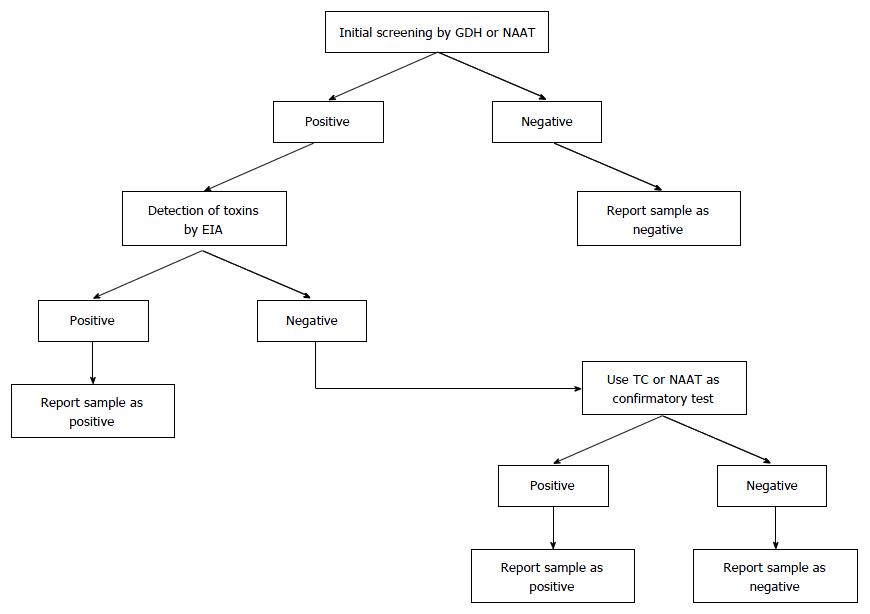
Nowadays, many infections are diagnosed via molecular tests. The new generation of NAATs amplify and detect pathogen-specific DNA or RNA sequences. Advantages of NAATs include high sensitivity, high specificity, and speed. Because no viable cells are needed, sampling, handling, transportation, and storage aspects are simplified. Furthermore, no culture is required. The role of NAAT in the diagnosis process for CDI may be supportive[84], part of a two or three-step algorithm according to ESCMID guidelines (Figure 1)[23], or as a stand-alone test in cases of documented diarrhea[24].
Despite the evident advantages, important issues to be considered before introducing CDI NAATs in the clinical laboratory are: the requirement of trained personnel and higher costs, and the probability of false-positive results because of the high sensitivity of the test and the detection of strains that do not produce toxins. Especially in stool samples from diarrhea cases due to other pathogens, false-positive results may not be recognized as such because of concomitant asymptomatic C. difficile carriage. Stool samples that were positive for both NAAT and toxin test had more bacteria and toxins than stool samples that were only NAAT positive[85]. The latter is an argument against the use of NAAT as a stand-alone test.
Single-plex NAATs: The FDA of the United States of America has cleared a set of commercially available single-plex NAATs. Table 2 summarizes the sensitivities and specificities of these tests, which often is over 90% or even close to 100%; these values depend on the test that was used as a reference test, which was often TC.
| Assay | Sensitivity | Specificity | Ref. |
| Cepheid Xpert C. difficile | 90%-100% | 92.9%-98.6% | [87,89,91] |
| IMDx C. difficile for Abbott m2000 Assay | 62.1%-92.8% | 99.4%-100% | [92,93] |
| BD Max Cdiff Assay | 81.6%-96.9% | 95%-95.8% | [92,93] |
| Portrait Toxigenic C. difficile Assay | 98.2% | 92.8% | [95] |
| Quidel Lyra Direct C. difficile Assay | 82.1%-85.7% | 96.9%-98.3% | [96] |
| Verigene C. difficile nucleic acid test | 95.2%-98.7% | 87.5%-99.4% | [97,128] |
| Simplexa C. difficile Universal Direct real time PCR | 87%-98% | 100% | [99,128] |
| AmpliVue C. difficile assay | 91%-96% | 89%-100% | [99,100] |
| Illumigene C. difficile assay | 93.3%-100% | 95.1%-100% | [95,101] |
| BD GeneOhm Cdiff assay | 89.6%-97.4% | 96.7%-98.5% | [95,103] |
| ProGastro Cd assay | 77.93%-100% | 93.4%-99.2% | [103,104] |
Cepheid Xpert®C. difficile and Xpert C. difficile/Epi: The Cepheid Xpert®C. difficile (Sunnyvale, CA) is a real-time PCR assay that detects the tcdB gene and thus allows CDI diagnosis, but without strain specification[86]. The multiplex RT-PCR assay Xpert C. difficile/Epi not only detects the tcdB gene but also the binary toxin genes (cdtA and cdtB), and a single-nucleotide deletion of the tcdC. Therefore, the Xpert C. difficile/Epi identifies ribotype 027[87]. Furthermore, Xpert C. difficile/Epi detects ribotype 033, a strain of veterinary importance that has been reported in cattle, veal calves, piglets, horses, and soil from various geographical locations worldwide[88]. The binary toxins of ribotype 033 were correctly amplified in all isolates (n = 52) included in a study. However, since ribotype 033 lacks the tcdA and tcdB genes, the GeneXpert Dx system reports that the sample is negative for CDI, and it is necessary to access the raw data in the instrument to obtain the amplification information[88]. Using TC as a reference, the sensitivity of the Cepheid Xpert C. difficile/Epi assay was 90%; its specificity, 92.9%; its PPV 71.4%; and its NPV 97.9%[89].
The introduction of the Cepheid Xpert C. difficile assay in a tertiary hospital significantly increased the rate of detection of toxigenic C. difficile from 4.7% to 9.9%. The increase was mainly due to cases that yielded indeterminate results with the C. Diff Quik Chek, but were positive with the Xpert C. difficile assay[90].
When the performance of the Xpert C. difficile Epi assay as a confirmatory test was compared to TC in a two-step algorithm with a GDH assay as a first step, there was a moderate agreement (kappa score 0.48) of 72.6%. The GDH-TC algorithm had a sensitivity of 57% and specificity of 97%, whereas the sensitivity of the GDH-Xpert algorithm was 100% and its specificity 97%. Furthermore, 42 out of 45 stool samples that were ribotype 027 positive were confirmed by PCR-ribotyping and sequencing, indicating a good epidemic value of the assay[91].
IMDx C. difficile for Abbott m2000 assay: The IMDx C. difficile for Abbott m2000 assay (IMDx) is a real-time PCR that detects not only C. difficile tcdA and tcdB genes, but also the rare variant strains rare toxin A+B- and toxin B variant (tcdBv) gene, which occurs in A-B+ strains. Lysis of the sample, target amplification, and detection are performed in the m2000 RealTime System (Abbott Laboratories, Abbott Park, IL)[92].
In a prospective analysis of 111 stool specimens and a retrospective analysis of 88 stool samples, in which the IMDx was compared to another FDA-cleared NAAT (the GeneOhm Cdiff Assay), the sensitivity was strain dependent: 100% for NAP1 strains and 90.3% for non-NAP1 strain with a limit of detection of 2250 colony-forming units[92]. However, when IMDx and 2 other molecular assays were compared to TC, IMDx had the lowest sensitivity (62.1%) and the highest specificity (99.4%)[93].
BD Max Cdiff assay: The BD Max Cdiff assay detects and amplifies the tcdB gene in a real-time PCR assay performed on the BD Max System (BD Diagnostics, Sparks, MD). After the addition of the sample, this hands-free platform combines DNA extraction and amplification. To extract genetic material a 10-μL loop is immersed in the specimen; next, the loop content is dispersed in BD Max Sample Buffer. The DNA extraction utilizes magnetic beads, which are eluted before a lyophilized amplification mix is added. The results are reported only as positive or negative for C. difficile[92].
Compared to the GeneOhm Cdiff Assay, BD Max Cdiff Assay had a sensitivity of 96.9% and a specificity of 95%[92]. In the same study, ribotyping was assessed, but there were no significant differences between the sensitivities and specificities of different ribotypes[94]. When the BD Max Cdiff Assay and 2 other molecular assays were compared to TC, the BD Max Cdiff Assay had a sensitivity of 81.6% and a specificity of 95.8%[93].
Portrait Toxigenic C. difficile assay: The Portrait Toxigenic C. difficile assay (Great Basin, West Valley City, UT) amplifies a 78-nucleotide fragment of the tcdB gene. The assay uses isothermal helicase-dependent amplification, followed by detection with an immobilized capture probe on a sliding array. Each reaction mixture contains three controls: a sample processing control, a hybridization control, and a detection control. Results for the specimen are reported only when the detection criteria for all controls are met[95].
A multicenter evaluation that included 49 stool specimens from 4 clinical sites compared the Portrait Toxigenic C. difficile Assay to TC on the same specimens. The sensitivity ranged from 92.9% to 100%, with an overall sensitivity of 98.2%. The specificity ranged from 88.9% to 96.9%, with an overall specificity of 92.8%[95]. When comparing Portrait Toxigenic C. difficile Assay results with those from other FDA-cleared tests, the concordance were as follows: 97.5% with Xpert C. difficile, 96.4% with GeneOhm Cdiff, and 93.8% with Illumigene C. difficile[95].
Quidel Lyra Direct C. difficile assay: The Quidel Lyra Direct C. difficile assay (Quidel, San Diego, CA) uses qualitative real-time PCR technology. Specimens are tested in a standard TaqMan real-time PCR assay utilizing primers/probes that detect but do not distinguish the tcdA and tcdB genes. The Lyra assay may be performed on any of three open-platform, real-time thermocyclers: SmartCycler II (Cepheid, Sunnyvale, CA), ABI 7500 Fast DX (Applied Biosystems, Carlsbad, CA), and ABI QuantStudio DX (Applied Biosystems, Carlsbad, CA). The Lyra assay has a running time of about 3 h[96]. Depending on the platform used, the sensitivity and specificity may differ; the ABI 7500 instrument is the most sensitive and the ABI QuantStudio DX is the most specific. The overall sensitivity is 85.7% and the overall specificity is 98.3% when compared to toxigenic culture[96].
Verigene C. difficile nucleic acid test: The Verigene C. difficile nucleic acid test is a multiplex qualitative assay that amplifies DNA by PCR in a nanoparticle-based microarray that targets the tcdA and tcdB genes and differentiates the hypervirulent strain 027/NAP1/BI via the binary toxin genes and the base pair deletion at position 117 in the regulator tcdC gene[97]. The Verigene system contains two modules: the Verigene Processor SP performs nucleic acid extraction, PCR amplification, and hybridization of amplicons; The Verigene Reader scans the test cartridge, realizes the optical analysis, and generates the results[97].
When compared to TC, the Verigene assay has sensitivity of 98.7% and a specificity of 87.5%[97]. With regard to strain typing, the assay assigns correctly 89.7% of hypervirulent strains compared with ribotyping[97]. When compared to fecal culture as a reference method, the Verigene C. difficile nucleic acid test was sensitive (96.7%), specific (97.4%), and accurate (97.1%)[98].
Simplexa C. difficile Universal Direct real-time PCR: This assay uses fluorescent bifunctional probes-primers to amplify a tcdB fragment. Samples in Tris-EDTA buffer are heat-treated; the lysate is used directly to perform the test. The system can accommodate a maximum of 94 samples and has an assay time of 91 min. When compared to another FDA-cleared assay, the Meridian Illumigene Asssay, the Simplexa C. difficile Assay had a sensitivity of 98% and a specificity of 100%; the concordance between the two systems was 98.7%[99].
AmpliVue C. difficile assay: The AmpliVue C. difficile assay uses helicase-dependent, isothermal amplification of a highly conserved 83-bp fragment of the tcdA gene. The assay includes a disposable detection device that allows for visual evaluation of amplification results. The AmpliVue system can perform a maximum of 24 assays, and has a total running time of 73 min. The AmpliVue assay had a sensitivity of 96% and specificity of 100% when compared to the FDA-cleared assay Meridian Illumigene Assay[99]. Compared to TC, the sensitivity and specificity were 91% and 89%, respectively[100].
Illumigene C. difficile assay: The Illumigene C. difficile assay uses loop-mediated isothermal DNA amplification technology to target a partial DNA conserved region of tcdA common to A+B+ and A-B- strains. The total time of analysis is 68 min and the maximum number of samples per run is 10[99]. When compared to TC, the sensitivity and specificity of the Illumigene assay were 100%[101].
BD GeneOhm Cdiff assay: The BD GeneOhm Cdiff assay (BD Diagnostics, San Diego, CA) amplifies the tcdB gene from stool samples with C. difficile, which is detected and analyzed with a SmartCycler instrument (Cepheid, Sunnyvale, CA)[102]. The performance of the BD GeneOhm Cdiff PCR assay has been compared to the Tox A/B II ELISA and a two-step method composed of the C. Diff Chek-60 GDH antigen assay followed by cytotoxin neutralization on 105 true positive samples. The detection rate was 66.7% for the Tox A/B II ELISA assay, 82.9% for the 2-step method, and 91.4% for the BD GeneOhm Cdiff PCR assay. The overall concordance between the BD GeneOhm Cdiff PCR assay and the Tox A/B II ELISA was 91.3%, while the concordance between the BD GeneOhm Cdiff and the two-step method was 93%. The BD GeneOhm Cdiff PCR and the two-step algorithm had similar performance but were more sensitive than Tox A/B II. Compared to the two-step algorithm, BD GeneOhm Cdiff PCR is faster but almost five times more expensive; BD GeneOhm Cdiff PCR is also six times more expensive than the Tox A/B assay[102]. When compared to TC, BD GeneOhm Cdiff PCR has a sensitivity of 89.6% and a specificity of 96.7%[103].
ProGastro Cd assay: ProGastro Cd assay (Prodesse, Waukesha, WI) is a Taqman real-time PCR assay that detects the tcdB gene. Amplification is performed on the Cepheid SmartCycler II (Sunnyvale, CA). Stool samples are processed to obtain genetic material using the NucliSENS easy MAG platform (bioMérieux, Inc., Durham, NC)[104].
There was a 95.7% agreement between TC and the ProGastro Cd assay. When comparted to TC, the ProGastro Cd assay had a sensitivity that ranged from 77.3% to 100% and a specificity between 99.2% and 93.4%[103,104]. A two-step algorithm, with the GDH-based C. Diff Quik Chek Complete as step 1 and the ProGastro Cd assay as step 2, yielded an estimated sensitivity of 97.9% and a specificity of 95.4%.
Multiplex platforms: There are two types of multiplex platforms: the first type includes the so-called "syndromic" platforms, i.e., a platform to detect pathogens associated with a particular symptom. In this case, a variety of syndromic platforms test for the main causative agents of diarrhea, independently of the kind of microorganism (bacteria, viruses or protozoa). The second type are pathogen class specific multiplex molecular assays[105].
Currently, there are two FDA-cleared syndromic multiplex assays that include the dectection of C. difficile: Luminex xTAG GPP (Luminex Molecular Diagnostics Inc., Toronto, Canada) and BioFire FilmArray GI Panel (BioFire Diagnostics, Salt Lake City, UT)[106]. In addition, there is a non-FDA cleared, but "Conformité Européene" (CE) marked multiplex assay, the Gastrofinder Smart 17 Fast (PathoFinder, Maastricht, The Netherlands)[105]. There are six additional commercially available platforms that include C. difficile among their targets, but none of these platforms are FDA cleared or CE marked[105].
Luminex xTAG pathogen panel: This assay simultaneously detects Salmonella sp., Shigella sp., Shiga toxin-producing E. coli (STEC) stx1/stx2, Vibrio cholerae, Yersinia enterocolitica, C. difficile toxin A/B, Campylobacter sp., E. coli O157, Enterotoxigenic E. coli (ETEC) LT/ST, adenovirus 40/41, rotavirus A, norovirus GI/GII, Giardia lamblia, Cryptosporidium sp. and Entamoeba histolytica[107]. The assay performs a multiplex reverse transcriptase PCR, using tagged and biotinylated primers. Amplicons are detected by hybridization to the pathogen-specific complementary antitag sequence coupled to specific beads and binding of the streptavidin-phycoerythrin reporter to the biotinylated primers[106,107].
The performance of the Luminex xTAG panel was evaluated with 185 stool samples from 176 patients. In 11% of the samples, multiple pathogens, including ETEC, G. lamblia, norovirus, Shigella sp., Campylobacter sp., Salmonella sp., adenovirus, and C. difficile, were detected[107]. There was a 100% sensitivity, specificity, PPV, and NPV. Another study evaluated the assay with a total of 254 clinical specimens[108]. Depending on the target organism, the sensitivity ranged from 90% to 100% with an overall sensitivity of 94.5% and a specificity of 99.1%. With respect to C. difficile, the sensitivity, specificity, PPV, and NPV were 91%, 100%, 100%, and 99%, respectively.
In a site-specific clinical evaluation, the Luminex xTAG Gastrointestinal assay showed a sensitivity of 98.7% and a specificity of 99.8%. C. difficile, Salmonella spp., and Cryptosporidium spp. accounted for 67% of the targets detected. Specifically, C. difficile was detected in 23 samples, all of which were confirmed with the Cepheid Xpert C. difficile assay. Furthermore, the multiplex detection led to savings in the hospital ward as compared to traditional methods[109].
FilmArray GI panel: The FilmArray uses nested multiplex PCR that is executed in two stages. First, the FilmArray performs a reverse transcription PCR. Next the diluted products are combined with a fluorescent double-stranded DNA-binding dye. This solution is aliquoted into an array with wells that contain primers to amplify internal sequences of the first product, so that in each well an individually nested PCR is performed. Results are obtained after fluorescence analysis[106,110].
The assay detects Campylobacter (C. jejuni, C. coli and C. upsaliensis), C. difficile, Plesiomonas shigelloides, Salmonella, Y. enterocolitica, Vibrio (V. parahaemolyticus, V. vulnificus, and V.cholerae), E. coli O157, Enteroaggregative E. coli, Enteropathogenic E. coli, ETEC, STEC, Shigella/Enteroinvasive E. coli, Adenovirus F 40/41, Astrovirus, Norovirus GI/GII, Rotavirus A, Sapovirus (I, II, IV, and V), Cryptosporidium, Cyclospora cayetanensis, E. histolytica and G. lamblia. For the identification of C. difficile, the FilmArray GI Panel targets both tcdA and tcdB[111].
In a study that evaluated the BioFire FilmArray GI Panel, C. difficile was the most frequently detected pathogen (83 out of 378 samples, 22%). The sensitivity of the BioFire FilmArray Gastrointestinal Panel to detect C. difficile was 95% and the specificity 99% using the Illumigene as a reference. In 91 episodes for which specific testing for C. difficile was ordered, 42 episodes (46%) were C. difficile positive according to standard testing and 40 (44%) according to the FilmArray GI Panel in 40 (44%)[112].
In a multicenter evaluation, the BioFire FilmArray GI Panel detected C. difficile in 204 out of 832 positive samples, contrary to 165 positive samples detected by the comparator (PCR of toxin A and B genes), resulting in a positive percent agreement of 98.8% and negative percent agreement of 97.1%[111].
There are seven non FDA-approved assays on the market that include C. difficile among their targets (Table 3). The majority of these assays, employ real-time PCR technology. To our knowledge, only three of them have been evaluated regarding the sensitivity and specificity at detection C. difficile.
| Assay | Company | Pathogens detected | Technology | Ref. |
| Gastrofinder Smart 17 Fast | PathoFinder | 9 bacteria, 4 viruses and 4 parasites | Multiplex Real-time PCR | [129] |
| EasyScreen Enteric assays | Genetic Signature | 7 bacteria, 8 viruses and 5 parasites | 3base Technology | [130] |
| RIDA®GENE | R-BioPharma AG | 11 bacteria, 4 viruses and 4 parasites | Multiplex Real-time PCR | [131] |
| FTD® Bacterial Gastroenteritis | Fast-Track Diagnostics | 9 bacteria, 5 viruses and 3 parasites | Multiplex Real-time PCR | [132] |
| CLART EnteroBac panel | Genomica | 19 bacteria | Low-density microarray | [133] |
| Faecal Bacteria | AusDiagnostics | 8 bacteria, 4 viruses and 3 parasites | Multiplex Tandem PCR technology | [134] |
| Seeplex Diarrhea ACE | Seegene | 10 bacteria and 4 viruses | Dual priming oligonucleotide technology | [135] |
The RIDA® GENE CD PCR assay (R-Biopharm AG, Darmstadt, Germany), when compared to TC as a reference, had a sensitivity of 98.1%, a specificity of 100%, a PPV of 100%, and a NPV of 98.1%[113]. Ylisiurua et al[113] found that the RIDA® GENE CD PCR assay outperforms competing molecular C. difficile assays, such as GeneOhm™ Cdiff assay (Becton Dickinson) and Xpert®C. difficile test (Cepheid).
The EasyScreen Enteric Bacterial Detection Kit is a multiplex assay with calculated analytical sensitivities that range from 2.5 to 12.5 copies of targeted pathogen, free of cross-reactivity to non-target microorganisms. For C. difficile, the calculated sensitivity was 100% and the specificity 81.2%. The EasyScreen assay correctly identified all C. difficile-containing samples (12 out of 18), including the ribotype 027 and 078 strains (1 of each)[114].
The Seeplex® Diarrhea ACE is another multiplex molecular assay that was compared to BD GeneOhm, with TC as a reference. There was a positivity rate of 35.4% (86/243). The concordance rate between the BD GeneOhm and Seeplex® Diarrhea ACE assay was 96% (234/243) with no significant differences between them. The sensitivity, specificity, PPV, and the NPV of the Seeplex® Diarrhea ACE assay were 90.0% (63/70), 97.1% (168/173), 92.6% (38/43), and 96.0% (168/175), respectively[115].
CDI is accompanied by intestinal inflammation. Inflammation biomarkers include cytokines, calprotectin, and fecal lactoferrin[116]. These biomarkers are not disease specific, but may be indicators of severity[117]. For example, fecal lactoferrin has been evaluated in both infectious diarrhea and inflammatory bowel disease. Fecal lactoferrin, blood biomarkers, white blood cell count and low serum albumin level, were significantly associated with severe CDI and stool toxin[116]. Furthermore, age, Charlson co-morbidity index, intensive care treatment, increased peripheral white blood cell count, elevated lactoferrin, decreased albumin, and elevated creatinine were significantly associated with death within 100 d of CDI diagnosis[118].
Calprotectin is a protein found in the cytoplasm of neutrophils and can be detected in stool in cases of intestinal inflammation, such as in inflammatory bowel disease and infectious diarrhea[117,119]. A large proportion of individuals with nosocomial diarrhea (diagnosed by PCR) have elevated levels of calprotectin[120]. High fecal levels of calprotectin have been associated with C. difficile strain 027[121] and complicated/recurrent CDI in a cohort of older adults[119]. Though elevated stool calprotectin has a low sensitivity as a diagnostic test (38.5%), its specificity for complicate or recurrent CDI was 91.9%, and thus provides valuable information for adequate treatment decision-making[119].
So far, there is no specific biomarker that detects CDI or any other pathogenic agent of clinically significant diarrhea. Anikst et al[122] suggests the implementation of measures to avoid unnecessary testing for C. difficile in order to diminish CDI overdiagnosis. The identification of such a biomarker, will be helpful to improve CDI diagnosis.
C. difficile has a worldwide distribution; its toxigenic strains are responsible for CDI. Despite increasing knowledge on risk factors that favor CDI and measures to reduce propagation, there is an increase in the prevalence of CDI in many countries[123-127]. One of the challenges at managing of CDI is the initial diagnosis of the disease. To date, there is no single test that accurately and rapidly diagnoses CDI. Multistep testing is recommended for a diagnosis with acceptable sensitivity and specificity. The inclusion of NAATs in the diagnostic algorithm combines high sensitivity with a short turnaround time. However, test results should be interpreted with caution and should consider clinical suspicion, the presence of risk factors, and a correct interpretation of test results. A better understanding of the pathogenesis of C. difficile will help both physicians and laboratories to develop the best strategy to overcome current issues with CDI diagnosis.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Mexico
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Lightner AL S- Editor: Yu J L- Editor: A E- Editor: Liu WX
| 1. | Heinlen L, Ballard JD. Clostridium difficile infection. Am J Med Sci. 2010;340:247-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 2. | Sarker MR, Paredes-Sabja D. Molecular basis of early stages of Clostridium difficile infection: germination and colonization. Future Microbiol. 2012;7:933-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Ghose C. Clostridium difficile infection in the twenty-first century. Emerg Microbes Infect. 2013;2:e62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 4. | Chen S, Sun C, Wang H, Wang J. The Role of Rho GTPases in Toxicity of Clostridium difficile Toxins. Toxins (Basel). 2015;7:5254-5267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 5. | Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1026] [Cited by in RCA: 1096] [Article Influence: 68.5] [Reference Citation Analysis (0)] |
| 6. | Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP. Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. MBio. 2015;6:e00551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 206] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 7. | Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 606] [Cited by in RCA: 647] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 8. | Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18:247-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 845] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 9. | Solomon K. The host immune response to Clostridium difficile infection. Ther Adv Infect Dis. 2013;1:19-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Rupnik M, Dupuy B, Fairweather NF, Gerding DN, Johnson S, Just I, Lyerly DM, Popoff MR, Rood JI, Sonenshein AL. Revised nomenclature of Clostridium difficile toxins and associated genes. J Med Microbiol. 2005;54:113-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Rupnik M, Grabnar M, Geric B. Binary toxin producing Clostridium difficile strains. Anaerobe. 2003;9:289-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 12. | Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:15-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 319] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 13. | Bartlett JG, Gerding DN. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis. 2008;46 Suppl 1:S12-S18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 391] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 14. | Larcombe S, Hutton ML, Lyras D. Involvement of Bacteria Other Than Clostridium difficile in Antibiotic-Associated Diarrhoea. Trends Microbiol. 2016;24:463-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 15. | Burnham CA, Carroll KC. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev. 2013;26:604-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 300] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 16. | Varughese CA, Vakil NH, Phillips KM. Antibiotic-associated diarrhea: a refresher on causes and possible prevention with probiotics--continuing education article. J Pharm Pract. 2013;26:476-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Sayedy L, Kothari D, Richards RJ. Toxic megacolon associated Clostridium difficile colitis. World J Gastrointest Endosc. 2010;2:293-297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (1)] |
| 18. | Jalan KN, Sircus W, Card WI, Falconer CW, Bruce CB, Crean GP, McManus JP, Small WP, Smith AN. An experience of ulcerative colitis. I. Toxic dilation in 55 cases. Gastroenterology. 1969;57:68-82. [PubMed] |
| 19. | Autenrieth DM, Baumgart DC. Toxic megacolon. Inflamm Bowel Dis. 2012;18:584-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 20. | Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2023] [Cited by in RCA: 1976] [Article Influence: 197.6] [Reference Citation Analysis (1)] |
| 21. | McDonald LC, Killgore GE, Thompson A, Owens RC, Kazakova SV, Sambol SP, Johnson S, Gerding DN. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433-2441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1625] [Cited by in RCA: 1549] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 22. | Valiente E, Cairns MD, Wren BW. The Clostridium difficile PCR ribotype 027 lineage: a pathogen on the move. Clin Microbiol Infect. 2014;20:396-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 23. | Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect Control Hosp Epidemiol. 2010;31:431-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2285] [Cited by in RCA: 2201] [Article Influence: 220.1] [Reference Citation Analysis (0)] |
| 24. | Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478-498; quiz 499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1179] [Cited by in RCA: 1187] [Article Influence: 98.9] [Reference Citation Analysis (0)] |
| 25. | Debast SB, Bauer MP, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20 Suppl 2:1-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 793] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 26. | Rineh A, Kelso MJ, Vatansever F, Tegos GP, Hamblin MR. Clostridium difficile infection: molecular pathogenesis and novel therapeutics. Expert Rev Anti Infect Ther. 2014;12:131-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 27. | Lancaster JW, Matthews SJ. Fidaxomicin: the newest addition to the armamentarium against Clostridium difficile infections. Clin Ther. 2012;34:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 28. | Crawford T, Huesgen E, Danziger L. Fidaxomicin: a novel macrocyclic antibiotic for the treatment of Clostridium difficile infection. Am J Health Syst Pharm. 2012;69:933-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 29. | Alcalá L, Martín A, Marín M, Sánchez-Somolinos M, Catalán P, Peláez T, Bouza E. The undiagnosed cases of Clostridium difficile infection in a whole nation: where is the problem? Clin Microbiol Infect. 2012;18:E204-E213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 30. | Alcalá L, Reigadas E, Marín M, Martín A, Catalán P, Bouza E. Impact of clinical awareness and diagnostic tests on the underdiagnosis of Clostridium difficile infection. Eur J Clin Microbiol Infect Dis. 2015;34:1515-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 31. | Dubberke ER, Burnham CA. Diagnosis of Clostridium difficile Infection: Treat the Patient, Not the Test. JAMA Intern Med. 2015;175:1801-1802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 32. | Planche T, Wilcox MH. Diagnostic pitfalls in Clostridium difficile infection. Infect Dis Clin North Am. 2015;29:63-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 33. | Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32:920-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1858] [Cited by in RCA: 2105] [Article Influence: 75.2] [Reference Citation Analysis (1)] |
| 34. | Caroff DA, Edelstein PH, Hamilton K, Pegues DA. The Bristol stool scale and its relationship to Clostridium difficile infection. J Clin Microbiol. 2014;52:3437-3439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 35. | Kundrapu S, Sunkesula VC, Jury LA, Sethi AK, Donskey CJ. Utility of perirectal swab specimens for diagnosis of Clostridium difficile infection. Clin Infect Dis. 2012;55:1527-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 36. | Elinav E, Planer D, Gatt ME. Prolonged ileus as a sole manifestation of pseudomembranous enterocolitis. Int J Colorectal Dis. 2004;19:273-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 37. | Jackson M, Olefson S, Machan JT, Kelly CR. A High Rate of Alternative Diagnoses in Patients Referred for Presumed Clostridium difficile Infection. J Clin Gastroenterol. 2016;50:742-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 38. | Galdys AL, Nelson JS, Shutt KA, Schlackman JL, Pakstis DL, Pasculle AW, Marsh JW, Harrison LH, Curry SR. Prevalence and duration of asymptomatic Clostridium difficile carriage among healthy subjects in Pittsburgh, Pennsylvania. J Clin Microbiol. 2014;52:2406-2409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 39. | Zacharioudakis IM, Zervou FN, Pliakos EE, Ziakas PD, Mylonakis E. Colonization with toxinogenic C. difficile upon hospital admission, and risk of infection: a systematic review and meta-analysis. Am J Gastroenterol. 2015;110:381-390; quiz 391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 40. | Tschudin-Sutter S, Carroll KC, Tamma PD, Sudekum ML, Frei R, Widmer AF, Ellis BC, Bartlett J, Perl TM. Impact of Toxigenic Clostridium difficile Colonization on the Risk of Subsequent C. difficile Infection in Intensive Care Unit Patients. Infect Control Hosp Epidemiol. 2015;36:1324-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 41. | Hung YP, Lee JC, Lin HJ, Liu HC, Wu YH, Tsai PJ, Ko WC. Clinical impact of Clostridium difficile colonization. J Microbiol Immunol Infect. 2015;48:241-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 42. | Rousseau C, Poilane I, De Pontual L, Maherault AC, Le Monnier A, Collignon A. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis. 2012;55:1209-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 142] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 43. | Adlerberth I, Huang H, Lindberg E, Åberg N, Hesselmar B, Saalman R, Nord CE, Wold AE, Weintraub A. Toxin-producing Clostridium difficile strains as long-term gut colonizers in healthy infants. J Clin Microbiol. 2014;52:173-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 44. | Tang P, Roscoe M, Richardson SE. Limited clinical utility of Clostridium difficile toxin testing in infants in a pediatric hospital. Diagn Microbiol Infect Dis. 2005;52:91-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 45. | Luo RF, Spradley S, Banaei N. Alerting physicians during electronic order entry effectively reduces unnecessary repeat PCR testing for Clostridium difficile. J Clin Microbiol. 2013;51:3872-3874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 46. | Green DA, Stotler B, Jackman D, Whittier S, Della-Latta P. Clinical characteristics of patients who test positive for Clostridium difficile by repeat PCR. J Clin Microbiol. 2014;52:3853-3855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 47. | Luo RF, Banaei N. Is repeat PCR needed for diagnosis of Clostridium difficile infection? J Clin Microbiol. 2010;48:3738-3741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Dubberke ER, Carling P, Carrico R, Donskey CJ, Loo VG, McDonald LC, Maragakis LL, Sandora TJ, Weber DJ, Yokoe DS. Strategies to prevent Clostridium difficile infections in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol. 2014;35 Suppl 2:S48-S65. [PubMed] |
| 49. | Lister M, Stevenson E, Heeg D, Minton NP, Kuehne SA. Comparison of culture based methods for the isolation of Clostridium difficile from stool samples in a research setting. Anaerobe. 2014;28:226-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (2)] |
| 50. | George WL, Sutter VL, Citron D, Finegold SM. Selective and differential medium for isolation of Clostridium difficile. J Clin Microbiol. 1979;9:214-219. [PubMed] |
| 51. | Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505-2512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 607] [Cited by in RCA: 547] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 52. | Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol. 1982;15:443-446. [PubMed] |
| 53. | Perry JD, Asir K, Halimi D, Orenga S, Dale J, Payne M, Carlton R, Evans J, Gould FK. Evaluation of a chromogenic culture medium for isolation of Clostridium difficile within 24 hours. J Clin Microbiol. 2010;48:3852-3858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 54. | Han SB, Chang J, Shin SH, Park KG, Lee GD, Park YG, Park YJ. Performance of chromID Clostridium difficile agar compared with BBL C. difficile selective agar for detection of C. difficile in stool specimens. Ann Lab Med. 2014;34:376-379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 55. | Yang JJ, Nam YS, Kim MJ, Cho SY, You E, Soh YS, Lee HJ. Evaluation of a chromogenic culture medium for the detection of Clostridium difficile. Yonsei Med J. 2014;55:994-998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 56. | Carson KC, Boseiwaqa LV, Thean SK, Foster NF, Riley TV. Isolation of Clostridium difficile from faecal specimens--a comparison of chromID C. difficile agar and cycloserine-cefoxitin-fructose agar. J Med Microbiol. 2013;62:1423-1427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 57. | Planche T, Wilcox M. Reference assays for Clostridium difficile infection: one or two gold standards? J Clin Pathol. 2011;64:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 58. | She RC, Durrant RJ, Petti CA. Evaluation of enzyme immunoassays to detect Clostridium difficile toxin from anaerobic stool culture. Am J Clin Pathol. 2009;131:81-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 59. | Thonnard J, Carreer F, Avesani V, Delmée M. Toxin A detection on Clostridium difficile colonies from 24-h cultures. Clin Microbiol Infect. 1996;2:50-54. [PubMed] |
| 60. | Reigadas E, Alcalá L, Marín M, Muñoz-Pacheco P, Catalán P, Martin A, Bouza E. Clinical significance of direct cytotoxicity and toxigenic culture in Clostridium difficile infection. Anaerobe. 2016;37:38-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 61. | Rupnik M. How to detect Clostridium difficile variant strains in a routine laboratory. Clin Microbiol Infect. 2001;7:417-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 62. | Rupnik M. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol Rev. 2008;32:541-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 63. | Freeman J, Wilcox MH. The effects of storage conditions on viability of Clostridium difficile vegetative cells and spores and toxin activity in human faeces. J Clin Pathol. 2003;56:126-128. [PubMed] |
| 64. | Maniar AC, Williams TW, Hammond GW. Detection of Clostridium difficile toxin in various tissue culture monolayers. J Clin Microbiol. 1987;25:1999-2000. [PubMed] |
| 65. | Strachan AJ, Evans NE, Williams OM, Spencer RC, Greenwood R, Probert CJ. Comparison of a frozen human foreskin fibroblast cell assay to an enzyme immunoassay and toxigenic culture for the detection of toxigenic Clostridium difficile. Diagn Microbiol Infect Dis. 2013;75:42-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 66. | Girinathan BP, Braun SE, Govind R. Clostridium difficile glutamate dehydrogenase is a secreted enzyme that confers resistance to H2O2. Microbiology. 2014;160:47-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 67. | Shetty N, Wren MW, Coen PG. The role of glutamate dehydrogenase for the detection of Clostridium difficile in faecal samples: a meta-analysis. J Hosp Infect. 2011;77:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 68. | Carman RJ, Wickham KN, Chen L, Lawrence AM, Boone JH, Wilkins TD, Kerkering TM, Lyerly DM. Glutamate dehydrogenase is highly conserved among Clostridium difficile ribotypes. J Clin Microbiol. 2012;50:1425-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 69. | Crobach MJ, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2016;22 Suppl 4:S63-S81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 394] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 70. | Kim H, Kim WH, Kim M, Jeong SH, Lee K. Evaluation of a rapid membrane enzyme immunoassay for the simultaneous detection of glutamate dehydrogenase and toxin for the diagnosis of Clostridium difficile infection. Ann Lab Med. 2014;34:235-239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 71. | Sharp SE, Ruden LO, Pohl JC, Hatcher PA, Jayne LM, Ivie WM. Evaluation of the C.Diff Quik Chek Complete Assay, a new glutamate dehydrogenase and A/B toxin combination lateral flow assay for use in rapid, simple diagnosis of clostridium difficile disease. J Clin Microbiol. 2010;48:2082-2086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 72. | Davies KA, Berry CE, Morris KA, Smith R, Young S, Davis TE, Fuller DD, Buckner RJ, Wilcox MH. Comparison of the Vidas C. difficile GDH Automated Enzyme-Linked Fluorescence Immunoassay (ELFA) with Another Commercial Enzyme Immunoassay (EIA) (Quik Chek-60), Two Selective Media, and a PCR Assay for gluD for Detection of Clostridium difficile in Fecal Samples. J Clin Microbiol. 2015;53:1931-1934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 73. | Goldenberg SD, Cliff PR, French GL. Glutamate dehydrogenase for laboratory diagnosis of Clostridium difficile infection. J Clin Microbiol. 2010;48:3050-3051; author reply 3051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 74. | Ota KV, McGowan KL. Clostridium difficile testing algorithms using glutamate dehydrogenase antigen and C. difficile toxin enzyme immunoassays with C. difficile nucleic acid amplification testing increase diagnostic yield in a tertiary pediatric population. J Clin Microbiol. 2012;50:1185-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 75. | Cheng JW, Xiao M, Kudinha T, Xu ZP, Sun LY, Hou X, Zhang L, Fan X, Kong F, Xu YC. The Role of Glutamate Dehydrogenase (GDH) Testing Assay in the Diagnosis of Clostridium difficile Infections: A High Sensitive Screening Test and an Essential Step in the Proposed Laboratory Diagnosis Workflow for Developing Countries like China. PLoS One. 2015;10:e0144604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 76. | Bartsch SM, Umscheid CA, Nachamkin I, Hamilton K, Lee BY. Comparing the economic and health benefits of different approaches to diagnosing Clostridium difficile infection. Clin Microbiol Infect. 2015;21:77.e1-77.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 77. | Goldenberg SD, Cliff PR, Smith S, Milner M, French GL. Two-step glutamate dehydrogenase antigen real-time polymerase chain reaction assay for detection of toxigenic Clostridium difficile. J Hosp Infect. 2010;74:48-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 78. | Kim H, Riley TV, Kim M, Kim CK, Yong D, Lee K, Chong Y, Park JW. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J Clin Microbiol. 2008;46:1116-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 61] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 79. | Androga GO, Hart J, Foster NF, Charles A, Forbes D, Riley TV. Infection with Toxin A-Negative, Toxin B-Negative, Binary Toxin-Positive Clostridium difficile in a Young Patient with Ulcerative Colitis. J Clin Microbiol. 2015;53:3702-3704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 80. | Cairns MD, Preston MD, Lawley TD, Clark TG, Stabler RA, Wren BW. Genomic Epidemiology of a Protracted Hospital Outbreak Caused by a Toxin A-Negative Clostridium difficile Sublineage PCR Ribotype 017 Strain in London, England. J Clin Microbiol. 2015;53:3141-3147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 81. | Quinn CD, Sefers SE, Babiker W, He Y, Alcabasa R, Stratton CW, Carroll KC, Tang YW. C. Diff Quik Chek complete enzyme immunoassay provides a reliable first-line method for detection of Clostridium difficile in stool specimens. J Clin Microbiol. 2010;48:603-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 82. | Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211-3217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 262] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 83. | Fenner L, Widmer AF, Goy G, Rudin S, Frei R. Rapid and reliable diagnostic algorithm for detection of Clostridium difficile. J Clin Microbiol. 2008;46:328-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 110] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 84. | Eckert C, Jones G, Barbut F. Diagnosis of Clostridium difficile infection: the molecular approach. Future Microbiol. 2013;8:1587-1598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 85. | Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern Med. 2015;175:1792-1801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 445] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 86. | Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM. Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difficile/Epi, and Illumigene C. difficile assays. J Clin Microbiol. 2012;50:1331-1335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 87. | Huang H, Weintraub A, Fang H, Nord CE. Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of clostridium difficile infections. J Clin Microbiol. 2009;47:3729-3731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 88. | Androga GO, McGovern AM, Elliott B, Chang BJ, Perkins TT, Foster NF, Riley TV. Evaluation of the Cepheid Xpert C. difficile/Epi and meridian bioscience illumigene C. difficile assays for detecting Clostridium difficile ribotype 033 strains. J Clin Microbiol. 2015;53:973-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 89. | Shin S, Kim M, Kim M, Lim H, Kim H, Lee K, Chong Y. Evaluation of the Xpert Clostridium difficile assay for the diagnosis of Clostridium difficile infection. Ann Lab Med. 2012;32:355-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 90. | Williamson DA, Basu I, Freeman J, Swager T, Roberts SA. Improved detection of toxigenic Clostridium difficile using the Cepheid Xpert C difficile assay and impact on C difficile infection rates in a tertiary hospital: a double-edged sword. Am J Infect Control. 2013;41:270-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 91. | Babady NE, Stiles J, Ruggiero P, Khosa P, Huang D, Shuptar S, Kamboj M, Kiehn TE. Evaluation of the Cepheid Xpert Clostridium difficile Epi assay for diagnosis of Clostridium difficile infection and typing of the NAP1 strain at a cancer hospital. J Clin Microbiol. 2010;48:4519-4524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 92. | Stellrecht KA, Espino AA, Maceira VP, Nattanmai SM, Butt SA, Wroblewski D, Hannett GE, Musser KA. Premarket evaluations of the IMDx C. difficile for Abbott m2000 Assay and the BD Max Cdiff Assay. J Clin Microbiol. 2014;52:1423-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 93. | Yoo J, Lee H, Park KG, Lee GD, Park YG, Park YJ. Evaluation of 3 automated real-time PCR (Xpert C. difficile assay, BD MAX Cdiff, and IMDx C. difficile for Abbott m2000 assay) for detecting Clostridium difficile toxin gene compared to toxigenic culture in stool specimens. Diagn Microbiol Infect Dis. 2015;83:7-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 94. | Chiang D, Ng S, La MV, Jureen R, Lin RT, Teo JW. Performance assessment of the BD MAX Cdiff assay in comparison to Xpert C. difficile assay in a setting with very low prevalence of toxigenic Clostridium difficile PCR ribotype 027. Anaerobe. 2014;30:156-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 95. | Buchan BW, Mackey TL, Daly JA, Alger G, Denys GA, Peterson LR, Kehl SC, Ledeboer NA. Multicenter clinical evaluation of the portrait toxigenic C. difficile assay for detection of toxigenic Clostridium difficile strains in clinical stool specimens. J Clin Microbiol. 2012;50:3932-3936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 96. | Beck ET, Buchan BW, Riebe KM, Alkins BR, Pancholi P, Granato PA, Ledeboer NA. Multicenter evaluation of the Quidel Lyra Direct C. difficile nucleic acid amplification assay. J Clin Microbiol. 2014;52:1998-2002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 97. | Carroll KC, Buchan BW, Tan S, Stamper PD, Riebe KM, Pancholi P, Kelly C, Rao A, Fader R, Cavagnolo R. Multicenter evaluation of the Verigene Clostridium difficile nucleic acid assay. J Clin Microbiol. 2013;51:4120-4125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 98. | Tojo M, Nagamatsu M, Hayakawa K, Mezaki K, Kirikae T, Ohmagari N. Evaluation of an automated rapid diagnostic test for detection of Clostridium difficile. PLoS One. 2014;9:e106102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 99. | Deak E, Miller SA, Humphries RM. Comparison of Illumigene, Simplexa, and AmpliVue Clostridium difficile molecular assays for diagnosis of C. difficile infection. J Clin Microbiol. 2014;52:960-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 100. | Neuendorf M, Guadarrama-Gonzalez R, Lamik B, MacKenzie CR. A prospective study of two isothermal amplification assays compared with real-time PCR, CCNA and toxigenic culture for the diagnosis of Clostridium difficile infection. BMC Microbiol. 2016;16:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 101. | Barkin JA, Nandi N, Miller N, Grace A, Barkin JS, Sussman DA. Superiority of the DNA amplification assay for the diagnosis of C. difficile infection: a clinical comparison of fecal tests. Dig Dis Sci. 2012;57:2592-2599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 102. | Kvach EJ, Ferguson D, Riska PF, Landry ML. Comparison of BD GeneOhm Cdiff real-time PCR assay with a two-step algorithm and a toxin A/B enzyme-linked immunosorbent assay for diagnosis of toxigenic Clostridium difficile infection. J Clin Microbiol. 2010;48:109-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 103. | Selvaraju SB, Gripka M, Estes K, Nguyen A, Jackson MA, Selvarangan R. Detection of toxigenic Clostridium difficile in pediatric stool samples: an evaluation of Quik Check Complete Antigen assay, BD GeneOhm Cdiff PCR, and ProGastro Cd PCR assays. Diagn Microbiol Infect Dis. 2011;71:224-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 104. | Stamper PD, Babiker W, Alcabasa R, Aird D, Wehrlin J, Ikpeama I, Gluck L, Carroll KC. Evaluation of a new commercial TaqMan PCR assay for direct detection of the clostridium difficile toxin B gene in clinical stool specimens. J Clin Microbiol. 2009;47:3846-3850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 105. | Zhang H, Morrison S, Tang YW. Multiplex polymerase chain reaction tests for detection of pathogens associated with gastroenteritis. Clin Lab Med. 2015;35:461-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 106. | Binnicker MJ. Multiplex Molecular Panels for Diagnosis of Gastrointestinal Infection: Performance, Result Interpretation, and Cost-Effectiveness. J Clin Microbiol. 2015;53:3723-3728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 107. | Beckmann C, Heininger U, Marti H, Hirsch HH. Gastrointestinal pathogens detected by multiplex nucleic acid amplification testing in stools of pediatric patients and patients returning from the tropics. Infection. 2014;42:961-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 108. | Navidad JF, Griswold DJ, Gradus MS, Bhattacharyya S. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. J Clin Microbiol. 2013;51:3018-3024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 131] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 109. | Patel A, Navidad J, Bhattacharyya S. Site-specific clinical evaluation of the Luminex xTAG gastrointestinal pathogen panel for detection of infectious gastroenteritis in fecal specimens. J Clin Microbiol. 2014;52:3068-3071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 110. | Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6:e26047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 283] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 111. | Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015;53:915-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 370] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 112. | Stockmann C, Rogatcheva M, Harrel B, Vaughn M, Crisp R, Poritz M, Thatcher S, Korgenski EK, Barney T, Daly J. How well does physician selection of microbiologic tests identify Clostridium difficile and other pathogens in paediatric diarrhoea? Insights using multiplex PCR-based detection. Clin Microbiol Infect. 2015;21:179.e9-179.15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 113. | Ylisiurua P, Koskela M, Vainio O, Tuokko H. Comparison of antigen and two molecular methods for the detection of Clostridium difficile toxins. Scand J Infect Dis. 2013;45:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 114. | Siah SP, Merif J, Kaur K, Nair J, Huntington PG, Karagiannis T, Stark D, Rawlinson W, Olma T, Thomas L. Improved detection of gastrointestinal pathogens using generalised sample processing and amplification panels. Pathology. 2014;46:53-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 115. | Shin BM, Mun SJ, Yoo SJ, Kuak EY. Comparison of BD GeneOhm Cdiff and Seegene Seeplex ACE PCR assays using toxigenic Clostridium difficile culture for direct detection of tcdB from stool specimens. J Clin Microbiol. 2012;50:3765-3767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 116. | Boone JH, DiPersio JR, Tan MJ, Salstrom SJ, Wickham KN, Carman RJ, Totty HR, Albert RE, Lyerly DM. Elevated lactoferrin is associated with moderate to severe Clostridium difficile disease, stool toxin, and 027 infection. Eur J Clin Microbiol Infect Dis. 2013;32:1517-1523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 117. | Swale A, Miyajima F, Roberts P, Hall A, Little M, Beadsworth MB, Beeching NJ, Kolamunnage-Dona R, Parry CM, Pirmohamed M. Calprotectin and lactoferrin faecal levels in patients with Clostridium difficile infection (CDI): a prospective cohort study. PLoS One. 2014;9:e106118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 118. | Boone JH, Archbald-Pannone LR, Wickham KN, Carman RJ, Guerrant RL, Franck CT, Lyerly DM. Ribotype 027 Clostridium difficile infections with measurable stool toxin have increased lactoferrin and are associated with a higher mortality. Eur J Clin Microbiol Infect Dis. 2014;33:1045-1051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 119. | Rao K, Santhosh K, Mogle JA, Higgins PD, Young VB. Elevated fecal calprotectin associates with adverse outcomes from Clostridium difficile infection in older adults. Infect Dis (Lond). 2016;48:663-669. [PubMed] [DOI] [Full Text] |
| 120. | Popiel KY, Gheorghe R, Eastmond J, Miller MA. Usefulness of Adjunctive Fecal Calprotectin and Serum Procalcitonin in Individuals Positive for Clostridium difficile Toxin Gene by PCR Assay. J Clin Microbiol. 2015;53:3667-3669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 121. | Peretz A, Tkhawkho L, Pastukh N, Brodsky D, Halevi CN, Nitzan O. Correlation between fecal calprotectin levels, disease severity and the hypervirulent ribotype 027 strain in patients with Clostridium difficile infection. BMC Infect Dis. 2016;16:309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 122. | Anikst VE, Gaur RL, Schroeder LF, Banaei N. Organism burden, toxin concentration, and lactoferrin concentration do not distinguish between clinically significant and nonsignificant diarrhea in patients with Clostridium difficile. Diagn Microbiol Infect Dis. 2016;84:343-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 123. | Desai K, Gupta SB, Dubberke ER, Prabhu VS, Browne C, Mast TC. Epidemiological and economic burden of Clostridium difficile in the United States: estimates from a modeling approach. BMC Infect Dis. 2016;16:303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 124. | Bouza E. Consequences of Clostridium difficile infection: understanding the healthcare burden. Clin Microbiol Infect. 2012;18 Suppl 6:5-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 125. | Yasunaga H, Horiguchi H, Hashimoto H, Matsuda S, Fushimi K. The burden of Clostridium difficile-associated disease following digestive tract surgery in Japan. J Hosp Infect. 2012;82:175-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 126. | Choi HY, Park SY, Kim YA, Yoon TY, Choi JM, Choe BK, Ahn SH, Yoon SJ, Lee YR, Oh IH. The epidemiology and economic burden of Clostridium difficile infection in Korea. Biomed Res Int. 2015;2015:510386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 127. | Slimings C, Armstrong P, Beckingham WD, Bull AL, Hall L, Kennedy KJ, Marquess J, McCann R, Menzies A, Mitchell BG. Increasing incidence of Clostridium difficile infection, Australia, 2011-2012. Med J Aust. 2014;200:272-276. [PubMed] |
| 128. | Gilbreath JJ, Verma P, Abbott AN, Butler-Wu SM. Comparison of the Verigene Clostridium difficile, Simplexa C. difficile Universal Direct, BD MAX Cdiff, and Xpert C. difficile assays for the detection of toxigenic C. difficile. Diagn Microbiol Infect Dis. 2014;80:13-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 129. | PathoFinder. GastroFinder SMART 17 FAST, (2016). Accesed October. 2016; Available from: http: //www.pathofinder.com/products/smartfinder/gastrofinder-smart-17-fast. |
| 130. | Signature G. EasyScreen Enteric assays, 2016. Accesed October. 2016; Available from: http: //geneticsignatures.com/pathogen-detection-europe-israel/easyscreen-products/enteric-pathogen-screening-kits/. |
| 131. | R-BioPharma. R-Biopharm RIDA®GENE, 2016. Accesed October. 2016; Available from: http://www.r-biopharm.com/products/clinical-diagnostics/molecular-diagnostics. |
| 132. | Diagnostics F-T. FTD® Bacterial Gastroenteritis, 2016. Accesed October. 2016; Available from: http://www.fast-trackdiagnostics.com/products/. |
| 133. | Genomica. CLART® EnteroBac, 2016. Accesed October. 2016; Available from: http://genomica.es/en/in_vitro_diagnostics_clart_enterobac.cfm. |
| 134. | AusDiagnostics. Faecal Bacteria, 2016. Accesed October. 2016; Available from: http://www.ausdiagnostics.com/faecal-pathogens.html. |
| 135. | Seegene. Seeplex Diarrhea ACE, 2016. Accesed October. 2016; Available from: http://www.seegene.com/neo/en/products/Gastrointestinal/seeplex_DIARRHEA.php. |