Published online Feb 21, 2017. doi: 10.3748/wjg.v23.i7.1189
Peer-review started: October 10, 2016
First decision: November 9, 2016
Revised: November 24, 2016
Accepted: December 16, 2016
Article in press: December 19, 2016
Published online: February 21, 2017
Processing time: 134 Days and 6.7 Hours
To explore novel therapeutic target of cisplatin resistance in human gastric cancer.
The sensitivity of SGC7901 cells and cisplatin-resistant SGC7901 cells (SGC7901/DDP) for cisplatin were detected by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. High-quality total RNA which isolated from SGC7901/DDP cells and SGC7901 cells were used for mRNA microarray analysis. Results were analyzed bioinformatically to predict their roles in the development of cisplatin resistance and the expression of 13 dysregulated mRNAs we selected were validated by quantitative real-time polymerase chain reaction (qRT-PCR).
SGC7901/DDP cells highly resistant to cisplatin demonstrated by MTT assay. A total of 1308 mRNAs (578 upregulated and 730 downregulated) were differentially expressed (fold change ≥ 2 and P-value < 0.05) in the SGC7901/DDP cells compared with SGC7901 cells. The expression of mRNAs detected by qRT-PCR were consistent with the microarray results. Gene Ontology, Kyoto Encyclopedia of Genes and Genomes pathway and protein-protein interaction analysis demonstrated that the differentially expressed mRNAs were enriched in PI3K-Akt, Notch, MAPK, ErbB, Jak-STAT, NF-kappaB signaling pathways which may be involved in cisplatin resistance. Several genes such as PDE3B, VEGFC, IGFBP3, TLR4, HIPK2 and EGF may associated with drug resistance of gastric cancer cells to cisplatin.
Exploration of those altered mRNAs may provide more promising strategy in diagnosis and therapy for gastric cancer with cisplatin resistance.
Core tip: We tested the sensitivity of human gastric cancer cells SGC7901/DDP and SGC7901 for cisplatin and compared their mRNA expression profile using a human mRNA microarray, and then performed bioinformatics analysis to depict comprehensively the properties of the differentially expressed mRNAs. Results demonstrated that the dysregulated mRNA were enriched in functions and pathways that may be involved in cisplatin resistance. Exploration of the dysregulated genes could suggest a promising strategy in diagnosis and therapy of gastric cancer with cisplatin resistance.
- Citation: Xie XQ, Zhao QH, Wang H, Gu KS. Dysregulation of mRNA profile in cisplatin-resistant gastric cancer cell line SGC7901. World J Gastroenterol 2017; 23(7): 1189-1202
- URL: https://www.wjgnet.com/1007-9327/full/v23/i7/1189.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i7.1189
Gastric cancer is the fourth most common cancer and the second leading cause of cancer death globally[1], and more than two thirds of patients when diagnosed with unresectable disease[2]. The 5-year overall survival rate of patients with advanced gastric cancer approximately 25%[3]. Currently, platinum-based chemotherapy regimen is the standout chemotherapy frequently used for advanced gastric cancer[4,5], and median overall survival and progression free survival was significantly longer in cisplatin-containing combination therapy compared to non-cisplatin containing regimens[6,7]. However, cisplatin-based chemotherapeutic agents are often limited in chemotherapy due to drug resistance[8,9].
Cisplatin resistance of gastric cancer is multifactorial, accumulating evidence have suggested that the aberrant expression of proteins which associated with decreased cellular accumulation, increased DNA repair capacity, increased drug inactivation[10] play important role in the acquisition of cisplatin resistance. Previous researches have shown that abnormal expression of copper transporter 1 (CTR1) and MRP2 lead to cisplatin resistance by reducing the concentration of cisplatin in cells[11-13]. Moreover, the upregulation of excision repair cross complementing 1 (ERCC1)[14], X-ray repair cross complementing 1 (XRCC1)[15] and breast cancer 1 (BRCA1)[16] have shown to be involved in cisplatin resistance by removal of Pt-DNA adducts[17,18]. Other studies have shown that downregulation of the human epidermal growth factor receptor II (ErbB2) can significantly enhanced the apoptosis-inducing effects of cisplatin in gastric cancer[19,20].
The mechanisms of cisplatin resistance are quite complex and have not been fully revealed till now, so investigation of the molecular mechanisms and biomarkers is urgently needed. This study aims to analyze mRNA expression profiles in SGC7901/DDP cells to explore more chemotherapeutic molecular targets and to guide appropriate chemotherapy for gastric cancer with cisplatin resistance.
The human cisplatin-resistant gastric cancer cell line SGC7901/DDP and its parental cells SGC7901 were purchased from KeyGEN Biotechnology Company (Nanjing, Jiangsu, China). Cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, United States) containing 10% fetal calf serum (Gibco, NY, United States) supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were cultured in a humidified atmosphere with 5% CO2 at 37 °C. Cisplatin (Sigma, CA, United States) with final concentration of 800 ng/mL was added to the culture media for SGC7901/DDP cells to maintain the cisplatin-resistant phenotype.
SGC7901/DDP and SGC7901 cells were suspended at a density of 1 × 105 cells/mL and planted into 96-well culture plate. After 24 hours, the cells were treated with freshly prepared DDP. The final concentrations were 133.34 μmol /L, 66.67 μmol/L, 6.67 μmol/L, 0.67 μmol/L and 0.067 μmol/L, because the human peak plasma concentration for DDP has been reported as 6.67 μmol/L[21]. Cell viability was examined after 48 h and was determined by adding 20 μL MTT (5 mg/mL) to each well and incubated for a further 4 h. The resulting formazan crystal was dissolved by addition of 150 μL dimethyl sulfoxide (DMSO) (sigma, Germany) each well, and then plates were shaken for 10 minutes. The absorbance at 490 nm was measured by spectrophotometer (ELx 800; BioTek; Winooski, VT, United States). The inhibition of growth (IC50) for DDP was calculated by the cells relative viability. Each experiment was performed in triplicate.
Cells were harvested when they had grown to 80%-90% confluency and were still in logarithmic phase. Total RNA was extracted from the three matched pairs of SGC7901/DDP and SGC7901 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The quality of total RNA was measured by NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Waltham, MA, United States). Total RNA from three paired samples were amplified and transcribed into fluorescent cDNA, and then the fluorescent labeled samples were hybridized to the Agilent LncRNA-mRNA Human Gene Expression Microarray V4.0 (Capital Bio Corp, Beijing, China) which contains 25069 human mRNA according to the manufacturer’s recommendations. The microarray was scanned by an Agilent Microarray Scanner. Image processing was conducted using Agilent Feature Extraction software and raw microarray signals normalized using Agilent Gene-Spring software. The normalized mRNA expression profiles data output was received in Excel spreadsheets. The two group of samples data were analyzed by t-test to get the P-values. FC values representing the differently expressed mRNAs between SGC7901/DDP and their parental cells. Cluster 3.0 software was performed to show differential expression patterns of mRNAs.
Bioinformatics analysis were generated using KOBAS software and STRING 9.1 software. KOBAS software was used to analyze Ontology, Disease and pathways of the dysregulated mRNAs. KOBAS associated with 1 ontology database (Gene Ontology), 5 disease databases (OMIM, KEGG DISEASE, PID Reactome, FunDO, GAD, NHGRI) and 7 pathway databases (KEGG PATHWAY, PID Curated, PID BioCarta, BioCyc, eactome, Panther). The entire analysis process includes two steps: first, bring the input gene ID map to the gene in the databases, and then annotate pathways, disease and function of these genes involved in. Second step, compare the first step results with background (usually the entire genome of the gene, or the entire probe on the chip), and unearth statistically significant enrichment pathways, disease or function. Fisher’s exact test and χ2 test were used as statistical tests and the FDR was performed to correct the P-value[22]. Additionally, we used STRING 9.1 software to decipher the protein-protein interaction (PPI) network of the differentially expressed proteins. The PPI network may help in understanding the molecular mechanism of cisplatin resistance. All mRNA microarray data were given by Capital Bio Corp.
To validate the reliability of microarray analysis, we performed quantitative real-time PCR (qRT-PCR). The reverse transcription production cDNA was synthesized using oligo-dT primers and Superscript II reverse transcriptase. PCR was performed with SYBR® Premix Ex TaqTM (TaKaRa Bio; Japan) by a Light Cycler PCR system (Agilent Technologies, Palo Alto, CA, United States) according to the manufacturer’s instructions. After amplification, melting curves were analyzed. Beta-actin snRNA used as endogenous control, each sample was done in triplicate. The relative expression levels of target mRNAs were calculated using the 2-∆∆Ct method (where ∆∆Ct is the difference in threshold cycles for the ∆Ct of SGC7901/DDP sample and SGC7901 sample, and ∆Ct is the difference between the target gene and endogenous control beta-actin). Sequences of primers for qRT-PCR are provided in supporting Table 1.
| ID | Primer | Sequence (5'to3') | Base (bp) | Tm (°C) | GC |
| HIPK2 | Forward | CCCCGTGTACGAAGGTATGG | 20 | 59.90 | 60% |
| Reverse | GGGATGTTCTTGCTCTGGCT | 20 | 60.03 | 55% | |
| PDE3B | Forward | TGAGAGTTATGGCTGCCTGT | 20 | 58.72 | 50% |
| Reverse | CTGAGGGGCATTTGTAGCCA | 20 | 60.30 | 55% | |
| FGF2 | Forward | TCCACCTATAATTGGTCAAAGTGGT | 25 | 59.99 | 40% |
| Reverse | CATCAGTTACCAGCTCCCCC | 20 | 59.82 | 60% | |
| TWIST1 | Forward | ATTCAAAGAAACAGGGCGTGG | 21 | 59.39 | 47.6% |
| Reverse | CAGAGGTGTGAGGATGGTGC | 20 | 60.39 | 40% | |
| ZEB2 | Forward | GCCTCTGTAGATGGTCCAGTGA | 22 | 61.21 | 54.6% |
| Reverse | ATCGCGTTCCTCCAGTTTTCT | 21 | 60.00 | 47.6% | |
| VEGFC | Forward | CCCGCCTCTCCAAAAAGCTA | 20 | 60.04 | 55% |
| Reverse | CGGGTGTCAGGTAAAAGCCT | 20 | 59.96 | 55% | |
| SPHK1 | Forward | GCTGCGAAGTTGAGCGAAAA | 20 | 60.04 | 50% |
| Reverse | CGTTCCCTACAGTGGCCTG | 19 | 60.08 | 63.2% | |
| BAX | Forward | GCCCTTTTGCTTCAGGGTTT | 20 | 59.24 | 50% |
| Reverse | CATCCTCTGCAGCTCCATGT | 20 | 59.82 | 55% | |
| PTEN | Forward | CAGGATACGCGCTCGGC | 17 | 60.73 | 70.6% |
| Reverse | ACAGCGGCTCAACTCTCAAA | 20 | 57.89 | 50% | |
| HTRA1 | Forward | AGCCAAAGAGCTGAAGGACC | 20 | 59.96 | 55% |
| Reverse | GACATCATTGGCGGAGACCA | 20 | 60.11 | 55% | |
| CCL5 | Forward | TGCTGCTTTGCCTACATTGC | 20 | 59.76 | 50% |
| Reverse | CTTGTTCAGCCGGGAGTCAT | 20 | 60.04 | 55% | |
| TGM2 | Forward | CCTCTGTCTCTCCGGGAACC | 20 | 61.32 | 65% |
| Reverse | TGGCAACCAGGGGTCCTAT | 19 | 60.23 | 57.9% | |
| TLR4 | Forward | CTCGGTCAGACGGTGATAGC | 20 | 59.97 | 60% |
| Reverse | TTTAGGGCCAAGTCTCCACG | 20 | 59.68 | 55% | |
| ACTB | Forward | CTCACCATGGATGATGATATCGC | 23 | 59.13 | 47.8% |
| Reverse | AGGAATCCTTCTGACCCATGC | 21 | 59.79 | 52.4% |
MTT test and qRT-PCR statistical analysis was performed using GraphPad Prism software (v. 5.0a; GraphPad Software, La Jolla, CA, United States). We used one-way analysis of variance (ANOVA) followed by Student’s t-test to assess the statistical significance of differences between different cell groups. The threshold for statistical significance was P-values < 0.05. Fold changes of mRNAs validated by qRT-PCR in SGC7901/DDP cells compared with SGC7901 cells are shown as mean ± SD.
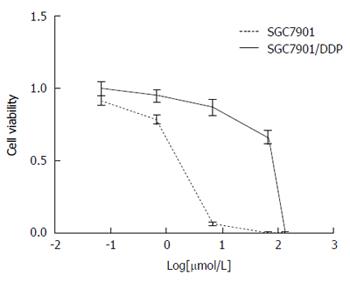
To determine the chemotherapy sensitivity of SGC7901/DDP and SGC7901 cell line to cisplatin, varying concentrations of cisplatin were added into the 96-well plates and incubated for 48 h. From these data, half maximal inhibitory concentration (IC50) cisplatin dose was calculated. IC50 cisplatin doses for SGC7901/DDP and SGC7901 (after 48 h in DDP-containing media) were 43.47 ± 0.21 μmol/L and 1.24 ± 0.02 μmol/L, respectively, and the resistance index for SGC7901/DDP cell lines was 35.12, confirming that these cells are refractory to cisplatin. Cell viability was checked by MTT assay (Figure 1).
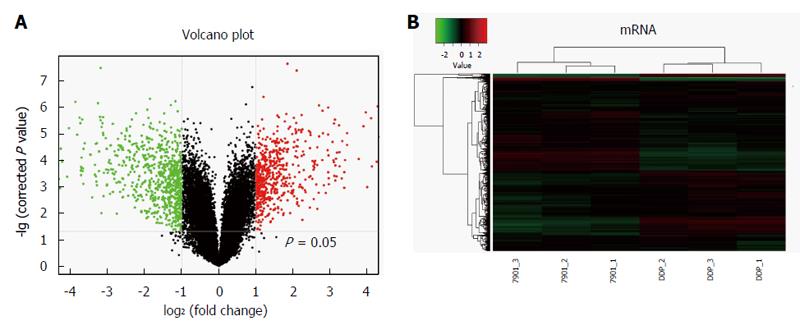
To show mRNA expression profile in cisplatin-resistant SGC7901/DDP cells, we used a stringency cutoff to identify significantly differently mRNAs (P < 0.05, FC ≥ 2) and two-dimensional hierarchical clustering 3.0 to represent expression profiles between samples (Figure 2). The results indicated that 1308 mRNAs were significantly differentially expressed in SGC7901/DDP cells compared with SGC7901 cells. Among these transcripts, 578 mRNAs were upregulated, and 730 mRNAs were downregulated.
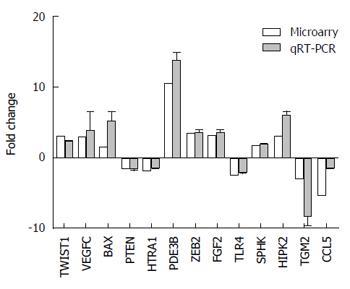
First, we concentrated on validating the microarray results. From the abnormally expressed (P < 0.05) mRNAs obtained from the microarray analyses, we selected 8 upregulated mRNAs (HIPK2, PDE3B, FGF2, TWIST1, ZEB2, VEGFC, SPHK1, BAX) and 5 downregulated (PTEN, HTRA1, CCL5, TGM2, TLR4) mRNAs for qRT-PCR validation. The relative fold-changes (SGC7901/DDP vs SGC7901) detected by qRT-PCR were consistent with the microarray results (Figure 3), indicating the dependability of our microarray platform.
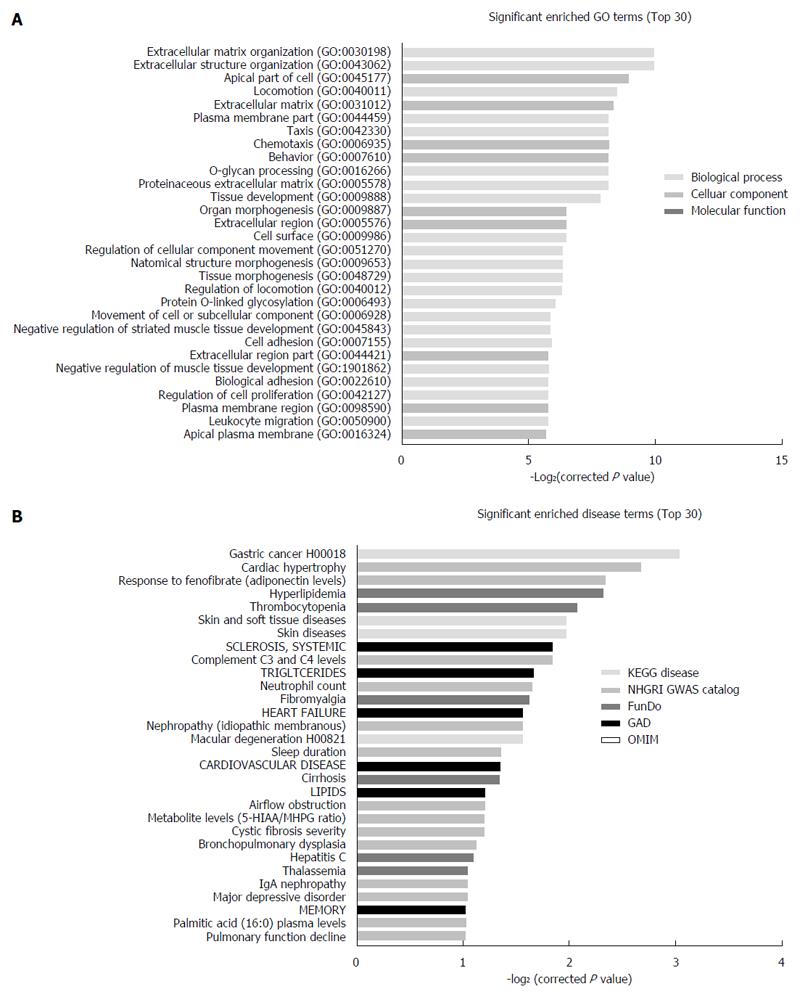
To depict comprehensively the properties of the differentially expressed mRNA in SGC7901/DDP cells, GO annotation and enrichment analysis was performed to evaluate which cellular components, molecular functions and biological processes may be are affected by this dysregulation. The GO enrichment analysis showed that the differentially expressed genes were involved in a variety of functions, including locomotion, chemotaxis, cell adhesion, regulation of cell migration, extracellular matrix disassembly, response to xenobiotic chemotaxis, localization of cell adhesion and blood vessel morphogenesis (Figure 4A).
Additionally, 59 human diseases were significant enriched (P < 0.05) in five human disease databases (KEGG DISEASE, FunDO, GAD, NHGRI GWAS Catalog and OMIM) (Figure 4B, Table 2). Furthermore, it is worth noting that in KEGG disease database, gastric cancer is the most highly enriched disease, and the input genes include DCC, CD44, CDH1, VEGFC, EGF, TGFA.
| Term | Database | P value | Input gene symbols |
| Gastric cancer | KEGG DISEASE | 0.0016 | DCC, CD44, CDH1, VEGFC, EGF, TGFA |
| Skin diseases | KEGG DISEASE | 0.0078 | DSP, TGM1, CCL5, IL31RA, SPINK5, HLA, FERMT1, KRT14, CTSC, COL17A1, LAMA3, REEP1, RIN2, ALOXE3, ABCC6, WNT10A, FBLN5 |
| Skin and soft tissue diseases | KEGG DISEASE | 0.0078 | DSP, TGM1, CCL5, IL31RA, SPINK5, HLA, FERMT1, KRT14, CTSC, COL17A1, LAMA3, REEP1, RIN2, ALOXE3, ABCC6, WNT10A, FBLN5 |
| Macular degeneration | KEGG DISEASE | 0.0140 | C3, FBLN5, CFH, TLR4 |
| Cancers of the digestive system | KEGG DISEASE | 0.0439 | DCC, CD44, CDH1, VEGFC, EGF, TGFA |
| Familial thoracic aortic aneurysm and dissection (TAAD) | KEGG DISEASE | 0.0459 | MYLK, TGFBR1 |
| Hypomagnesemia | KEGG DISEASE | 0.0459 | TRPM6, EGF |
| Multiple epiphyseal dysplasia (MED) | KEGG DISEASE | 0.0459 | COL9A3, MATN3 |
| Transient neonatal diabetes mellitus (TNDM) | KEGG DISEASE | 0.0459 | PLAGL1, ZFP57 |
| Non-syndromic autosomal dominant mental retardation | KEGG DISEASE | 0.0461 | EPB41L1, DOCK8, PACS1, SMARCA4 |
| Cardiac hypertrophy | NHGRI GWAS Catalog | 0.0028 | PLXNA2, GRIK2, COL17A1, JAG1, SNAP25, BTBD3, SLX4IP |
| Response to fenofibrate (adiponectin levels) | NHGRI GWAS Catalog | 0.0046 | OAS2, PMEPA1, SHANK2, SCUBE1, SLC30A4, PCK1 |
| Complement C3 and C4 levels | NHGRI GWAS Catalog | 0.0094 | HLA, CFHR3, CFH, C3 |
| Neutrophil count | NHGRI GWAS Catalog | 0.0119 | PLCB4, TGFA, FGGY, PDGFD, PSD3 |
| Nephropathy(idiopathic membranous) | NHGRI GWAS Catalog | 0.0137 | HLA, ITGB6, PLA2R1 |
| Sleep duration | NHGRI GWAS Catalog | 0.0195 | PLLP, TMC5, ADAMTS14 |
| Airflow obstruction | NHGRI GWAS Catalog | 0.0259 | HYKK, LEF1, SERPINB8, GPR126, MAP3K13, PTPRD |
| Cystic fibrosis severity | NHGRI GWAS Catalog | 0.0265 | HLA, EHF, AHRR |
| Metabolite levels (5-HIAA/ MHPG Ratio) | NHGRI GWAS Catalog | 0.0265 | PIEZO2, ROBO2, ADAM12 |
| Bronchopulmonary dysplasia | NHGRI GWAS Catalog | 0.0296 | PLXDC2, ZNF770, SPOCK1, TRPS1, RASGRF1, HIVEP3 |
| Major depressive disorder | NHGRI GWAS Catalog | 0.0346 | PCLO, SLC6A15, ENOX1, SYPL2, IGFBP1, IGFBP3, C12orf5, ATXN1, PIEZO2, TRPS1, RASGEF1B, FGF12, KCNH5 |
| IgA nephropathy | NHGRI GWAS Catalog | 0.0346 | HLA, ACOXL, TNFSF13 |
| Pulmonary function decline | NHGRI GWAS Catalog | 0.0368 | MUSK, CSMD1, RORA, FLRT2 |
| Palmitic acid (16:0) plasma levels | NHGRI GWAS Catalog | 0.0368 | SCD, CNN3, GRIK2, PTPRD |
| Male-pattern baldness | NHGRI GWAS Catalog | 0.0439 | AUTS2, EDA2R, AR |
| Response to citalopram treatment | NHGRI GWAS Catalog | 0.0439 | LAMA1, RORA, EGFLAM |
| Hyperlipidemia | FunDO | 0.0050 | IRS1, CCL5, C3, PAPPA, TXNIP, APOC1, F3, SCD |
| Thrombocytopenia | FunDO | 0.0068 | GATA1, CCL5, ITGB3, IL11, CXCL8, MPL |
| Fibromyalgia | FunDO | 0.0126 | MAOB, CXCL8, BDNF, IGFBP3 |
| Cirrhosis | FunDO | 0.0209 | RBP4, KRT18, IGFBP3, KRT8, EGF, F3, FGF2IGFBP1 |
| Hepatitis C | FunDO | 0.0321 | CD274, CCL5, RBP4, MKI67, CXCL8, KRT18, TLR4, KRT8, FGF2 |
| Thalassemia | FunDO | 0.0345 | LCN2, CXCL8, ANK2, KIR3DL1, MUC1 |
| Gingival overgrowth | FunDO | 0.0417 | EDN1, IL15, FGF7 |
| Pulmonary fibrosis | FunDO | 0.0474 | CSF1, BDNF, MMP7, EDN1, CCL5, ERBB3 |
| Ovary cancer | FunDO | 0.0477 | LCN2, IL15, CXCL8, FGF7, CASP1 |
| Esophageal tumor | FunDO | 0.0477 | CD274, TSPAN8, FRAT1, PDCD1LG2, FGF7 |
| Hyperlipidemia | GAD | 0.0093 | CCL5, HLA, CXCL8, CD22, TNFRSF1B, CD19 |
| Thrombocytopenia | GAD | 0.0114 | CSMD1, DOCK4, GALNTL6, SOBP, PLXDC2, SESN3, ADAMTS5, EHF, TMC5, LPL, CD109, FAM117B, PDE1C, TAGLN, PTN, FGD4, DYNC2H1, GNG4, MUSK, FBLN5, CCDC54, TTC9, PMEPA1, TLR4, ANK3, EDA2R, APOC1, BMP2, TOX3, NRG1, ITPK1, PTPRD, KLF6, PAM, PTPRU, LEPR, IKZF2, LHX5, MCTP2, ANKRD50, SEMA6D, PLXNA2, DPYD, GRIK2, SRGAP3, ACOXL, TDRKH, FAM135B, VEGFC, CHST2 |
| Fibromyalgia | GAD | 0.0136 | GLI3, CELF2, VWA3B, PLXDC2, EDNRA, EDN1, JUN, DOCK8, DCLK2, BTBD3, DCN, CD74, EGFLAM, TLL1, TLR4, BMP2, PTPRD, ANK2, PTPRU, JADE2, IGF2BP2, PAPPA, DOCK2, KLK4, FAM49A, RGS3, AATK, FN1, IGSF10, NCOA7, SCIN, TNS1, FAM135B, MUC16, ADAM19, ATXN1, MTUS2, NXNL2, KCNQ3, ANPEP, CDH2 |
| Cirrhosis | GAD | 0.0204 | IRS1, CCL5, ITGB3, NPR1, NPR3, APOC1, LPL |
| Hepatitis C | GAD | 0.0258 | DPYD, CELF4, CELF2, FAM117B, TDRKH, LPCAT4, FBLN5, SOBP, PMEPA1, CSMD1, STOX1, CACNB2, CADM1, VEGFC, SLC7A11, LPL, CD109, MCTP2, SLC24A2, PTPRD, ITPK1 |
| Thalassemia | GAD | 0.0362 | MCTP2, PSD3, CCDC54, ROBO2, ELOVL6 |
| Gingival overgrowth | GAD | 0.0419 | PLXNA2, ATXN1, IGF2BP2, ABCA13, FN1 |
| Pulmonary fibrosis | GAD | 0.0420 | CREG2, GALNTL6, LINC01550, KIF16B, SH3BGR, TRPS1, PDE1C, NCKAP5, TNFRSF21, RYR3, MAGEC2, EDIL3, CXCL16, MCF2, DTD1, GPC5, KLF6, IKZF2, KCNH5, AJAP1, BTBD3, PHACTR2, ITPK1, IGSF10, SRGAP3, C12orf75, ABI3BP, FOS, SCUBE1 |
| Ovary cancer | GAD | 0.0426 | CELF4, TRPS1, TWIST1, PQLC2L, MAL2, PSD3, RCAN2, SUPT3H, TGFA, TMEM131L, HIVEP3, CSMD1, ROBO2, CCDC54, PRNP, APOC1, HRK, GPC5, AR, FN1, ABCA13, F2RL2, KLF6, IGF2BP2, LEPREL1, GNG4, SNAP25, MCTP2, FAM49A, ANKRD50, CACNA2D1, PLXNA2, ELOVL6, RUNX2, SCN8A, ATXN1, ID2, SLC24A2, CMTM7, LINGO2 |
| Esophageal tumor | GAD | 0.048 | CACNA2D1, SLC46A3, CHST2, PKDCC, PPID, CDH2 |
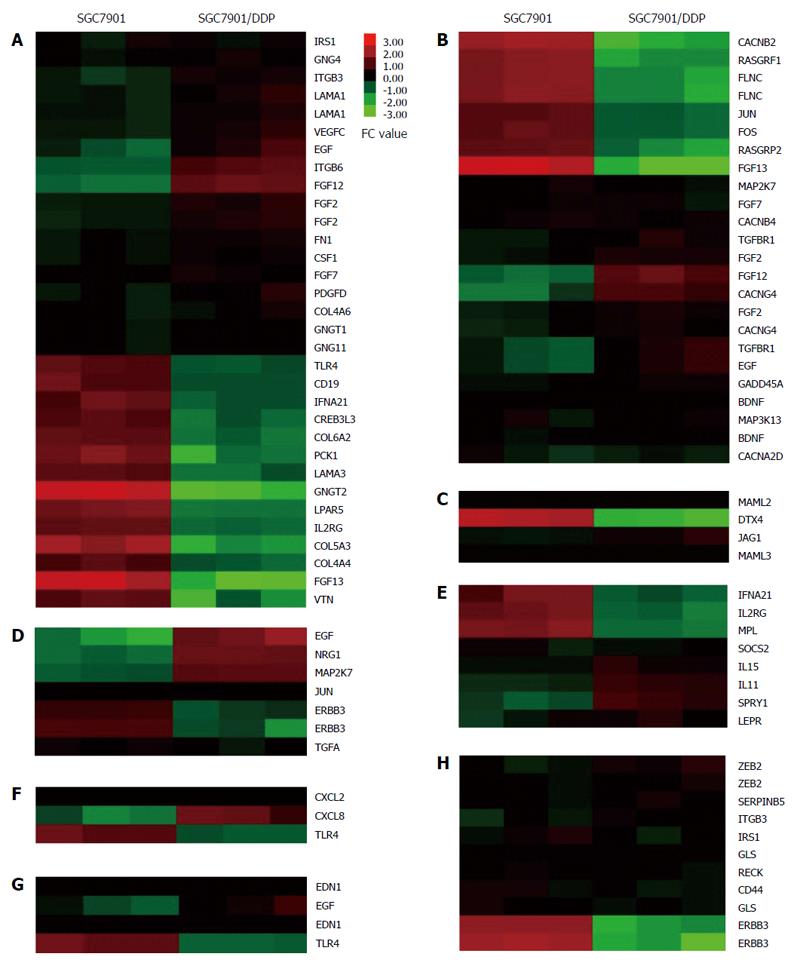
To determine which pathway might be involved in drug resistance formation, KEGG pathway analysis was used to authenticate pathways and understand biological functions of significantly differentially expressed genes. The result indicated that the differentially expressed mRNAs were enriched for 233 pathways, including the Rap1 signaling pathway, PI3K-Akt signaling pathway, ECM-receptor interaction, TNF signaling pathway, and pathways in cancer, among others (Figure 5, Table 3). Cluster 3.0 software were performed the heat-map.This finding identified many candidate pathways and input genes that may play an important role in resistance mechanism.
| Pathway | Input gene | Fold change | Regulation | Genomic coordinates | Cyto band |
| PI3K-Akt signaling pathway | LAMA1 | 2.60826 | Up | Chr18:6958512-6956742 | hs|18p11.31 |
| LAMA1 | 2.75269 | Up | Chr18:6942035-6941976 | hs|18p11.31 | |
| GNG4 | 2.09356 | Up | Chr1:235714443-235714384 | hs|1q42.3 | |
| ITGB3 | 2.96629 | Up | Chr17:45389027-45389086 | hs|17q21.32 | |
| ITGB6 | 7.72783 | Up | Chr2:160964233-160958330 | hs|2q24.2 | |
| VEGFC | 2.92538 | Up | Chr4:177604882-177604823 | hs|4q34.3 | |
| PDGFD | 2.42861 | Up | Chr11:103778445-103778386 | hs|11q22.3 | |
| IRS1 | 2.00967 | Up | Chr2:227596677-227596618 | hs|2q36.3 | |
| GNGT1 | 2.04779 | Up | Chr7:93536149-93540155 | hs|7q21.3 | |
| CSF1 | 2.25620 | Up | Chr1:110466137-110466196 | hs|1p13.3 | |
| EGF | 4.76437 | Up | Chr4:110932689-110932748 | hs|4q25 | |
| FGF2 | 3.02437 | Up | Chr4:123819331-123819390 | hs|4q28.1 | |
| FGF2 | 2.99240 | Up | Chr4:123819317-123819376 | hs|4q28.1 | |
| FN1 | 2.31254 | Up | Chr2:216288895-216288217 | hs|2q35 | |
| COL4A6 | 2.08497 | Up | Chrx:107399109-107399050 | hs|Xq22.3 | |
| FGF12 | 10.99211 | Up | Chr3:191860574-191860515 | hs|3q28 | |
| GNG11 | 2.01984 | Up | Chr7:93555764-93555823 | hs|7q21.3 | |
| FGF7 | 2.19252 | Up | Chr15:49776810-49776869 | hs|15q21.2 | |
| LAMA3 | 2.56116 | Down | Chr18:21534735-21534794 | hs|18q11.2 | |
| IFNA21 | 2.30808 | Down | Chr9:21166331-21166272 | hs|9p21.3 | |
| CREB3L3 | 2.40183 | Down | Chr19:4172219-4172278 | hs|19p13.3 | |
| TLR4 | 2.13271 | Down | Chr9:120476856-120476915 | hs|9q33.1 | |
| COL6A2 | 2.89458 | Down | Chr21:47546086-47546145 | hs|21q22.3 | |
| CD19 | 2.09302 | Down | Chr16:28950600-28950659 | hs|16p11.2 | |
| LPAR5 | 3.83177 | Down | Chr12:6728794-6728735 | hs|12p13.31 | |
| COL4A4 | 2.11177 | Down | Chr2:227867523-227867464 | hs|2q36.3 | |
| PCK1 | 4.49558 | Down | Chr20:56141030-56141089 | hs|20q13.31 | |
| VTN | 3.82587 | Down | Chr17:26694806-26694747 | hs|17q11.2 | |
| GNGT2 | 16.48365 | Down | Chr17:47284034-47283975 | hs|17q21.32 | |
| IL2RG | 2.87954 | Down | Chrx:70328539-70328480 | hs|Xq13.1 | |
| COL5A3 | 7.53410 | Down | Chr19:10070602-10070543 | hs|19p13.2 | |
| FGF13 | 17.08866 | Down | Chrx:137713947-137713888 | hs|Xq26.3 | |
| MAPK signaling pathway | FLNC | 4.57879 | Down | Chr7:128498538-128498597 | hs|7q32.1 |
| FLNC | 4.81302 | Down | Chr7:128498476-128498535 | hs|7q32.1 | |
| CACNB2 | 7.83293 | Down | Chr10:18787305-18787364 | hs|10p12.31 | |
| RASGRF1 | 4.87152 | Down | Chr15:79254554-79254495 | hs|15q25.1 | |
| FOS | 2.17501 | Down | Chr14:75748214-75748273 | hs|14q24.3 | |
| JUN | 2.04000 | Down | Chr1:59246570-59246511 | hs|1p32.1 | |
| RASGRP2 | 3.10358 | Down | Chr11:64508971-64508912 | hs|11q13.1 | |
| FGF13 | 17.08866 | Down | Chrx:137713947-137713888 | hs|Xq26.3 | |
| TGFBR1 | 2.93035 | Up | Chr9:101916322-101916381 | hs|9q22.33 | |
| TGFBR1 | 4.76437 | Up | Chr4:110932689-110932748 | hs|4q25 | |
| EGF | 4.76437 | Up | Chr4:110932689-110932748 | hs|4q25 | |
| FGF12 | 10.99211 | Up | Chr3:191860574-191860515 | hs|3q28 | |
| MAP3K13 | 2.25019 | Up | Chr3:185161379-185165590 | hs|3q27.2 | |
| FGF2 | 3.02437 | Up | Chr4:123819331-123819390 | hs|4q28.1 | |
| FGF2 | 2.99240 | Up | Chr4:123819317-123819376 | hs|4q28.1 | |
| MAP2K7 | 2.08267 | Up | Chr19:7979302-7979361 | hs|19p13.2 | |
| FGF7 | 2.19252 | Up | Chr15:49776810-49776869 | hs|15q21.2 | |
| CACNG4 | 8.83585 | Up | Chr17:65028139-65028198 | hs|17q24.2 | |
| CACNG4 | 2.94145 | Up | Chr17:65029115-65029174 | hs|17q24.2 | |
| CACNB4 | 2.14311 | Up | Chr2:152694239-152694180 | hs|2q23.3 | |
| GADD45A | 2.56659 | Up | Chr1:68153371-68153430 | hs|1p31.3 | |
| BDNF | 2.32411 | Up | Chr11:27679959-27679900 | hs|11p14.1 | |
| BDNF | 2.30323 | Up | Chr11:27677072-27677013 | hs|11p14.1 | |
| CACNA2D1 | 2.09452 | Up | Chr7:81579504-81579445 | hs|7q21.11 | |
| Notch signaling pathway | MAML2 | 2.03379 | Up | Chr11:95712434-95712375 | hs|11q21 |
| JAG1 | 3.20086 | Up | Chr20:10619120-10619061 | hs|20p12.2 | |
| MAML3 | 2.57919 | Up | Chr4:140810806-140810747 | hs|4q31.1 | |
| DTX4 | 9.99859 | Down | Chr11:58975615-58975674 | hs|11q12.1 | |
| ErbB signaling pathway | EGF | 4.76437 | Up | Chr4:110932689-110932748 | hs|4q25 |
| NRG1 | 2.77996 | Up | Chr8:32474390-32585512 | hs|8p12 | |
| MAP2K7 | 2.08267 | Up | Chr19:7979302-7979361 | hs|19p13.2 | |
| JUN | 2.04000 | Down | Chr1:59246570-59246511 | hs|1p32.1 | |
| ERBB3 | 5.29571 | Down | Chr12:56482380-56482439 | hs|12q13.2 | |
| ERBB3 | 8.12050 | Down | Chr12:56496160-56496219 | hs|12q13.2 | |
| TGFA | 2.37427 | Down | Chr2:70675378-70675319 | hs|2p13.3 | |
| Jak-STAT signaling pathway | IL11 | 4.21849 | Up | Chr19:55875847-55875788 | hs|19q13.42 |
| IL15 | 2.92970 | Up | Chr4:142654431-142654490 | hs|4q31.21 | |
| SOCS2 | 2.00180 | Up | Chr12:93969799-93969858 | hs|12q22 | |
| SPRY1 | 5.95682 | Up | Chr4:124324494-124324553 | hs|4q28.1 | |
| LEPR | 2.90187 | Up | Chr1:66102129-66102188 | hs|1p31.3 | |
| IL2RG | 2.87954 | Down | Chrx:70328539-70328480 | hs|Xq13.1 | |
| MPL | 3.41581 | Down | Chr1:43819826-43819885 | hs|1p34.2 | |
| NF-kappaB signaling pathway | CXCL2 | 2.03846 | Up | Chr4:74963044-74962985 | hs|4q13.3 |
| CXCL8 | 9.97781 | Up | Chr4:74609265-74609324 | hs|4q13.3 | |
| TLR4 | 2.13271 | Down | Chr9:120476856-120476915 | hs|9q33.1 | |
| HIF-1 signaling pathway | EDN1 | 2.39081 | Up | Chr6:12296672-12296731 | hs|6p24.1 |
| EDN1 | 2.46437 | Up | Chr6:12296218-12296277 | hs|6p24.1 | |
| EGF | 4.76437 | Up | Chr4:110932689-110932748 | hs|4q25 | |
| TLR4 | 2.13271 | Down | Chr9:120476856-120476915 | hs|9q33.1 | |
| MicroRNAs in cancer | IRS1 | 2.00967 | Up | Chr2:227596677-227596618 | hs|2q36.3 |
| ZEB2 | 3.32563 | Up | Chr2:145146320-145146261 | hs|2q22.3 | |
| ZEB2 | 2.70558 | Up | Chr2:145182422-145182363 | hs|2q22.3 | |
| CD44 | 2.02409 | Up | Chr11:35253812-35253871 | hs|11p13 | |
| RECK | 2.25018 | Up | Chr9:36124319-36124378 | hs|9p13.3 | |
| ITGB3 | 2.96629 | Up | Chr17:45389027-45389086 | hs|17q21.32 | |
| SERPINB5 | 2.61864 | Up | Chr18:61172218-61172277 | hs|18q21.33 | |
| GLS | 2.36144 | Up | Chr2:191829716-191829775 | hs|2q32.2 | |
| GLS | 2.07371 | Up | Chr2:191827822-191827881 | hs|2q32.2 | |
| ERBB3 | 5.29571 | Down | Chr12:56482380-56482439 | hs|12q13.2 | |
| ERBB3 | 8.12050 | Down | Chr12:56496160-56496219 | hs|12q13.2 |
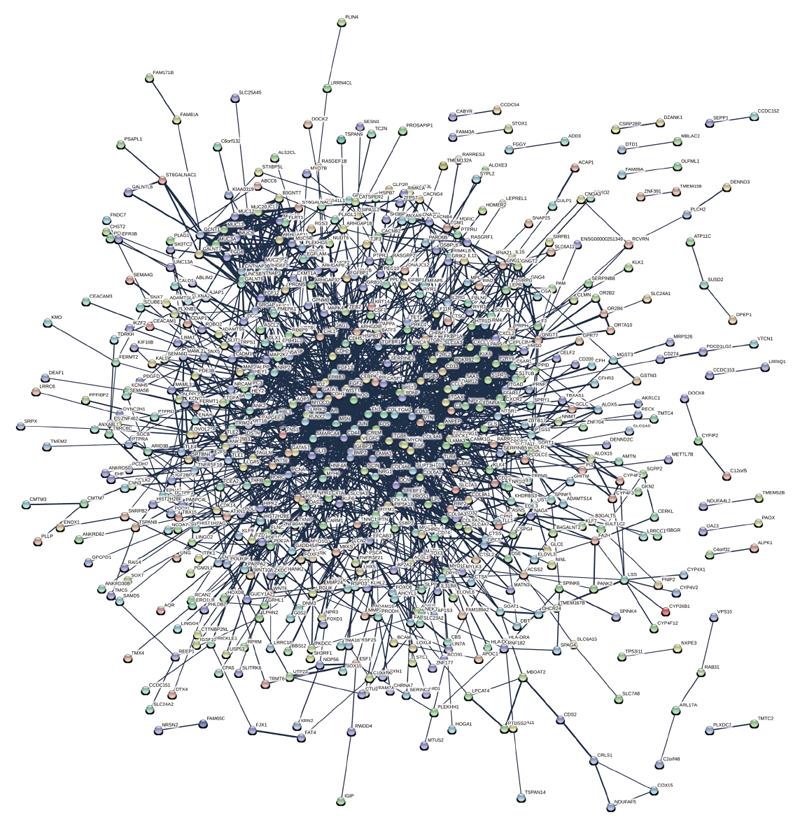
The STRING 9.1 software (Search Tool for the Retrieval of Interacting Genes) was used to perceive functional relations and generate networks of differential expression of proteins (Figure 6). For all of the 1002 differentially expressed proteins, we extracted a network containing 443 upregulated and 559 downregulated proteins which functionally associated with each other. We found that interacting proteins which participate in angiogenesis, toll-like receptor signaling pathway and cell adhesion had a high level of co-expression.
Cisplatin is widely used against a variety of solid neoplasms, including testicular, ovarian, colorectal, bladder, head and neck cancers and gastric cancer[23]. However, the repeated clinical expose to cisplatin often results in the tumor cells evading the apoptosis program initiated by cisplatin. Therefore, there is a need to explore the molecular mechanisms of cisplatin resistance, in order to overcome drug resistance in tumor therapy. Recently, several studies have indicated that many proteins are involved in the recognition of Pt-DNA adducts and cisplatin-induced apoptosis program[24,25]. In this study, we used microarray, GO, KEGG pathway and protein-protein interaction (PPI) analysis to explore the roles of differentially expressed mRNAs in cisplatin resistance and to support other studies.
Many genes which shown differentially expression in the microarray analysis have been demonstrated to be associated with cisplatin resistance in human cancer (Table 4), such as PDE3B, which was substantially upregulated (P value = 0.00029, Fold Chang (FC) = 10.45) in SGC7901/DDP cells. Treatment with a combination of a PDE3B inhibitor and DDP can significantly increase the number of apoptotic and cell growth-suppressive cancer cells in cisplatin resistant squamous cell carcinoma (SCC) and Hela cells[26]. Research shows that VEGFC, which is upregulated in our data (P value = 0.00013 FC = 2.93), enhanced cell invasion and cisplatin resistance in gastric cancer[27]. In non-small cell lung cancer, loss of IGFBP-3 expression may activate the PI3K/AKT pathway and induce resistance to cisplatin[28]. In support of this association, our results showed that this mRNA is downregulated (P = 0.00007, FC = 2.93) in SGC7901/DDP cells.
| Gene symbol | P value | FC (abs) | Regulation | Genename | Ref. |
| FGF7 | 0.00035 | 2.19252 | Up | Fibroblast growth factor 7 | PMID: 22990650 |
| HIPK2 | 2.63E-06 | 4.06213 | Up | Homeodomain interacting protein kinase 2 | PMID: 24846322 |
| EDN1 | 9.94E-05 | 2.46437 | Up | Endothelin 1 | PMID: 21220476 |
| CBS | 0.00108 | 2.29340 | Up | Cystathionine-beta-synthase | PMID: 24236104 |
| PDE3B | 0.00029 | 10.44998 | Up | Phosphodiesterase 3B, cgmp-inhibited | PMID: 24133626 |
| E2F5 | 0.00041 | 2.42888 | Up | E2F transcription factor 5, p130-binding | PMID: 22193543 |
| PIN1 | 0.00104 | 2.13293 | Up | Peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | PMID: 26820938 |
| EGF | 0.00346 | 4.76437 | Up | Epidermal growth factor | PMID: 27086487 |
| CSF1 | 0.00025 | 2.25620 | Up | Colony stimulating factor 1 (macrophage) | PMID: 22005523 |
| PCNA | 0.00103 | 2.17028 | Up | Proliferating cell nuclear antigen | PMID: 24474685 |
| HIPK2 | 2.63E-06 | 4.06213 | Up | Homeodomain interacting protein kinase 2 | PMID: 24846322 |
| ENTPD6 | 0.00011 | 2.43726 | Up | Ectonucleoside triphosphate diphosphohydrolase 6 (putative) | PMID: 21519793 |
| AKR1C1 | 0.00097 | 2.29646 | Up | Aldo-keto reductase family 1, member C1 | PMID: 23165153, PMID: 17266043 |
| ASNS | 0.00172 | 2.19491 | Up | Asparagine synthetase (glutamine-hydrolyzing) | PMID: 23956056, PMID: 17409444 |
| BDNF | 0.00062 | 2.32411 | Up | Brain-derived neurotrophic factor | PMID: 22276165, PMID: 17044982 |
| CABYR | 0.01089 | 2.55664 | Up | Calcium binding tyrosine-(Y)-phosphorylation regulated | PMID: 24362251 |
| FGF2 | 2.15E-06 | 2.99240 | Up | Fibroblast growth factor 2 (basic) | PMID: 12894531 |
| SLC7A11 | 1.95E-05 | 2.93256 | Up | Solute carrier family 7 member 11 | PMID: 24516043 |
| TUBB3 | 0.00046 | 2.00213 | Up | Tubulin, beta 3 class III | PMID: 25107571 |
| TWIST1 | 0.00180 | 2.96340 | Up | Twist family bhlh transcription factor 1 | PMID: 22673193, PMID: 22245869 |
| JAG1 | 9.41E-05 | 3.20086 | Up | Jagged 1 | PMID: 24659709 |
| ANXA11 | 0.00031 | 2.36619 | Down | Annexin A11 | PMID: 19484149, PMID: 17982121 |
| CCL5 | 2.67E-05 | 5.05630 | Down | Chemokine (C-C motif) ligand 5 | PMID: 26983899 |
| FGF13 | 0.00044 | 17.08866 | Down | Fibroblast growth factor 13 | PMID: 24113164 |
| IGFBP3 | 7.48E-05 | 2.92508 | Down | Insulin-like growth factor binding protein 3 | PMID: 20023704 |
| KLK6 | 0.00066 | 2.24596 | Down | Kallikrein-related peptidase 6 | PMID: 23307575 |
| SLC7A8 | 4.50E-05 | 5.36735 | Down | Solute carrier family 7 member 8 | PMID: 23462296 |
| TGM2 | 2.88E-05 | 6.24520 | Down | Transglutaminase 2 | PMID: 21424127, PMID: 24828664 |
| TLR4 | 0.00114 | 2.13271 | Down | Toll-like receptor 4 | PMID: 21616060, PMID: 22583829 |
| XAF1 | 0.02405 | 3.20613 | Down | XIAP associated factor 1 | PMID: 25824780, PMID: 25240826 |
| TCEA2 | 0.00061 | 3.65969 | Down | Transcription elongation factor A (SII), 2 | PMID: 16142353 |
GO enrichment analysis exhibits many functions which the differently expressed mRNAs are involved in, including locomotion, chemotaxis, cell adhesion, regulation of cell migration, extracellular matrix disassembly, response to xenobiotic chemotaxis, localization of cell adhesion and blood vessel morphogenesis. Functional annotation showed that the differently expressed mRNAs mainly regulate cellular biological behaviors in the progress of regulation of transcription. How the underlying targets of each GO term are implicated in the cisplatin resistance needs further investigation in the future.
Our KEGG pathway analysis showed that the differently expressed mRNAs are enriched in pathways of ECM-receptor interaction, PI3K-Akt, Rap1, MAPK, Notch1, ErbB, ABC transporters, Jak-STAT, NF-κB, HIF-1 and TGF-β. All of those pathways have been confirmed to be involved in cisplatin resistance in different experiments described previously. For example, the inhibition of PI3K-Akt signaling pathway may increase the sensitivity of gastric cancer cells to cisplatin chemotherapy[29]. Another study found that Janus kinase 2 (JAK2) signal transducer and activator of transcription 3 (STAT3) signaling pathways were activated by overexpressed AKT in cisplatin resistant human gastric cancer cells[30]. A study revealed that the canonical NF-κB signaling pathway was involved in APRIL-mediated cisplatin resistance in gastric cancer[31]. Our data are consistent with these previous studies, and these pathways and input genes deserve our attention in gastric cancer cisplatin resistance.
Although protein expression is generally stable when organs mature, under various pathological and physiological conditions, gene expression may change and ultimately result in aberrant protein levels. Therefore, research on proteomics is helpful to illustrate some biological mechanisms, including cisplatin resistance. Protein-protein interaction network analysis might uncover previously unknown molecular mechanisms of cisplatin resistance. Hub proteins of subnetworks which interact with many partners might associate with drug resistance. For example, studies have shown that dysregulation of the genes PDE3B, TLR4, and HIPK2 is associated with cisplatin resistance in human SCC cells, ovarian granulosa tumor cells and bladder cancer cells, respectively[26,32,33]. Moreover, hub proteins and their partners may have similar biological functions. Since downregulation of EGF has been shown to substantially overcome resistance to cisplatin in ovarian cancer[34], we predict that the proteins EDN1 and DCN, whose hub protein is EGF, may contribute to cisplatin resistance in a similar fashion. We also found that ZEB2, which over-expressed in SGC7901/DDP compared with SGC7901 has a similar expression profile to TWIST1, suggesting that ZEB2 may play an important role in cisplatin resistance by regulating the expression of TWIST1. Nevertheless, more evidence and research is needed.
In conclusion, our study identified mRNAs differentially expressed between gastric cancer cell lines SGC7901/DDP and SGC7901. These results provide a global view of the function of the differentially expressed mRNAs. Several molecular and pathway abnormalities detected in our study have previously been reported to be associated with drug resistance in gastric cancer. The dysregulated mRNAs identified participate in cisplatin resistance through diverse mechanisms, and further investigation is required to confirm the role in drug resistance of these transcripts, pathways and the interaction networks of the proteins they code for.
Cisplatin-contained chemotherapy is one of the most frequently used for advanced gastric cancer; however, this chemotherapeutic agent is often limited due to drug resistance and result unsatisfactory prognosis. Research increasingly suggests that abnormal expression of biological pathway and proteins associated with cisplatin resistance. This demonstrated that more bioinformatics study is needed to predict targets for gastric cancer with cisplatin.
Bioinformatics analysis demonstrated that some mRNAs which related to the biological behavior abnormal expression in SGC7901/DDP cells. These mRNAs have already been shown to play important roles in the process of cisplatin resistance of various cancers, including gastric cancer.
The authors performed bioinformatics analysis of mRNA expression profile in SGC7901/DDP cells compared with SGC7901 cells, and found that many mRNAs and pathways in SGC7901/DDP cells expressed abnormally, these may participate in and predict cisplatin resistance in gastric cancer.
These results suggest that targeting the differently expression mRNA may provide more selective approaches to reverse cisplatin resistance of therapeutic targets.
The definition of cisplatin resistance: in the clinic, if a patient who have disease recurrence within the first months after the recent cisplatin dose, the patient is considered cisplatin resistance; in cells, generally, resistance index > 20 exhibited high resistance, resistance index 5-15 is moderate resistance, resistance index < 5 represent low or no resistance. Correct P: Using Benjamini Hochberg FDR method for correction of p values. Fold change (FC): gene expression in SGC7901 / DDP cells compared with SGC7901 cells.
The paper is a good study on mRNAs expression profile in SGC7901/DDP cells. The investigators shown that many mRNAs was abnormal expressed in SGC7901/DDP cells and these mRNAs enriched in many biological process which have already been shown to play important roles in the process of cisplatin resistance in human cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Ghosh RD S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 502] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 2. | Wöhrer SS, Raderer M, Hejna M. Palliative chemotherapy for advanced gastric cancer. Ann Oncol. 2004;15:1585-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 3. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10002] [Cited by in RCA: 10452] [Article Influence: 696.8] [Reference Citation Analysis (0)] |
| 4. | Al-Batran SE, Hartmann JT, Probst S, Schmalenberg H, Hollerbach S, Hofheinz R, Rethwisch V, Seipelt G, Homann N, Wilhelm G. Phase III trial in metastatic gastroesophageal adenocarcinoma with fluorouracil, leucovorin plus either oxaliplatin or cisplatin: a study of the Arbeitsgemeinschaft Internistische Onkologie. J Clin Oncol. 2008;26:1435-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 573] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 5. | Kang YK, Kang WK, Shin DB, Chen J, Xiong J, Wang J, Lichinitser M, Guan Z, Khasanov R, Zheng L. Capecitabine/cisplatin versus 5-fluorouracil/cisplatin as first-line therapy in patients with advanced gastric cancer: a randomised phase III noninferiority trial. Ann Oncol. 2009;20:666-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 591] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 6. | Koizumi W, Narahara H, Hara T, Takagane A, Akiya T, Takagi M, Miyashita K, Nishizaki T, Kobayashi O, Takiyama W. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): a phase III trial. Lancet Oncol. 2008;9:215-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1320] [Cited by in RCA: 1421] [Article Influence: 83.6] [Reference Citation Analysis (0)] |
| 7. | Wagner AD, Unverzagt S, Grothe W, Kleber G, Grothey A, Haerting J, Fleig WE. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010;CD004064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 350] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 8. | Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1204] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 9. | Garcia JA, Dreicer R. Systemic chemotherapy for advanced bladder cancer: update and controversies. J Clin Oncol. 2006;24:5545-5551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Wilson TR, Longley DB, Johnston PG. Chemoresistance in solid tumours. Ann Oncol. 2006;17 Suppl 10:x315-x324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 11. | Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92:1295-1302. [PubMed] |
| 12. | Hioki M, Gotohda N, Konishi M, Nakagohri T, Takahashi S, Kinoshita T. Predictive factors improving survival after gastrectomy in gastric cancer patients with peritoneal carcinomatosis. World J Surg. 2010;34:555-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Yang T, Chen M, Chen T, Thakur A. Expression of the copper transporters hCtr1, ATP7A and ATP7B is associated with the response to chemotherapy and survival time in patients with resected non-small cell lung cancer. Oncol Lett. 2015;10:2584-2590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Metzger R, Leichman CG, Danenberg KD, Danenberg PV, Lenz HJ, Hayashi K, Groshen S, Salonga D, Cohen H, Laine L. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol. 1998;16:309-316. [PubMed] |
| 15. | Xu W, Chen Q, Wang Q, Sun Y, Wang S, Li A, Xu S, Røe OD, Wang M, Zhang R. JWA reverses cisplatin resistance via the CK2-XRCC1 pathway in human gastric cancer cells. Cell Death Dis. 2014;5:e1551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Shim HJ, Yun JY, Hwang JE, Bae WK, Cho SH, Lee JH, Kim HN, Shin MH, Kweon SS, Lee JH. BRCA1 and XRCC1 polymorphisms associated with survival in advanced gastric cancer treated with taxane and cisplatin. Cancer Sci. 2010;101:1247-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 17. | Liu J, Deng N, Xu Q, Sun L, Tu H, Wang Z, Xing C, Yuan Y. Polymorphisms of multiple genes involved in NER pathway predict prognosis of gastric cancer. Oncotarget. 2016;7:48130-48142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Sargent RG, Meservy JL, Perkins BD, Kilburn AE, Intody Z, Adair GM, Nairn RS, Wilson JH. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 2000;28:3771-3778. [PubMed] |
| 19. | Funato T, Kozawa K, Fujimaki S, Miura T, Kaku M. Increased sensitivity to cisplatin in gastric cancer by antisense inhibition of the her-2/neu (c-erbB-2) gene. Chemotherapy. 2001;47:297-303. [PubMed] |
| 20. | Zhou N, Qu Y, Xu C, Tang Y. Upregulation of microRNA-375 increases the cisplatin-sensitivity of human gastric cancer cells by regulating ERBB2. Exp Ther Med. 2016;11:625-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Sileni VC, Fosser V, Maggian P, Padula E, Beltrame M, Nicolini M, Arslan P. Pharmacokinetics and tumor concentration of intraarterial and intravenous cisplatin in patients with head and neck squamous cancer. Cancer Chemother Pharmacol. 1992;30:221-225. [PubMed] |
| 22. | Zhou C, Zhang T, Liu F, Zhou J, Ni X, Huo R, Shi Z. The differential expression of mRNAs and long noncoding RNAs between ectopic and eutopic endometria provides new insights into adenomyosis. Mol Biosyst. 2016;12:362-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | Muggia FM, Bonetti A, Hoeschele JD, Rozencweig M, Howell SB. Platinum Antitumor Complexes: 50 Years Since Barnett Rosenberg’s Discovery. J Clin Oncol. 2015;33:4219-4226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 24. | Ge X, Liu X, Lin F, Li P, Liu K, Geng R, Dai C, Lin Y, Tang W, Wu Z. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget. 2016;7:24466-24482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116-1120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 876] [Cited by in RCA: 855] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 26. | Uzawa K, Kasamatsu A, Baba T, Usukura K, Saito Y, Sakuma K, Iyoda M, Sakamoto Y, Ogawara K, Shiiba M. Targeting phosphodiesterase 3B enhances cisplatin sensitivity in human cancer cells. Cancer Med. 2013;2:40-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Cho HJ, Kim IK, Park SM, Baek KE, Nam IK, Park SH, Ryu KJ, Choi J, Ryu J, Hong SC. VEGF-C mediates RhoGDI2-induced gastric cancer cell metastasis and cisplatin resistance. Int J Cancer. 2014;135:1553-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 28. | Cortés-Sempere M, de Miguel MP, Pernía O, Rodriguez C, de Castro Carpeño J, Nistal M, Conde E, López-Ríos F, Belda-Iniesta C, Perona R. IGFBP-3 methylation-derived deficiency mediates the resistance to cisplatin through the activation of the IGFIR/Akt pathway in non-small cell lung cancer. Oncogene. 2013;32:1274-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Fu X, Feng J, Zeng D, Ding Y, Yu C, Yang B. PAK4 confers cisplatin resistance in gastric cancer cells via PI3K/Akt- and MEK/Erk-dependent pathways. Biosci Rep. 2014; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 30. | Zhang LL, Zhang J, Shen L, Xu XM, Yu HG. Overexpression of AKT decreases the chemosensitivity of gastric cancer cells to cisplatin in vitro and in vivo. Mol Med Rep. 2013;7:1387-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Zhi X, Tao J, Xiang G, Cao H, Liu Z, Yang K, Lv C, Ni S. APRIL induces cisplatin resistance in gastric cancer cells via activation of the NF-κB pathway. Cell Physiol Biochem. 2015;35:571-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 32. | Lin J, Zhang Q, Lu Y, Xue W, Xu Y, Zhu Y, Hu X. Downregulation of HIPK2 increases resistance of bladder cancer cell to cisplatin by regulating Wip1. PLoS One. 2014;9:e98418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 33. | Woods DC, White YA, Dau C, Johnson AL. TLR4 activates NF-κB in human ovarian granulosa tumor cells. Biochem Biophys Res Commun. 2011;409:675-680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Tang XH, Li M, Deng S, Lu MS. Cross-reacting material 197, a heparin-binding EGF-like growth factor inhibitor, reverses the chemoresistance in human cisplatin-resistant ovarian cancer. Anticancer Drugs. 2014;25:1201-1210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |