Published online Feb 28, 2017. doi: 10.3748/wjg.v23.i8.1434
Peer-review started: September 22, 2016
First decision: October 20, 2016
Revised: November 3, 2016
Accepted: December 8, 2016
Article in press: December 8, 2016
Published online: February 28, 2017
Processing time: 158 Days and 23 Hours
To understand the molecular mechanism of esophageal cancer development and provide molecular markers for screening high-risk populations and early diagnosis.
Two-dimensional electrophoresis combined with mass spectrometry were adopted to screen differentially expressed proteins in nine cases of fetal esophageal epithelium, eight cases of esophageal cancer, and eight cases of tumor-adjacent normal esophageal epithelium collected from fetuses of different gestational age, or esophageal cancer patients from a high-risk area of esophageal cancer in China. Immunohistochemistry (avidin-biotin-horseradish peroxidase complex method) was used to detect the expression of peroxiredoxin (PRX)6 in 91 cases of esophageal cancer, tumor-adjacent normal esophageal tissue, basal cell hyperplasia, dysplasia, and carcinoma in situ, as well as 65 cases of esophageal epithelium from fetuses at a gestational age of 3-9 mo.
After peptide mass fingerprint analysis and search of protein databases, 21 differential proteins were identified; some of which represent a protein isoform. Varying degrees of expression of PRX6 protein, which was localized mainly in the cytoplasm, were detected in adult and fetal normal esophageal tissues, precancerous lesions, and esophageal cancer. With the progression of esophageal lesions, PRX6 protein expression showed a declining trend (P < 0.05). In fetal epithelium from fetuses at gestational age 3-6 mo, PRX6 protein expression showed a declining trend with age (P < 0.05). PRX6 protein expression was significantly higher in well-differentiated esophageal cancer tissues than in poorly differentiated esophageal cancer tissues (P < 0.05).
Development and progression of esophageal cancer result from interactions of genetic changes (accumulation or superposition). PRX6 protein is associated with fetal esophageal development and cancer differentiation.
Core tip: This was a retrospective study to identify 21 significantly differentially expressed proteins that may be related to the development and growth of fetal esophageal epithelium or the development and progression of esophageal cancer. Peroxiredoxin (PRX)6 protein was localized mainly in the cytoplasm, and detected in adult and fetal normal esophageal tissues, precancerous lesions, and esophageal cancer. With the progression of esophageal lesions, PRX6 protein expression showed a declining trend. In epithelium from fetuses at gestational age 3-6 mo, PRX6 expression showed a declining trend with age. PRX6 protein expression was significantly higher in well-differentiated than poorly differentiated esophageal cancer tissues. PRX6 protein is associated with fetal esophageal development and esophageal cancer differentiation.
- Citation: Guo JH, Xing GL, Fang XH, Wu HF, Zhang B, Yu JZ, Fan ZM, Wang LD. Proteomic profiling of fetal esophageal epithelium, esophageal cancer, and tumor-adjacent esophageal epithelium and immunohistochemical characterization of a representative differential protein, PRX6. World J Gastroenterol 2017; 23(8): 1434-1442
- URL: https://www.wjgnet.com/1007-9327/full/v23/i8/1434.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i8.1434
Esophageal cancer is one of the most common thoracic malignancies that pose a serious threat to human health. There are about 400000 newly diagnosed cases of esophageal cancer and about 300000 related deaths worldwide each year, with the majority of patients diagnosed in developing countries. China is one of the countries or regions with a high incidence and mortality rates of esophageal cancer. Since the 1990s, the overall incidence and mortality rates of esophageal cancer have declined in China; however, esophageal cancer is still the leading cause of cancer-related death in Northern Henan Province and Chaoshan Region of Guangdong Province; both of which are high-risk areas for esophageal cancer. Squamous cell carcinoma (SCC) is the most important histological subtype of esophageal cancer in China. Esophageal cancer, especially esophageal SCC, is still a key research field for thoracic surgical research in China. Although China has a high incidence of esophageal cancer, > 90% of patients are diagnosed at a middle or late stage. As a result, the 5-year survival rate for Chinese esophageal cancer patients has not obviously declined over recent decades. In patients with early esophageal cancer who have undergone surgical or minimally invasive endoscopic treatment, the 5-year survival rate can reach > 90%. Therefore, early diagnosis is important for its prognosis. Clarifying the molecular mechanisms underlying the development and progression of esophageal cancer and identifying biological markers for screening high-risk populations are therefore of great clinical importance for early diagnosis. In the present study, we collected fetal esophageal tissues, esophageal cancer tissues, different degrees of precancerous lesions, and tumor-adjacent normal esophageal epithelial tissues from fetuses of different gestational age, or esophageal cancer patients in a high-risk area for esophageal cancer in China. We utilized proteomic technology to analyze the differential protein expression profiles in the development and growth of fetal esophageal epithelium and the development and progression of esophageal cancer, with the aim of clarifying the molecular mechanism of esophageal cancer development.
Esophageal cancer specimens were collected from 91 patients who underwent surgery at the Yaocun Esophageal Cancer Hospital (Linzhou, Henan Province, China) from 2000 to 2005. No patients underwent chemotherapy or radiotherapy. There were 51 men and 40 women, with a median age of 58 years (range, 35-78 years).
Tumor-adjacent normal esophageal mucosal epithelial tissues were collected under a microscope. A solitary lesion was considered only when the lesion was separated from other lesions by ≥ 1 cm of morphologically normal esophageal epithelium. Tumor-adjacent esophageal epithelium was regarded as normal regardless of whether it was completely or only partially normal.
Fetal esophageal tissue specimens were collected from 65 fetuses (30 male and 35 female), and they had a gestational age of 3 (n = 7), 4 (n = 13), 5 (n = 9), 6 (n = 10), 7 (n = 10), 8 (n = 10), or 9 (n = 6) mo. All fetuses were legally aborted fetuses or fetuses induced with misoprostol in the Family Planning Service Center of Hui County (Henan Province, China). Their parents had no prior history of usage of special drugs. The age of fetuses was determined based on the date of the last menstrual period, the crown-heal length, and morphology of the hand and foot[1,2].
WK600 nitric oxide laser gas analyzer (Weck, United States), Voyager DE Pro MALDI-TOF mass spectrometer (Applied Biosystems, United States), and GS-800 Calibrated Densitometer (Bio-Rad, United States) were used. Mouse anti-peroxiredoxin (PRX)6 monoclonal antibody was purchased from Chemicon (United States); ABC kit was purchased from Vector Laboratories (United States); DAB substrate kit was purchased from Beijing Zhong Shan-Golden Bridge Biological Technology Co. Ltd. (China); and lyophilized bovine serum albumin was purchased from Sigma (United States).
Proteomic analysis was performed based on the study flow chart presented in Figure 1.
Solid-phase isoelectric focusing electrophoresis: Following removal of cover film, dried immobilized pH gradient (IPG, pH 3-10) strips (17 cm long) were plated in a fixed-length strip holder with the gel side down. After dispelling the air, 2 mL cover fluid (mineral oil) was applied to minimize evaporation. The holder was covered and placed on the IPG electrode platform, and isoelectric focusing electrophoresis was performed according to the protocol described in Table III-1. After electrophoresis was completed, Milli-Q water-wetted filter paper was used to wipe the mineral oil, and the strips were placed in a rehydration tray with the gel side up. Eight milliliters of equilibrium buffer I consisting of 6 mol/L urea, 2% SDS, 0.375 mol/L Tris-HCl (pH 8.8), 20% glycerol and 2% dithiothreitol was added. After shaking on a horizontal rotator at 15 rpm for 15 min, the strips were taken out and put into equilibrium buffer II consisting of 6 mol/L urea, 2% SDS, 0.375 mol/L Tris-HCl (pH 8.8), 20% glycerol and 2.5% iodoacetamide for 15 min. The strips were finally dried and used for the second dimension electrophoresis.
SDS-PAGE: SDS-PAGE was performed using Tris-glycine-SDS buffer (5 × : Tris 15.1 g, glycine 94 g, and SDS 5 g, dissolved in 1000 mL Milli-Q water). SDS-PAGE gels were prepared according to Table III-2. The gel was poured and allowed to polymerize. Pre-stained protein markers (10-15 μL) were spotted on 0.25 cm2 Whatman filter paper (3 mm), which was subsequently placed on the “+” end of the IPG strip. IPG strips were imbedded in the second dimension and sealed using 0.5% agarose sealing solution (low melting point agarose 0.5 g and 10 μL 1% bromophenol blue, dissolved in 100 mL 1 × electrophoresis buffer). Electrophoresis was carried out at a constant current of 10 mA per gel for 30 min, which was then raised to 40 mA. After electrophoresis was completed, gels were removed and stained.
Silver nitrate staining and coomassie blue staining: Silver nitrate staining was performed as described previously[3]. For Coomassie blue staining, SDS-PAGE gels were placed in Coomassie blue solution (Coomassie blue R-250 1.16 g, absolute ethanol 500 mL, and glacial acetic acid 100 mL, dissolved in 1000 mL Milli-Q water) and incubated at room temperature for 2 h. The gels were then put into a destaing solution (30% absolute ethanol and 10% glacial acetic acid) until the background staining was removed.
Image scanning and analysis: Stained SDS-PAGE gels were scanned using a GS-800 calibrated densitometer at an optical resolution of 300 dpi and pixel of 8 bits. Images were analyzed with PDQuest 7.1.1 software. Image analysis included detecting spots on gels, editing spot and editing match, and correction of molecular weight (Mr) and isoelectric point (pI) of protein spots.
Tryptic digestion of in-gel proteins: For 2-D gels used for mass spectrometry (MS) analysis, the loading amount of protein was raised to 1 mg. 2-D gel electrophoresis was performed as described above. To ensure the integrity and purity of protein spots, they were cut and subjected to tryptic digestion at 2 wk after staining[4].
MALDI-TOF-MS analysis: MALDI-TOF-MS analysis of tryptic-digested fragments was performed by the Mass Spectrometry Laboratory of Shanghai Jikang Biochemical Technology Co. Ltd. (China). α-Cyano-4-hydroxycinnamic acid was dissolved in 50% acetonitrile containing 0.1% trifluoroacetic acid to prepare a saturated solution, and 1 μL of the solution was mixed with 1 μL tryptic-digested fragments. The mixture (1 μL) was loaded onto the mass spectrometer. Positive-ion mass spectra were measured. Two peaks of self-digested trypsin (842.51 and 2211.1046) were used as internal standards.
The avidin-biotin-horseradish peroxidase complex (ABC) method was used. Paraffin embedded sections were dewaxed, rehydrated through graded ethanol solutions, and washed three times with PBS. After endogenous peroxidase activity was blocked with 0.3% H2O2, sections were incubated with normal horse/goat serum (1:50) for 20 min, followed by incubation with a primary antibody diluted with 2% BSA (sPLA2, 1:200; PRX6, 1:500) at 4 °C overnight. Sections were then incubated with a secondary antibody diluted with 2% BSA (1:200) for 45 min and ABC (1:1:50) for 30 min at room temperature. Following diaminobenzidine coloration, sections were counterstained with hematoxylin for 15-30 s and mounted. A negative control was run by replacing the primary antibody with PBS, and p53-positive esophageal cancer tissue was used as a positive control[5]. Staining intensity was scored as follows: 0 (no staining); 1 (light yellow granules on the membrane or in the cytoplasm); 2 (yellow granules); and 3 (brown granules). Five hundred cells were counted in five microscopic fields per slide to calculate the percentage of stained cells, which was scored as follows: 0 (< 5%); 1 (5%-25%); 2 (25%-75%); and 3 (> 75%). Negative, positive and strongly positive staining was considered when the sum of the score of staining intensity score and the score of the percentage of stained cells was 0, 1-4, and > 4, respectively.
Differential protein spots that were present on 2-D gels for esophageal cancer and fetal esophageal tissues but not on gels for normal esophageal epithelial tissues were chosen for further analysis. To ensure its reproducibility, 2-D gel electrophresis for each sample was repeated at least twice.
Statistical analyses were performed using SPSS version 10.0 software. Kruskal-Wallis and Mann-Whitney tests were used for comparing intergroup differences, with statistical significance set at α = 0.05.
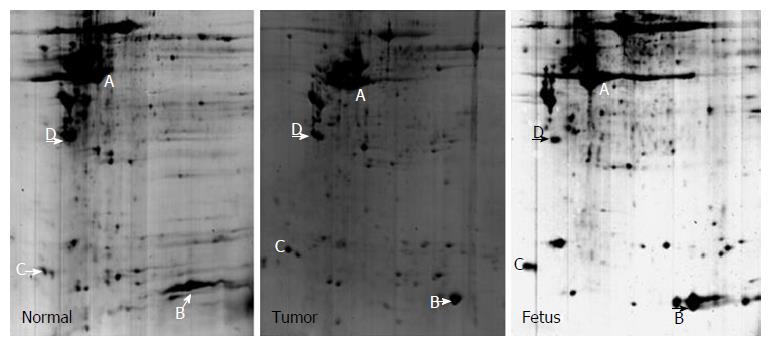
By comparing different 2-D gel images for different samples, four landmark protein spots (A-D) that were present on all gel images were selected (Figures 2 and 3). Comparison of these spots with protein markers showed that their Mr was 40.2, 14.2, 16.8 and 28 kD, respectively, and PDQuest version 7.1.1 software determined their PI as 5.9, 7.1, 3.4 and 4.2, respectively.
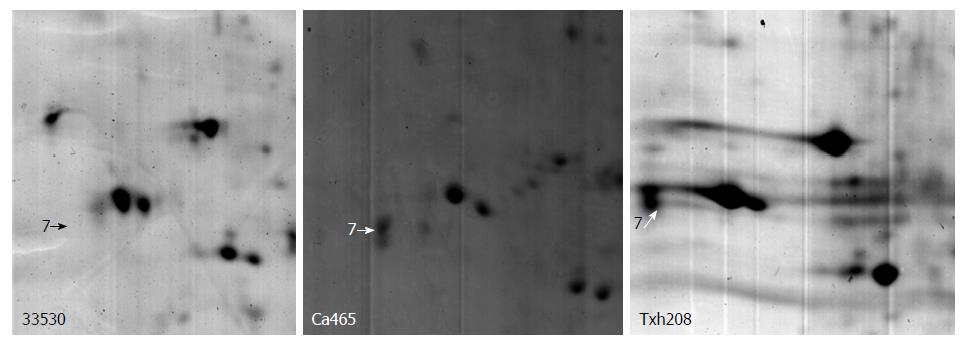
Protein spots that were significantly differentially expressed on 2-D gel images were cut and then subjected to in-gel tryptic digestion and MALDI-TOF analysis to obtain the peptide mass fingerprint (PMF). PMF analysis and search of protein databases were then performed to identify differentially expressed protein spots. Table 1 lists the names of proteins, their theoretical and observed pI and Mr, number of matched peptide fragments, sequence coverage and MOWSE score. When searching protein databases, we restricted the above parameters to improve the accuracy of the search. After PMF analysis and search of protein databases, 21 differential proteins were identified; some of which represent a protein isoform.
| Spot | Protein name and theoretical pI and Mr | Observed pI and Mr | No. of matched peptide fragments | Sequence coverage | MOWSE score |
| 2 | USP30 6.87/45546 | 9.3/32 | 2 | 4 | 21 |
| 4 | Thyroid hormone receptor interactor 4.56/11 8489 | 7.8/31 | 3 | 56 | 51 |
| 5 | Cardiac muscle 5.34/22563 | 6.25/29 | 5 | 24 | 66 |
| 6 | Chain A, crystal structure of a recombinant glutathione transferase 5.09/23202 | 6.2/24 | 5 | 43 | 85 |
| 7 | Dermatopontin precursor 4.7/23989 | 5.3/21.5 | 5 | 29 | 82 |
| 10 | Heat shock protein 27 7.83/22313 | 7.1/20.5 | 5 | 29 | 79 |
| 11 | S100 calcium-binding protein A9 5.71/13234 | 6.3/15 | 5 | 40 | 74 |
| 12 | G-gamma-hemoglobin 6.17/11033 | 7.5/16 | 3 | 45 | 65 |
| 13 | MB protein 9.24/10863 | 8.4/15.5 | 3 | 32 | 65 |
| 14 | Immunoglobulin heavy chain variable region 9.78/13026 | 8.6/14 | 3 | 34 | 70 |
| 15 | PREDICTED: hypothetical protein 5.27/6706 | 8.1/12 | 2 | 44 | 52 |
| 16 | Immunoglobulin heavy chain variable region 9.04/13053 | 7.1/11 | 2 | 26 | 49 |
| 17 | Heat shock 27kDa protein 1 5.98/22768 | 9.2/16 | 6 | 33 | 80 |
| 18 | hCG1659706 10.28/5287 | 9.5/21 | 2 | 64 | 37 |
| 19 | Smooth muscle protein 8.56/22461 | 9.4/19 | 6 | 26 | 86 |
| 21 | hCG1997574 9.07/29453 | 7.3/32 | 7 | 31 | 93 |
| 22 | Heat shock 27 kDa protein 1 5.98/22768 | 6.8/32 | 6 | 35 | 84 |
| 23 | Disabled homolog 1 (Drosophila) 9.40/22903 | 7.15/31 | 6 | 26 | 66 |
| 24 | Phosphoglycerate mutase 1 (brain), 6.40/24669 | 7.3/31 | 8 | 40 | 117 |
| 25 | Peroxiredoxin 6 6.00/25019 | 7.1/27.8 | 8 | 41 | 116 |
| 26 | Chain A, human triosephosphate isomerase of new crystal form 6.50/26666 | 7.3/30 | 10 | 38 | 160 |
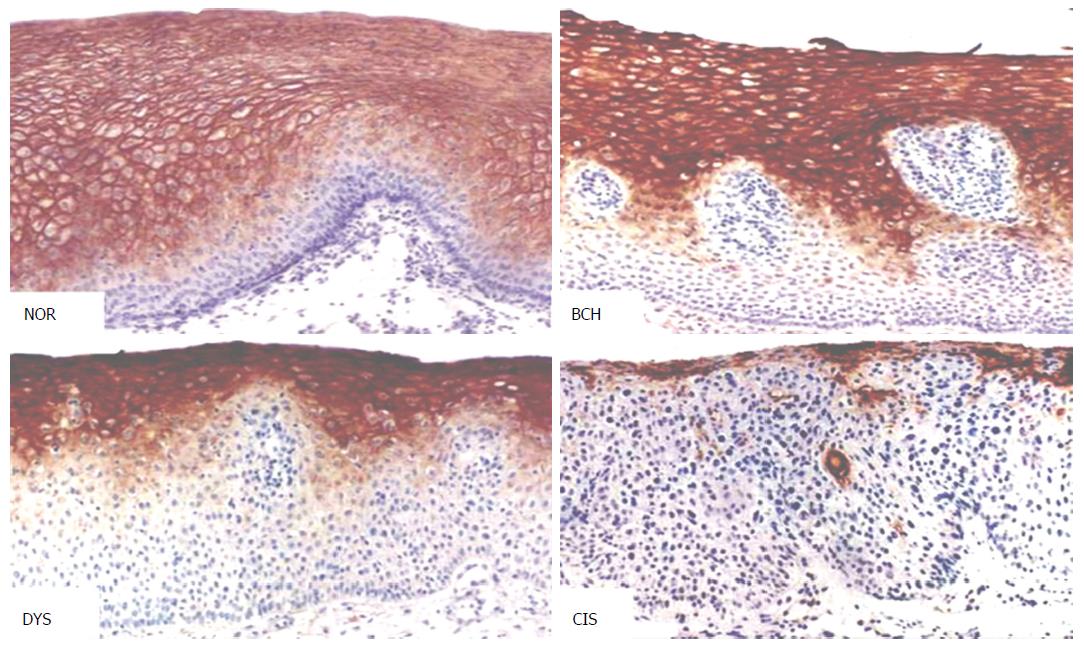
PRX6 protein was stained brown and mainly localized on the cell membrane, and occasionally in the cytoplasm and nucleus (Figures 4 and 5).
In morphologically normal adult esophageal epithelial tissues, moderate staining of PRX6 protein was observed. With the progression of esophageal lesions, the positive rate of PRX6 protein expression showed a declining trend, especially prominent in dysplasia (DYS) and carcinoma in situ (CIS) (Figure 4), in which weak staining of PRX6 protein predominated. In tumor-adjacent esophageal epithelial tissues, the rate of strongly positive staining (≥ 4 points) decreased from 43% in normal epithelium (NOR) to 35% in basal cell hyperplasia. In DYS, moderately or weakly positive staining of PRX6 protein (< 4 points) predominated. The majority of CIS lesions (79%) showed no staining for PRX6 protein, with weak staining predominating. Interestingly, cancer cells in SCC showed varying degrees of PRX6 staining, with strongly positive staining (≥ 4 points) predominating. In seven cases of well-differentiated SCC, varying degrees of PRX6 staining were observed. With the decrease in the differentiation degree of SCC, PRX6 protein expression gradually declined (P < 0.05) (Table 2).
| Tissue | PRX6 | ||
| No. of cases | Negative | Positive | |
| Tumor-adjacent | |||
| NOR | 37 | 3 (8) | 34 (92) |
| BCH | 37 | 3 (8) | 34 (92) |
| DYS | 29 | 18 (62) | 11 (38) |
| CIS | 28 | 22 (79) | 6 (21) |
| Esophageal cancer | |||
| W-SCC | 7 | 0 | 7 (100) |
| M-SCC | 16 | 3 (19) | 13 (81) |
| P-SCC | 6 | 5 (83) | 1 (17) |
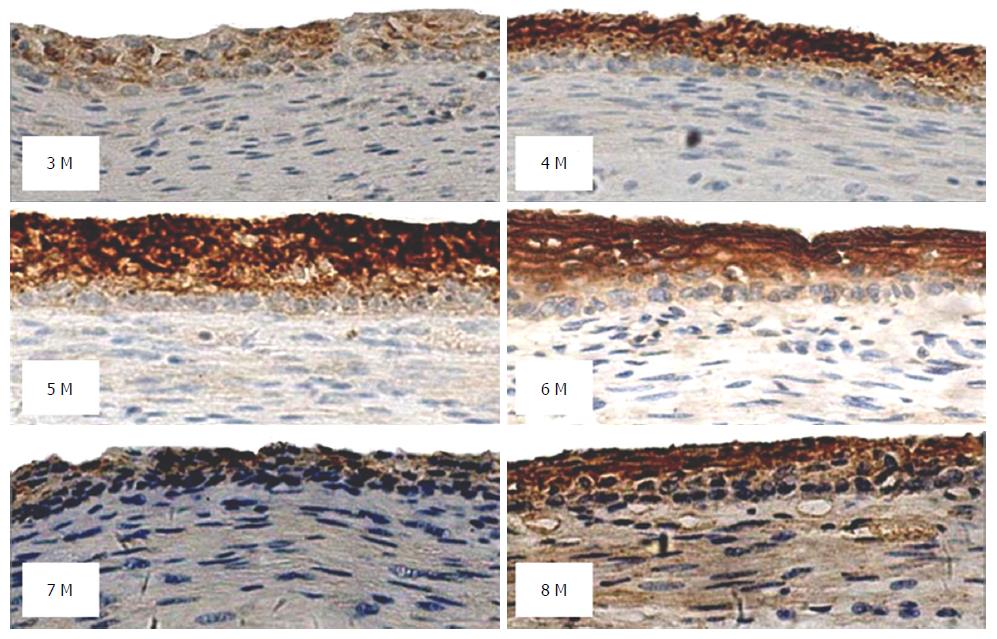
In fetal esophageal epithelial tissues, the positive rate of PRX6 protein was 87%. In esophageal epithelial tissues from fetuses at a gestational age of 4-6 mo, strongly positive staining of PRX6 protein (≥ 4 points) predominated. In esophageal epithelial tissues from fetuses at a gestational age of 3, 7 or 8 mo, moderately or weakly positive staining (< 4 points) predominated. In esophageal epithelial tissues from fetuses at a gestational age of 8 mo, both strongly positive staining and negative staining were observed (Figure 5). Statistical analysis showed that PRX6 protein expression in esophageal epithelial tissues from fetuses at a gestational age of 4 or 7 mo differed significantly compared with that from fetuses of other gestation age groups (P < 0.05, Table 3).
| Age (mo) | PRX6 | ||
| No. of cases | Negative | Positive | |
| 3 | 6 | 0 | 6 (100) |
| 4 | 4 | 0 | 4 (100) |
| 5 | 6 | 1 (17) | 5 (83) |
| 6 | 6 | 0 | 6 (100) |
| 7 | 5 | 1 (20) | 4 (80) |
| 8 | 6 | 1 (17) | 5 (83) |
| 9 | 6 | 2 (33) | 4 (67) |
The development and progression of esophageal cancer is a multistage process involving many genes[6-9]. Proteomics is the study of the entire set of proteins expressed by a single cell or tissue. With the aid of proteomics, changes in protein molecules directly related to tumor development and progression can be identified, thus allowing to find markers for early diagnosis of tumors and monitoring of curative effects. This study utilized the 2-DE-based proteomic technology to identify differentially expressed proteins in fetal esophageal tissues, esophageal cancer tissues, and tumor-adjacent normal esophageal epithelial tissues from fetuses of different gestational age, or esophageal cancer patients from a high-risk area for esophageal cancer in China. Twenty-one significantly differentially expressed proteins were identified, which may be related to the development and growth of fetal esophageal epithelium or the development and progression of esophageal cancer.
PRX6 protein belongs to the antioxidant PRX family, which is ubiquitously expressed in prokaryotes and eukaryotes[10] and has six mammalian members[11]. Chevallet et al[12] found that PRX6 expression continued to rise within 1-24 h after oxidative stress. Besides antioxidant properties, PRXs can also be radiation protective, stimulate cell proliferation, and participate in cell signal transduction. This study found that PRX6 expression was elevated in esophageal cancer tissues where cell metabolism and proliferation are active, suggesting that increased PRX6 may inhibit the excessive production of reactive oxygen species and maintain cell homeostasis to protect cells from damage. Similar to our results, PRX protein expression was also found to be elevated in malignant mesothelioma, lung cancer, and oral cancer[13-15]. In our previous proteomic study of esophageal SCC, PRX1 protein expression was upregulated, but PRX2 expression was downregulated[16], indicating that the development and progression of esophageal SCC involves changes in a variety of antioxidant proteins, and different antioxidant proteins or protein isoforms may have different functions in the development and progression of esophageal cancer. Further studies should be performed to clarify the exact mechanisms of these antioxidant proteins.
The most important finding of this study is that expression of PRX6 protein decreased with progression of esophageal lesions. In tumor-adjacent normal esophageal epithelial tissues, PRX6 protein was highly expressed; however, PRX6 protein expression was significantly reduced in CIS. PRX6 protein expression was positively correlated with the degree of tumor differentiation in esophageal SCC. With the decrease in tumor differentiation, PRX6 protein expression decreased. In the fetal esophageal epithelium, PRX6 protein expression showed a fluctuating pattern with the increase of gestational age. The intensity of PRX6 protein expression was positively correlated with the degree of differentiation of epithelial cells. PRX6 protein expression was highly expressed in well-differentiated epithelial cells, but lowly expressed or not expressed in lowly differentiated epithelial cells.
Both fetal esophageal epithelium and carcinoma tissues have active cell proliferation and metabolism, which may result in increased peroxide generation. PRX6 protein in fetal esophageal epithelium and carcinoma tissues may have antioxidant effects by eliminating peroxides produced by the cell metabolism, thus protecting cells from damage. Loss of expression of PRX6 protein in CIS may promote cell proliferation and facilitate the progression of precancerous lesions. However, the dynamic expression pattern of PRX6 protein in esophageal precancerous lesions and fetal esophageal epithelium suggests that PRX6 protein is associated with the multistage evolution of esophageal cancer and may inhibit cell proliferation and promote the differentiation of esophageal epithelial cells. Loss of expression of PRX6 protein in CIS is likely to promote cell proliferation and facilitate the progression of precancerous lesions. In poorly differentiated carcinoma tissue, loss of PRX6 protein may lead to rapid cell proliferation. Under certain conditions, PRX6 protein may also be involved in the induction of differentiation of cancer cells, thus resulting in the formation of highly differentiated carcinoma tissue. In fetal esophageal epithelium, PRX6 protein expression showed no significant correlation with gestational age, but was, to some extent, associated with the morphology of epithelial cells. In the base layer where cells are arranged neatly, the level of PRX6 protein expression was high. In contrast, when the arrangement of epithelial cells was disorderly, PRX6 protein exhibited low expression. These findings suggest that PRX6 protein plays an important role in the process of differentiation of fetal esophageal epithelial cells. We speculate that PRX6 protein may promote the trans-differentiation from precancerous lesions to normal esophageal epithelium. However, the precise role of PRX6 protein in cell differentiation requires further research.
This study utilized the 2-DE based proteomic technology to analyze the differential protein expression profiles in fetal esophageal tissues, esophageal cancer tissues, and tumor-adjacent normal esophageal epithelial tissues from fetuses of different gestational age, or esophageal cancer patients from a high-risk area for esophageal cancer in China. We identified 21 significantly differentially expressed proteins that may be related to the development and growth of fetal esophageal epithelium or the development and progression of esophageal cancer, such as dermatopontin, S100A9, HSP27, and PRX6. Our results suggest that the development and progression of esophageal cancer are a result of interactions of many genetic changes (accumulation or superposition). These proteins are related not only to esophageal cancer, but also to other tumors. The identification of proteins associated with esophageal cancer and fetal esophageal development can improve the understanding of the molecular mechanism of esophageal cancer development, and may help provide a fetal esophageal model for esophageal cancer. PRX6 protein plays an important role in the induction of differentiation of esophageal epithelial cells, and PRX6 protein expression level may be used as an important marker for evaluating cell differentiation. In this regard, further research of molecular mechanism of PRX6 is of great significance.
The fetal esophageal epithelium is developed from unmatured monolayer columnar epithelium gradually into a matured stratified squamous epithelium, differentiated from immature gradually to mature, epithelial cells differentiated from the active division and proliferation epithelial stem cells gradually matured into stratified squamous epithelium; Adult esophageal epithelial carcinogenesis is the differentiation of mature epithelial tissue in the role of carcinogenic factors, the emergence of hyperplasia and abnormal differentiation eventually developed into cancer. Obviously, the two have the opposite differentiation process. The aims of this study were to: (1) find the protein molecules that are differentially or identically expressed in the process of maturation and differentiation of fetal esophageal epithelium and carcinogenesis of adult esophageal epithelium; and (2) to strengthen the understanding of the molecular mechanism of esophageal carcinogenesis, so as to provide an important theoretical basis for the screening and detection of early molecular markers in high-risk population.
Clarifying the molecular mechanisms underlying the development and progression of esophageal cancer and identifying biological markers for screening high-risk populations are of clinical importance for early diagnosis. Utilize the proteomic technology to analyze the differential protein expression profiles in the development and growth of fetal esophageal epithelium and the development and progression of esophageal cancer, with an aim to clarify the molecular mechanism of esophageal cancer development.
The authors identified 21 differentially expressed proteins by peptide mass fingerprinting and database analysis in the study, and some proteins contained different isoforms.
Peroxiredoxin (PRX)6 protein expression level may be used as an important marker for evaluating cell differentiation. In this regard, further research of molecular mechanism of PRX6 is of great significance.
MALDI-TOF-MS: matrix-assisted laser desorption/ionization time of flight mass spectrometry.
This is an excellent study. In this study, the authors analyzed the differential protein expression profiles in fetal.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A, A, A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: AndersenV, Kruis W, Sato H, Yamaoka Y S- Editor: Qi Y L- Editor: Kerr C E- Editor: Zhang FF
| 1. | Sheng KL, Zhu SY, Liu YS. Observation of human fetal eosphageal epihtelium. Acta Anat Sin. 1999;30:158-160. [DOI] [Full Text] |
| 2. | Johnson FP. The development of the mucous membrane of the esophagus, stomach and small intestine in the human embryo. Am J Anat. 1910;10:521-559. [RCA] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 108] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | He QY, Chen J, Kung HF, Yuen AP, Chiu JF. Identification of tumor-associated proteins in oral tongue squamous cell carcinoma by proteomics. Proteomics. 2004;4:271-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 4. | Srisomsap C, Sawangareetrakul P, Subhasitanont P, Panichakul T, Keeratichamroen S, Lirdprapamongkol K, Chokchaichamnankit D, Sirisinha S, Svasti J. Proteomic analysis of cholangiocarcinoma cell line. Proteomics. 2004;4:1135-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 5. | Wang LD, Shi ST, Zhou Q, Goldstein S, Hong JY, Shao P, Qiu SL, Yang CS. Changes in p53 and cyclin D1 protein levels and cell proliferation in different stages of human esophageal and gastric-cardia carcinogenesis. Int J Cancer. 1994;59:514-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Vizcaino AP, Moreno V, Lambert R, Parkin DM. Time trends incidence of both major histologic types of esophageal carcinomas in selected countries, 1973-1995. Int J Cancer. 2002;99:860-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 303] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 7. | Wang LD, Qin YR, Fan ZM, Kwong D, Guan XY, Tsao GS, Sham J, Li JL, Feng XS. Comparative genomic hybridization: comparison between esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-incidence area for both cancers in Henan, northern China. Dis Esophagus. 2006;19:459-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Roth MJ, Guo-Qing W, Lewin KJ, Ning L, Dawsey SM, Wesley MN, Giffen C, Yong-Qiang X, Maher MM, Taylor PR. Histopathologic changes seen in esophagectomy specimens from the high-risk region of Linxian, China: potential clues to an etiologic exposure? Hum Pathol. 1998;29:1294-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, Li JM, Kong GQ, Qi H, Cui J. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 338] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 10. | Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 496] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 11. | Shen C, Nathan C. Nonredundant antioxidant defense by multiple two-cysteine peroxiredoxins in human prostate cancer cells. Mol Med. 2002;8:95-102. [PubMed] |
| 12. | Chevallet M, Wagner E, Luche S, van Dorsselaer A, Leize-Wagner E, Rabilloud T. Regeneration of peroxiredoxins during recovery after oxidative stress: only some overoxidized peroxiredoxins can be reduced during recovery after oxidative stress. J Biol Chem. 2003;278:37146-37153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 131] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 13. | Kinnula VL, Lehtonen S, Sormunen R, Kaarteenaho-Wiik R, Kang SW, Rhee SG, Soini Y. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196:316-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 184] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 14. | Chang JW, Jeon HB, Lee JH, Yoo JS, Chun JS, Kim JH, Yoo YJ. Augmented expression of peroxiredoxin I in lung cancer. Biochem Biophys Res Commun. 2001;289:507-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Yanagawa T, Iwasa S, Ishii T, Tabuchi K, Yusa H, Onizawa K, Omura K, Harada H, Suzuki H, Yoshida H. Peroxiredoxin I expression in oral cancer: a potential new tumor marker. Cancer Lett. 2000;156:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 88] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Qi Y, Chiu JF, Wang L, Kwong DL, He QY. Comparative proteomic analysis of esophageal squamous cell carcinoma. Proteomics. 2005;5:2960-2971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |