Published online Dec 14, 2017. doi: 10.3748/wjg.v23.i46.8109
Peer-review started: October 28, 2017
First decision: November 21, 2017
Revised: October 28, 2017
Accepted: December 4, 2017
Article in press: December 4, 2017
Published online: December 14, 2017
Processing time: 45 Days and 24 Hours
Metabolic syndrome (MetS), as a chronic inflammatory disorder has a potential role in the development of inflammatory and cancerous complications of the colonic tissue. The interaction of DNA damage and inflammation is affected by the insulin-like growth factor 1 receptor (IGF1R) signaling pathway. The IGF1R pathway has been reported to regulate autophagy, as well, but sometimes through a bidirectional context. Targeting the IGF1R-autophagy crosstalk could represent a promising strategy for the development of new antiinflammatory and anticancer therapies, and may help for subjects suffering from MetS who are at increased risk of colorectal cancer. However, therapeutic responses to targeted therapies are often shortlived, since a signaling crosstalk of IGF1R with other receptor tyrosine kinases or autophagy exists, leading to acquired cellular resistance to therapy. From a pharmacological point of view, it is attractive to speculate that synergistic benefits could be achieved by inhibition of one of the key effectors of the IGF1R pathway, in parallel with the pharmacological stimulation of the autophagy machinery, but cautiousness is also required, because pharmacologic IGF1R modulation can initiate additional, sometimes unfavorable biologic effects.
Core tip: Targeting the insulin-like growth factor 1 receptor (IGF1R)-autophagy crosstalk could represent a promising strategy for the development of new antiinflammatory and anticancer therapies, and may help for subjects suffering from metabolic syndrome who are at increased risk of colorectal cancer. However, cautiousness is also required, because pharmacologic IGF1R modulation can initiate additional, sometimes unfavorable biologic effects.
- Citation: Sipos F, Székely H, Kis ID, Tulassay Z, Műzes G. Relation of the IGF/IGF1R system to autophagy in colitis and colorectal cancer. World J Gastroenterol 2017; 23(46): 8109-8119
- URL: https://www.wjgnet.com/1007-9327/full/v23/i46/8109.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i46.8109
Nowadays, metabolic syndrome (MetS) has being viewed as a chronic inflammatory disorder[1]. Although numerous molecular mechanisms still have to be clearly defined, the role of pro-inflammatory cytokines, reactive oxygen species and free fatty acid intermediaries have been suggested as key factors in modulating specific intracellular signaling pathways that appear to regulate insulin sensitivity[2]. In order to expand our understanding of MetS, it is important to link their potential role in the development of complications.
In mild, longstanding colonic inflammation, the expression of epithelial insulin-like growth factor 1 receptor (IGF1R) is elevated both on mRNA and protein levels[3]. This may allow epithelial cells bearing inflammation-associated genetic defects to pathologically survive and proliferate. In acute murine colitis, however, insulin-like growth factor 1 (IGF1)-primed macrophages were found to suppress immune inflammation in the intestine by producing interleukin-10[4]. Regarding colonic inflammation, the biologic role of the IGF1/IGF1R axis seems to be controversial.
Under inflammatory circumstances, immune and epithelial cells release reactive oxygen and nitrogen species, resulting in DNA damage[5]. The crosstalk between DNA damage and inflammation has a role in cancer development, therefore chronic inflammation represents a hallmark phenomenon of tumorigenesis[6]. The prevalence of MetS is increasing in parallel with growing incidence of cancerous diseases worldwide. Previous studies have reported that MetS is associated with the development of several types of tumors including colorectal cancer (CRC)[7-9]. The elevated risk of CRC in MetS patients[8] indicates that the consequence of MetS in cancer is an important issue needs to be resolved.
The binding of insulin to cell surface receptors like insulin receptor and IGF1R on cancer cells results in cell proliferation and survival[10]. Elevated serum insulin levels modify the IGF-IGF1R axis involved in cancer development and progression[11]. Based on these results, antiinflammatory and anti-cancer strategies blocking the aberrant activation of IGF1R are therapeutically relevant.
Autophagy is a fundamental eukaryotic cellular homeostatic process and integral component of the immune system influencing inflammation and immunity[12]. Regarding colorectal carcinogenesis it has a dual-faced role. The down-regulation of autophagy-associated genes promotes colorectal cancer development and invasion[13,14], while induction of autophagy redounds the proliferative arrest of human colon cancer cell lines[15,16]. Autophagy is considered to be a crucial approach of killing apoptosis-resistant tumor cells[17-19]. The insulin/IGF1/PI3K-Akt-mTOR (mammalian target of rapamycin) pathway has been reported to regulate autophagy through the insulin receptor[20,21]. Moreover, the autophagic lysosomal pathway can be suppressed by the activation of IGF1R-signaling[22,23]. Thus, targeting the IGF1R-autophagy crosstalk could represent a promising strategy for the development of new antiinflammatory and anticancer therapies, and may help for subjects suffering from MetS who are at increased risk of colorectal cancer.
The first identified member of the insulin/IGF family was insulin. Determination of the protein’s structure, functions, mode of action, role in glucose metabolism, and it’s implication in the etiology of diabetes mellitus resulted in the concession of three Nobel Prizes; in 1923 for the discovery of it’s capacity to treat diabetes (by Frederick Banting and J. J. Macleod); in 1958 for studies regarding the protein structure and sequence (by Frederick Sanger); and, in 1963 for the first determination of the three-dimensional structure (by Dorothy Hodgkin).
The existence of the IGFs was first proposed by Salmon and Daughaday (in 1957)[24], based on studies indicating that growth hormone-mediated stimulation of sulfate incorporation into the cartilage is mediated through a serum factor. This factor was originally termed “sulfation factor”, then “somatomedin”. The term “insulin-like” was later used, based on the partial structural homology with insulin and stimulation of glucose uptake into fat- and muscle cells[25].
IGF1 and IGF2 are growth factors with both mitogenic and metabolic functions, having a role in the growth, differentiation and survival of numerous cell types and tissues. IGFs are unique among growth factors, since they can act as systemically (like hormones), as locally (like autocrine/paracrine factors)[26].
The metabolic functions of the insulin/IGF axis are well known, since insulin is a key regulator controlling cellular glucose-, amino-, and fatty-acid uptake, as well as glycogen-, lipid-, and protein synthesis, and other related metabolic processes[26].
IGFs also display multiple functions. They are expressed ubiquitously, although in different amounts and ratios in a variety of tissues and cells, exerting auto-, para- and endocrine biological effects. They act mainly as growth hormones, regulating the growth of human cells and tissues, as well as influencing their lifespan. They have a substantial effect on maintaining tissue homeostasis and a differentiated phenotype in normal tissue, are involved in angiogenesis, cell adhesion, migration and wound healing[27].
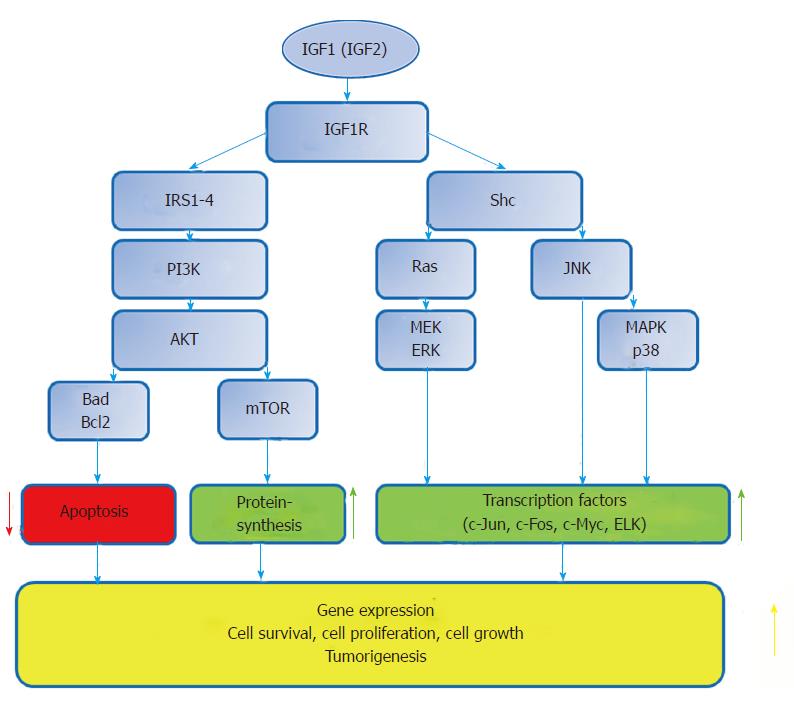
This network, along with a complex crosstalk with other signaling pathways has also a role in determining the balance between apoptosis and cell survival. The antiapoptotic and pro-survival effects are of major importance in the development and progression of some cancer types[28]. The variety of cellular responses to the insulin/IGF signal depend on the availability of growth factors, the ratios of the receptors and signalling molecules, the cell and tissue types as well as tissue microenvironment[28] (Figure 1).
Insulin receptors (IR) exist in IR-A, IR-B and IRR isoforms, while IGF receptors include IGF1R and IGF2R[29]. IGF1R, IR and IRR are composed of an extracellular ligand-binding domain and an intracellular protein kinase domain. Their structural similarity permits the formation of heterodimer receptors, formed by subunits of different receptor proteins. Heterodimers are spontaneously formed and represent the most abundant receptor subtype in many tissues. These receptors bind insulin and IGF ligands with different affinities, depending on their subunit composition. Ranking from high to low and very low affinity, IGF1R binds IGF1, IGF2 and insulin; IGF2R binds IGF2 and other ligands, such as mannose-6-phosphate, IGF1; IR binds insulin, IGF2 and IGF1. IR-A possesses higher IGF2 affinity than IR-B. IRR is an orphan receptor with unknown ligand binding; it participates primarily in signal transduction[29,30].
IGFs and IRs constitute a complex interacting receptor network. Depending on the availability of IGF/insulin ligands and the ratios of these receptors, IGFs can activate IR and, conversely, insulin activates IGF1R[28].
The endocrine actions of IGFs are regulated by the IGF-binding protein (IGFBP) system by modulating the amount of bioavailable IGFs in a positive or negative manner. The IGFBPs produced locally act as autocrine/paracrine regulators of IGF actions. IGFBPs may also fix IGFs in the extracellular matrices for future actions. Some IGFBPs act also by a mechanism independent of IGFs[26].
Binding of IGFs by inhibitory IGFBPs results in altered IGF actions, thus preventing the interaction of IGFs with the specific IGF receptors (until released) and protecting them from proteases within the circulation. Due to the significantly greater affinity of IGFBPs for IGFs as compared to the affinity of IGFs to their receptors only few IGF binds to receptors in the presence of an equimolar concentration of receptor and binding protein. In this regard the excess presence of IGFBPs in various tissues is an additional factor. By limiting the complex functions of the IGFs, it may be hypothesised that IGFBPs may to certain extent act as tumor suppressors[26,31].
Nevertheless, IGFBPs may promote IGF signalling. IGFBPs stabilise and allow slow release of IGFs for receptor interactions, thereby preventing receptor downregulation by high IGF exposure, and thusly promoting a prolonged and constant receptor activation[28].
IGFBP proteases presented in the circulation, as well may release IGFBP-bound ligands by degrading IGFBP into a form with a considerably lower affinity for IGFs compared with that of intact IGFBP. The amount of these proteases may be modified under certain physiological and pathological conditions, and their activity depends on the activators and inhibitors of proteases[26].
Autophagy, an evolutionarily highly conserved process of cellular self-digestion[32] is intensely implicated in the regulation of various functions, such as cell development, differentiation, survival, or senescence[33]. Additionally, it influences inflammation along with the innate and adaptive immunity[34]. Autophagy involves several sequential steps, including autophagosome nucleation, elongation, lipidation, and degradation, which are controlled by autophagy-related genes (Atgs)[32]. Intact basal autophagy serves constantly and constitutively as a critical adaptive and surveillance mechanism in maintaining cellular homeostasis[35]. However, to preserve cell viability autophagy is inducible in response to different cellular metabolic stress conditions, and, in case of protein aggregation and accumulation of misfolded proteins, when structural remodeling is warranted[36].
Defective autophagy has been ultimately related to several chronic inflammatory diseases including inflammatory bowel disease (IBD), or malignancies[35-38]. Regarding tumorigenesis, a Janus-faced role of autophagy has been proposed. It may be critical for cancer cell survival and progression, in particular under stressful situations, however, it may also elicit tumor death signaling pathways. The pro-survival or pro-death function of autophagy is context-dependent, and influenced by several intra- and extracellular factors, like involved tissues, surrounding microenvironment, genetic background, or stages of tumor development[37,39].
The connection of the IGF1/IGF1R system to the autophagy machinery is rather complicated. Insulin receptor substrate 1 (IRS1), an adaptor of IGF1R has a crucial role in the signal transduction of IGF1R. Tyrosine phosphorylation of IGF1R induces the binding of IRS1 to IGF1R, and phosphorylation of tyrosine residues in IRS1. This allows IRS1 to activate the PI3K pathway[40]. The PI3K-Akt-mTOR pathway has been documented to regulate autophagy via the insulin receptor[20]. In addition, IGF1R-mediated cell survival under hypoxia depends on enhanced autophagy caused by the suppression of the PI3K-Akt-mTOR signaling pathway[21]. The autophagic lysosomal pathway can also be suppressed by the activation of IGF1R-signaling[22,23].
In a recent study[41], it has been shown that fragments of IGF1R are localized separately from full-length IGF1R, colocalizing with LC-3 II, and activate the ubiquitously expressed Shc A adapter protein in dense organelles. The IGF1R fragments and Shc A have been found to be phosphorylated, indicating that after activation both the IGF1R and a key adapter protein are sequestered in autophagic vacuoles for degradation. Shc adapter protein transmits IGF1/IGF1R signaling via the mitogen activated protein kinase (MAPK) pathway, resulting finally in cell proliferation. Upon cathepsin inhibition autophagy seems to be involved in downregulation of IGF1–mediated cell proliferation[41].
The nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylase sirtuin 1 (SIRT1; silent mating type information regulation 2 homolog 1) has emerged as a significant target for epigenetic therapeutics of colon cancer since its increased expression is closely related to cancer progression. Additionally, SIRT1 represses p53 function via deacetylation, and so, promotes tumor growth[42]. IGF1R signaling can be improved by adipokines through SIRT1[43]. Moreover, SIRT1 overexpression stimulates epithelial wound healing via the downregulation of the IGFBP3 protein, the activation of the IGF1R/Akt pathway, and the posttranslational modification of p53 expression[44]. It has also been demonstrated that IGF1 and IGF1R expression levels can be negatively regulated by SIRT1 upon modulation of the AKT and ERK1/2 phosphorylation[45]. In turn, in human cancer cells aberrant cytoplasmic localization and protein stability of SIRT1 has been found to be regulated by the PI3K/IGF1R signaling[46]. SIRT1 can directly interact with and deacetylate several Atg proteins, including Atg5, Atg7, and Atg8, leading to the activation of these proteins[47,48]. By decreasing genetic stability and DNA mismatch repair, impaired SIRT1 and autophagy signaling pathway could increase the risk of genetic mutations and carcinogenesis. Further, the dysregulation of mTOR and AMP activated kinase (PRKA) pathways could remodel cell metabolism during the growth and metastasis of cancer cells. Moreover, these pathways may couple metabolic and epigenetic alterations that are essential to tumorigenic transformation[49]. Therefore, the modulation of the IGF1R/SIRT1/autophagy system is of great therapeutic interest in colon cancer.
The neural-specific deletion of sirtuin 6 (SIRT6) has been found to attenuate IGF1 level[50]. This finding may connect SIRT6 to IGF1 signaling, a conserved pathway with the ability to affect lifespan, metabolism, neurodegeneration, or cancer[51,52]. Recent evidences propose that autophagy may be associated with increased activation of SIRT6, because transcriptional factors like nuclear factor κ light chain enhancer of activated B cells (NF-κB), and activator protein 1 (AP-1), whose activity is negatively regulated by SIRT6, are shown to be positive regulators of autophagy[53,54]. These findings suggest that pharmacologic modulation of IGF1/SIRT6 might have a therapeutic value, as well.
The stress-induced protein TRB3 is a member of mammalian Tribbles homologs, which contain a Ser/thr protein kinase-like domain, but lack the ATP binding pocket and catalytic residues[55]. TRB3 coordinates crucial cellular processes, such as lipid and glucose metabolism, apoptosis, cell differentiation, and stress response[55]. In several human tumors and cancer cells metabolic stress conditions, including insulin/IGF1 enhance the expression of TRB3. In cancer cells TRB3 depletion protects against the tumor-promoting actions of insulin/IGF1. TRB3 interacts with p62, and interfers with the p62 cargo function, hence it results in p62 accumulation and p62-mediated autophagy dysfunction[56]. The interaction between TRB3 and sequestosome-1 (SQSTM1) has been found to be essential to mediate the insulin/IGF-1-related (metabolic stress-promoted) tumorigenesis by suppressing autophagic and proteasomal degradation[57].
Metabolic disorders display a strong inflammatory basis, and vice versa, inflammation is deeply associated with metabolic alterations[58,59]. At molecular level, metabolically-driven and immune-mediated disorders induce cellular stress responses[60], and, further, in several chronic diseases increased levels of pro-inflammatory cytokines, dysregulated autophagy, as well as alterations in the intestinal microbiome can be detected[61-63].
Intestinal epithelial cells (IECs) maintain homeostasis by creating an interface between the gut microbiota and the immune system. IECs directly sense enteric luminal bacteria, collaborate with intraepithelial lymphocytes and immune cells of the lamina propria[64]. Evidences suggest that the IGF/IGFR system plays a fundamental role in the gastrointestinal tract[65]. IBD patients often exhibit metabolic changes concomitantly with the altered adipokine levels and increased inflammatory parameters[66,67]. Relative insulin resistance (i.e. increased insulin levels with normal blood glucose levels) and changes of lipid metabolism are common phenomena in IBD[67]. Moreover, in IBD patients hyperinsulinemia was proved as an independent protective factor for a 6-month-maintenance of remission[68]. In mild and moderately active ulcerative colitis epithelial IGF1R expression was found to be elevated as compared to severely inflamed or normal mucosa[3]. In Crohn’s disease, elevated IGF1R expression was observed in submucosal fibroblast-like cells, subserosal adipocytes, and hypertrophic nervous plexi[69]. Intestinal fibrosis in form of fibrotic strictures is a well described complication of longstanding Crohn’s disease. IGF1 stimulates collagen I synthesis in intestinal fibroblasts via the IGF1/IGF1R/ERK1/2 pathway[70]. These may suggest a role for IGF1R in the maintenance of chronic inflammation and stricture formation in IBD.
It has recently been found that IGF1-induced collagen I expression of intestinal fibroblasts can be repressed by resveratrol either via activating SIRT1 or inhibiting the activation of IGF1R[70]. In SW620 cells, mTOR has also been proposed as a novel direct target of resveratrol action. In addition, mTOR inhibition is necessary for autophagy induction. Inhibition of mTOR by resveratrol was found to be independent of AMPK, SIRT1, PDE, and PI3K[71], raising a putative role of the IGF/IGF1R system while mediating this inhibitory effect.
Inflammation-associated catabolic states, including sepsis and cancer are characterized by accelerated proteolysis. Among the signaling pathways that could mediate proteolysis induced by acute inflammation, like in IBD, the transcription factor forkhead box O induced by glucocorticoids and inhibited by IGF1, is likely to play a key role. Lipopolysaccharide can stimulate the expression of several components of the autophagy. This induction is associated with a rapid increase of circulating levels of TNFα together with an activation of NF-κB followed by a decrease in circulating and tissue levels of IGF1[72]. In murine model of colitis, serum IGF1 level was found to be reduced in ileitis[73]. This reduction may be due to post-growth hormone receptor effects of IL-6 on IGF1 stability[74]. Furthermore, granulocyte-monocyte colony stimulating factor neutralization via STAT5 suppression and the deficiency of the CARD15 gene, an autophagy-activating sensor may also be involved in that phenomenon[73]. Therefore, targeted inhibition of IGF1 either by restoring tissue and circulating IGF1 levels, or modifying IGF1 stability all could have possible therapeutic potentials in IBD, partly due to alteration of the autophagy process (Table 1).
| Inducing effects/therapeutic agents | Corresponding cellular actions/processes | Final outcome | |
| Resveratrol | mTOR inhibition ↑ | Autophagy induction ↑ | IGF1-induced fibrosis ↓ |
| IGF1R inhibition ↑ | |||
| SIRT1 activation ↓ | |||
| Targeted inhibition of IGF1 | IGF1/IGF1R signaling ↓ | Altered autophagy machinery | Amelioration of colitis |
| Modifying IGF1 stability | IGF1/IGF1R signaling ↓ | Altered autophagy machinery | Amelioration of colitis |
| Chronic inflammation | IGF/IGF1R signaling ↑ | Altered autophagy machinery; | Pro-tumor effect ↑ |
| Survival and proliferation of cells bearing genetic errors ↑ | |||
| Chronic inflammation + small molecule RTK inhibitors | IGF/IGF1R signaling ↓ | Survival and proliferation of cells bearing genetic errors ↓ | Pro-tumor effect ↓ |
| Targeted inhibition of IGF1R | IGF1R signaling ↓ | Cell-protective autophagy ↑ | Efficacy of IGF1R targeting ↓ |
| Targeted inhibition of IGF1R + | IGF1R signaling ↓ | Cell-protective autophagy ↓ | Efficacy of IGF1R targeting ↑ |
| Autophagy disrupting agents | |||
| BCAA | IGF1R activation ↓ | Insulin-induced cell proliferation ↓ | Anti-tumor effect ↑ |
| IGF1R/EGFR inhibition + | IGF1R activation ↓ | Cell-protective autophagy ↓ | Anti-tumor effect ↑ |
| Increasing miR216b level + autophagy blocking | |||
Although combinations of surgery, radiotherapy and chemotherapy are used generally, innovative strategies are needed to improve the therapeutic outcome of CRC patients, especially with advanced stages of the disease. In the last decade new hypotheses have been considered on the mechanisms implicated in the early steps of CRC. Mainly, it has been postulated that mucosal inflammation, and epithelial injury can be considered as important determinants. Indeed, tissue injury caused by infectious, mechanical, or chemical agents may elicit a chronic immune response leading to cell proliferation, regeneration, and altered autophagy. When the immune response fails to resolve injury, the inflammatory milieu rich in cytokines, growth factors (including IGFs/IGFRs), and reactive oxygen species participates in making an attempt to repair, resulting finally in accumulation of genetic errors and a sustained inappropriate proliferation. Numerous evidence supports the contribution of inflammatory responses in the subsequent development of CRC[75]. The development of targeted therapies that block selectively molecular pathways driving CRC is in the focus of current research. Small molecule inhibitors specific for receptor tyrosine kinases (including IGFRs) have so far demonstrated promising effects[76,77].
IGFRs, expressed physiologically in the mucosal and muscular layers of the colon[78], are definitely overexpressed by colon cancer cells[79]. Abnormal activation of the IGF/IGF1R axis is a key element in MetS-related cancer development, since it affects the expression and function of many proteins being involved in regulation of autophagy and apoptosis, and is also involved in cancer cell survival, resistance to apoptosis, and cell-cycle progression[80].
The biologic function of autophagy in CRC is rather controversial. Indeed, the down-regulated expression of Atgs are associated with colorectal tumorigenesis[13,14], however, the induction of autophagy contributes to proliferative arrest of human colon cancer cells[15,16]. It has also been suggested that cytotoxic agents, including chemotherapeutics, induce autophagy in cancer cells[22,81].
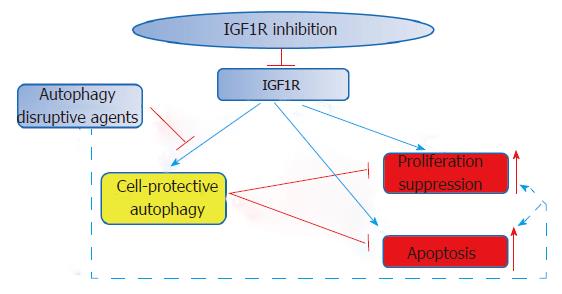
In general, treatment with anti-IGF1R monoclonal antibodies seems to be relatively well-tolerated; the main detected side effects include hyperglycemia, fatigue, and thrombocytopenia. Its beneficial clinical activities have been observed in a broad range of different tumor, including CRC[82]; nonetheless, in groups of unselected cancer patients clinical studies with pharmacological agents targeting the IGF pathway have so far demonstrated modest efficacy regarding the outcome. The complexity of the IGF/IGFR pathway may in part account for this failure. Similar to IGF1R interaction with IGF1, binding of IGF2 to IGF1R or IR-A can also stimulate IGF signaling. The situation is further complicated if cells contain hybrid heterodimeric receptors consisting of IGF1R and IR subunits, which can act as a major transducer of IGF signaling[83]. In case of triple negative breast cancer cells, IGF1R inhibition on the one part induces cell-protective autophagy, which may to some degree rescue cells from other actions of the same receptorial inhibition, like proliferation suppression and apoptosis, and thereby weakens the efficacy of IGF1R-targeting agents. However, autophagy-disrupting agents can enhance the effect of IGF1R inhibitors[84], which may constitute a potential therapeutic strategy for cancers, including CRC (Figure 2).
By defining a cut-off for IGF2 overexpression based on differential expression between colorectal tumors and normal tissue samples, an attractive patient selection biomarker for IGF pathway inhibitors were found[85]. Additionally, combined targeting of IGF/VEGF and autophagy systems may further improve clinical outcomes[86].
In vivo studies have reported that branched chain amino acid (BCAA) supplementation inhibits the activation of IGF1R[87-89]. BCAA has been found to enhance LC3-II and beclin 1 expressions, indicating its putative autophagy inductive effect. Moreover, BCAA also decreases the insulin-induced proliferation of HCT-116 colon cancer cells by inhibiting IGF1R and inducing autophagy[90]. These results suggest that an active intervention using BCAA might serve as a novel therapeutic approach for insulin-related CRC.
In case of cathepsin inhibition, higher levels of activated Shc and reduction of of activated MAPK can be found in epithelial-derived cells. The activated Shc trapped in autophagic vesicles is not able to activate downstream cytosolic proteins including MAPK. Further activation of MAPK by IGF-1 is also diminished. Cathepsin inhibition in cancer cells leads to accumulation of Shc proteins in autophagolysosomes and impairs MAPK signaling, identifying a novel mechanism by which protease inhibitors can block cell proliferation, and lead to tumor cell death[41].
Therapeutic responses to targeted therapies are often shortlived as tumor cells acquire resistance pathways. The IGF1R system plays a critical role in the regulation of cell growth and malignant transformation via the MAPK and PI3K/Akt pathways. Interactions of IGF1R with other receptor tyrosine kinases have been reported, and a signaling crosstalk of IGF1R/EGFR was also observed[77]. The use of monoclonal antibodies for EGFR blockade is a well-established strategy in CRC treatment. Nevertheless, the loss of EGFR signaling in CRC cells can be compensated simply via activation of alternative signaling pathways, controlled in part by IGF1R[91]. Moreover, studies indicate that the mechanism of resistance to anti-EGFR antibodies biochemically involves as the Ras/Raf/Mek/Erk, as the PI3K/Akt/mTOR pathways. In addition, recent data suggest that failure of anti-EGFR therapies is accompanied by inhibition of EGFR internalization, ubiquitination, degradation and prolonged downregulation[92,93].
Cetuximab, a monoclonal antibody blocking EGFR has been used for CRC treatment, but some CRCs failed to respond to anti-EGFR therapy. Anti-EGFR therapy, in vitro, has been found to activate dose-dependently Beclin-1 when HT29 and SW480 CRC cell lines were used. Moreover, microRNA (miR)-216b level was significantly downregulated in anti-EGFR-treated CRC cells[94]. According to these data, anti-EGFR antibodies may decrease miR-216b level in CRC cells, whith the subsequent upregulation of Beclin-1 that increases cancer cell autophagy in order to antagonize anti-EGFR-induced cell death.
One can speculate, that in CRC the outcome of combined anti-receptor tyrosine kinase therapies could be optimized by strategies that inhibit IGF1R/EGFR[95], increase miR-216b level, or block cell autophagy simultaneously.
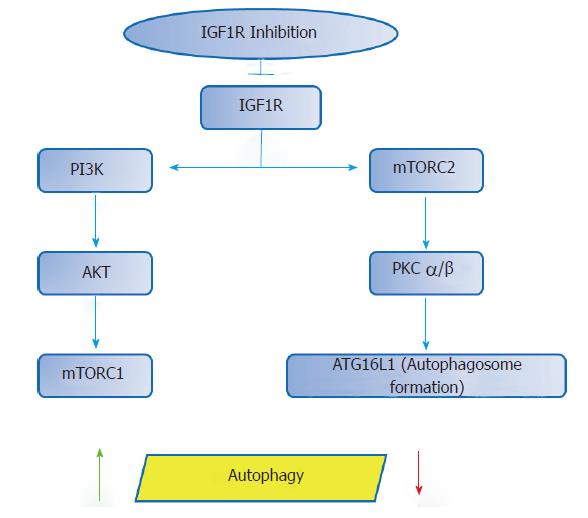
The possibility of targeting the IGF1R with several actions involved in carcinogenesis suggests that it may represent a potential therapeutic option. Even so, cautiousness is required, since pharmacologic modulation of the IGF1R can initiate additional biologic effects. According to recent data, IGF1R inhibition may lead to a decrease in mTORC2 function, which, in turn, reduces the activity of protein kinase C (PKC) alpha and beta, and thus, influences the autophagosome formation by modulating the cytoskeleton and the rate of endocytosis[96] Therefore, via IGF1R inhibition the process of autophagy could be affected bi-directionally (Figure 3).
From a pharmacological point of view, however, it is attractive to speculate that synergistic benefits could be achieved by inhibition of one of the key effectors of the IGF1R pathway, in parallel with the pharmacological stimulation of the autophagy machinery. Additionally, data also suggest that there may be benefits in using dual mTORC1/2 catalytic inhibitors for longer periods, as these may result in autophagy inhibition, which may decrease viability of at least some types of cancers[97-99].
The crosstalk between cell cycle progression and autophagy is not fully understood. According to earlier results, cells undergoing mitosis are more resistant to autophagy stimuli like mTOR inhibition[100]. The active ingredient of a gum resin from Boswellia serrata, 3-acetyl-11-keto-β-boswellic acid (AKBA), has recently gained attention as a chemopreventive compound due to its ability to target key oncogenic proteins[101,102]. AKBA has been shown to inhibit the growth of CRC cells partly by its ability to regulate cell epigenetic machinery[103]. Using a potent natural AKBA analog (BA145) robust autophagy was detected in pancreatic cancer cells in a time and dose dependent manner[104]. The BA145-triggered autophagy resulted in G2/M arrest of cell cycle along with inhibited cell growth. Induction of autophagy was associated with the BA145-mediated inhibition of mTOR, which, in turn led to feedback activation of Akt via IGF1R/PI3K signaling. This feedback activation of Akt, however, lessened the BA145-triggered autophagy and its related effects on cell cylce arrest and cell death, thus indicating the decreased effectiveness of a single target-based cancer therapy.
In summary, recent data suggest that inhibition of IGF/IGF1R system along with manipulation of the autophagy process could play an important role in suppressing insulin-related inflammatory and cancerous disorders of the colon. On the other hand, wariness is required, as well, since single or combined pharmacologic modulation of the IGF1R - autophagy machinery can initiate further, sometimes undesirable pathobiologic outcomes.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Hungary
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Zhong ZH S- Editor: Chen K L- Editor: A E- Editor: Ma YJ
| 1. | Lopez-Candales A, Hernández Burgos PM, Hernandez-Suarez DF, Harris D. Linking Chronic Inflammation with Cardiovascular Disease: From Normal Aging to the Metabolic Syndrome. J Nat Sci. 2017;3:Pii: e341. [PubMed] |
| 2. | Bastard JP, Maachi M, Lagathu C, Kim MJ, Caron M, Vidal H, Capeau J, Feve B. Recent advances in the relationship between obesity, inflammation, and insulin resistance. Eur Cytokine Netw. 2006;17:4-12. [PubMed] |
| 3. | Sipos F, Galamb O, Herszényi L, Molnár B, Solymosi N, Zágoni T, Berczi L, Tulassay Z. Elevated insulin-like growth factor 1 receptor, hepatocyte growth factor receptor and telomerase protein expression in mild ulcerative colitis. Scand J Gastroenterol. 2008;43:289-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Ge RT, Mo LH, Wu R, Liu JQ, Zhang HP, Liu Z, Liu Z, Yang PC. Insulin-like growth factor-1 endues monocytes with immune suppressive ability to inhibit inflammation in the intestine. Sci Rep. 2015;5:7735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Kawanishi S, Ohnishi S, Ma N, Hiraku Y, Murata M. Crosstalk between DNA Damage and Inflammation in the Multiple Steps of Carcinogenesis. Int J Mol Sci. 2017;18:Pii: E1808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 191] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 6. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47134] [Article Influence: 3366.7] [Reference Citation Analysis (5)] |
| 7. | Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2502] [Cited by in RCA: 2509] [Article Influence: 119.5] [Reference Citation Analysis (0)] |
| 8. | Giovannucci E, Michaud D. The role of obesity and related metabolic disturbances in cancers of the colon, prostate, and pancreas. Gastroenterology. 2007;132:2208-2225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 402] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 9. | Frezza EE, Wachtel MS, Chiriva-Internati M. Influence of obesity on the risk of developing colon cancer. Gut. 2006;55:285-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 242] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 10. | Hagiwara A, Nishiyama M, Ishizaki S. Branched-chain amino acids prevent insulin-induced hepatic tumor cell proliferation by inducing apoptosis through mTORC1 and mTORC2-dependent mechanisms. J Cell Physiol. 2012;227:2097-2105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Durai R, Yang W, Gupta S, Seifalian AM, Winslet MC. The role of the insulin-like growth factor system in colorectal cancer: review of current knowledge. Int J Colorectal Dis. 2005;20:203-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Műzes G, Constantinovits M, Fűri I, Tulassay Z, Sipos F. Interaction of autophagy and Toll-like receptors: a regulatory cross-talk--even in cancer cells? Curr Drug Targets. 2014;15:743-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573-18583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 316] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 14. | Cho DH, Jo YK, Kim SC, Park IJ, Kim JC. Down-regulated expression of ATG5 in colorectal cancer. Anticancer Res. 2012;32:4091-4096. [PubMed] |
| 15. | Xie CM, Chan WY, Yu S, Zhao J, Cheng CH. Bufalin induces autophagy-mediated cell death in human colon cancer cells through reactive oxygen species generation and JNK activation. Free Radic Biol Med. 2011;51:1365-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 205] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 16. | Huang S, Sinicrope FA. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6:256-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Pal S, Salunke-Gawalib S, Konkimallaa VB. Induction of Autophagic Cell Death in Apoptosis-resistant Pancreatic Cancer Cells using Benzo[α]phenoxazines Derivatives, 10-methyl-benzo[α]phenoxazine-5-one and benzo[α]phenoxazine-5-one. Anticancer Agents Med Chem. 2017;17:115-125. [PubMed] |
| 18. | Hu T, Wang L, Zhang L, Lu L, Shen J, Chan RL, Li M, Wu WK, To KK, Cho CH. Sensitivity of apoptosis-resistant colon cancer cells to tanshinones is mediated by autophagic cell death and p53-independent cytotoxicity. Phytomedicine. 2015;22:536-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Chitikova ZV, Gordeev SA, Bykova TV, Zubova SG, Pospelov VA, Pospelova TV. Sustained activation of DNA damage response in irradiated apoptosis-resistant cells induces reversible senescence associated with mTOR downregulation and expression of stem cell markers. Cell Cycle. 2014;13:1424-1439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 20. | Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1088] [Cited by in RCA: 1146] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 21. | Liu Q, Guan JZ, Sun Y, Le Z, Zhang P, Yu D, Liu Y. Insulin-like growth factor 1 receptor-mediated cell survival in hypoxia depends on the promotion of autophagy via suppression of the PI3K/Akt/mTOR signaling pathway. Mol Med Rep. 2017;15:2136-2142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Degtyarev M, De Mazière A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 359] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 23. | Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243-35246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 485] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 24. | SALMON WD Jr, DAUGHADAY WH. A hormonally controlled serum factor which stimulates sulfate incorporation by cartilage in vitro. J Lab Clin Med. 1957;49:825-836. [PubMed] |
| 25. | Brahmkhatri VP, Prasanna C, Atreya HS. Insulin-like growth factor system in cancer: novel targeted therapies. Biomed Res Int. 2015;2015:538019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 187] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 26. | Mohan S, Baylink DJ. IGF-binding proteins are multifunctional and act via IGF-dependent and -independent mechanisms. J Endocrinol. 2002;175:19-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 305] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 27. | Iams WT, Lovly CM. Molecular Pathways: Clinical Applications and Future Direction of Insulin-like Growth Factor-1 Receptor Pathway Blockade. Clin Cancer Res. 2015;21:4270-4277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 28. | Massoner P, Ladurner-Rennau M, Eder IE, Klocker H. Insulin-like growth factors and insulin control a multifunctional signalling network of significant importance in cancer. Br J Cancer. 2010;103:1479-1484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Pandini G, Frasca F, Mineo R, Sciacca L, Vigneri R, Belfiore A. Insulin/insulin-like growth factor I hybrid receptors have different biological characteristics depending on the insulin receptor isoform involved. J Biol Chem. 2002;277:39684-39695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 355] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 30. | Belfiore A, Frasca F, Pandini G, Sciacca L, Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr Rev. 2009;30:586-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 753] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 31. | Kasprzak A, Kwasniewski W, Adamek A, Gozdzicka-Jozefiak A. Insulin-like growth factor (IGF) axis in cancerogenesis. Mutat Res Rev Mutat Res. 2017;772:78-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 32. | Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1502] [Cited by in RCA: 1665] [Article Influence: 111.0] [Reference Citation Analysis (0)] |
| 33. | Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2780] [Cited by in RCA: 2972] [Article Influence: 141.5] [Reference Citation Analysis (0)] |
| 34. | Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 709] [Cited by in RCA: 705] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 35. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5292] [Article Influence: 311.3] [Reference Citation Analysis (0)] |
| 36. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5765] [Cited by in RCA: 5747] [Article Influence: 338.1] [Reference Citation Analysis (1)] |
| 37. | Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 528] [Cited by in RCA: 580] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 38. | Cadwell K, Patel KK, Komatsu M, Virgin HW 4th, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 182] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 39. | Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322-329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 365] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 40. | Kawaguchi T, Nagao Y, Matsuoka H, Ide T, Sata M. Branched-chain amino acid-enriched supplementation improves insulin resistance in patients with chronic liver disease. Int J Mol Med. 2008;22:105-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Soori M, Lu G, Mason RW. Cathepsin Inhibition Prevents Autophagic Protein Turnover and Downregulates Insulin Growth Factor-1 Receptor-Mediated Signaling in Neuroblastoma. J Pharmacol Exp Ther. 2016;356:375-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Ghosh A, Sengupta A, Seerapu GPK, Nakhi A, Shivaji Ramarao EVV, Bung N, Bulusu G, Pal M, Haldar D. A novel SIRT1 inhibitor, 4bb induces apoptosis in HCT116 human colon carcinoma cells partially by activating p53. Biochem Biophys Res Commun. 2017;488:562-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 43. | Reverchon M, Rame C, Bunel A, Chen W, Froment P, Dupont J. VISFATIN (NAMPT) Improves In Vitro IGF1-Induced Steroidogenesis and IGF1 Receptor Signaling Through SIRT1 in Bovine Granulosa Cells. Biol Reprod. 2016;94:54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 44. | Wang Y, Zhao X, Shi D, Chen P, Yu Y, Yang L, Xie L. Overexpression of SIRT1 promotes high glucose-attenuated corneal epithelial wound healing via p53 regulation of the IGFBP3/IGF-1R/AKT pathway. Invest Ophthalmol Vis Sci. 2013;54:3806-3814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 45. | Sansone L, Reali V, Pellegrini L, Villanova L, Aventaggiato M, Marfe G, Rosa R, Nebbioso M, Tafani M, Fini M. SIRT1 silencing confers neuroprotection through IGF-1 pathway activation. J Cell Physiol. 2013;228:1754-1761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 46. | Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6:599-612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 47. | Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374-3379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1134] [Cited by in RCA: 1182] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 48. | Pillai VB, Sundaresan NR, Gupta MP. Regulation of Akt signaling by sirtuins: its implication in cardiac hypertrophy and aging. Circ Res. 2014;114:368-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 215] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 49. | Yang J, Nishihara R, Zhang X, Ogino S, Qian ZR. Energy sensing pathways: Bridging type 2 diabetes and colorectal cancer? J Diabetes Complications. 2017;31:1228-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 50. | Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, Schneider JI, Chai H, Bronson RT, Tsai LH. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci USA. 2010;107:21790-21794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 145] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 51. | Kenyon CJ. The genetics of ageing. Nature. 2010;464:504-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1979] [Cited by in RCA: 2037] [Article Influence: 135.8] [Reference Citation Analysis (0)] |
| 52. | Bartke A, Brown-Borg H. Life extension in the dwarf mouse. Curr Top Dev Biol. 2004;63:189-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 257] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 53. | Xiao G. Autophagy and NF-kappaB: fight for fate. Cytokine Growth Factor Rev. 2007;18:233-243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 88] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 54. | Guo Y, Chang C, Huang R, Liu B, Bao L, Liu W. AP1 is essential for generation of autophagosomes from the trans-Golgi network. J Cell Sci. 2012;125:1706-1715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 55. | Schwarzer R, Dames S, Tondera D, Klippel A, Kaufmann J. TRB3 is a PI 3-kinase dependent indicator for nutrient starvation. Cell Signal. 2006;18:899-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Hua F, Li K, Yu JJ, Lv XX, Yan J, Zhang XW, Sun W, Lin H, Shang S, Wang F. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun. 2015;6:7951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 125] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 57. | Hua F, Li K, Yu JJ, Hu ZW. The TRIB3-SQSTM1 interaction mediates metabolic stress-promoted tumorigenesis and progression via suppressing autophagic and proteasomal degradation. Autophagy. 2015;11:1929-1931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 6392] [Article Influence: 355.1] [Reference Citation Analysis (1)] |
| 59. | Zietek T, Rath E. Inflammation Meets Metabolic Disease: Gut Feeling Mediated by GLP-1. Front Immunol. 2016;7:154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 60. | Rath E, Haller D. Inflammation and cellular stress: a mechanistic link between immune-mediated and metabolically driven pathologies. Eur J Nutr. 2011;50:219-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 61. | Brestoff JR, Artis D. Immune regulation of metabolic homeostasis in health and disease. Cell. 2015;161:146-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 356] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 62. | Monkkonen T, Debnath J. Inflammatory signaling cascades and autophagy in cancer. Autophagy. 2017;1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 63. | Øyri SF, Műzes G, Sipos F. Dysbiotic gut microbiome: A key element of Crohn’s disease. Comp Immunol Microbiol Infect Dis. 2015;43:36-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 64. | Clavel T, Haller D. Bacteria- and host-derived mechanisms to control intestinal epithelial cell homeostasis: implications for chronic inflammation. Inflamm Bowel Dis. 2007;13:1153-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 65. | Kuemmerle JF. Insulin-like growth factors in the gastrointestinal tract and liver. Endocrinol Metab Clin North Am. 2012;41:409-423, vii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 66. | Korkmaz H, Sahin F, Ipekci SH, Temel T, Kebapcilar L. Increased pulse wave velocity and relationship with inflammation, insulin, and insulin resistance in inflammatory bowel disease. Eur J Gastroenterol Hepatol. 2014;26:725-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 67. | Karrasch T, Obermeier F, Straub RH. Systemic metabolic signaling in acute and chronic gastrointestinal inflammation of inflammatory bowel diseases. Horm Metab Res. 2014;46:445-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 68. | Valentini L, Wirth EK, Schweizer U, Hengstermann S, Schaper L, Koernicke T, Dietz E, Norman K, Buning C, Winklhofer-Roob BM. Circulating adipokines and the protective effects of hyperinsulinemia in inflammatory bowel disease. Nutrition. 2009;25:172-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 69. | El Yafi F, Winkler R, Delvenne P, Boussif N, Belaiche J, Louis E. Altered expression of type I insulin-like growth factor receptor in Crohn’s disease. Clin Exp Immunol. 2005;139:526-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 70. | Li P, Liang ML, Zhu Y, Gong YY, Wang Y, Heng D, Lin L. Resveratrol inhibits collagen I synthesis by suppressing IGF-1R activation in intestinal fibroblasts. World J Gastroenterol. 2014;20:4648-4661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (3)] |
| 71. | Park D, Jeong H, Lee MN, Koh A, Kwon O, Yang YR, Noh J, Suh PG, Park H, Ryu SH. Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Sci Rep. 2016;6:21772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 187] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 72. | Schakman O, Dehoux M, Bouchuari S, Delaere S, Lause P, Decroly N, Shoelson SE, Thissen JP. Role of IGF-I and the TNFα/NF-κB pathway in the induction of muscle atrogenes by acute inflammation. Am J Physiol Endocrinol Metab. 2012;303:E729-E739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 73. | D’Mello S, Trauernicht A, Ryan A, Bonkowski E, Willson T, Trapnell BC, Frank SJ, Kugasathan S, Denson LA. Innate dysfunction promotes linear growth failure in pediatric Crohn’s disease and growth hormone resistance in murine ileitis. Inflamm Bowel Dis. 2012;18:236-245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 74. | De Benedetti F, Meazza C, Oliveri M, Pignatti P, Vivarelli M, Alonzi T, Fattori E, Garrone S, Barreca A, Martini A. Effect of IL-6 on IGF binding protein-3: a study in IL-6 transgenic mice and in patients with systemic juvenile idiopathic arthritis. Endocrinology. 2001;142:4818-4826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 90] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 75. | Mariani F, Sena P, Roncucci L. Inflammatory pathways in the early steps of colorectal cancer development. World J Gastroenterol. 2014;20:9716-9731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 105] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 76. | Hopfner M, Sutter AP, Huether A, Baradari V, Scherubl H. Tyrosine kinase of insulin-like growth factor receptor as target for novel treatment and prevention strategies of colorectal cancer. World J Gastroenterol. 2006;12:5635-5643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 77. | Kaulfuss S, Burfeind P, Gaedcke J, Scharf JG. Dual silencing of insulin-like growth factor-I receptor and epidermal growth factor receptor in colorectal cancer cells is associated with decreased proliferation and enhanced apoptosis. Mol Cancer Ther. 2009;8:821-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 78. | Alemán JO, Eusebi LH, Ricciardiello L, Patidar K, Sanyal AJ, Holt PR. Mechanisms of obesity-induced gastrointestinal neoplasia. Gastroenterology. 2014;146:357-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 152] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 79. | Ouban A, Muraca P, Yeatman T, Coppola D. Expression and distribution of insulin-like growth factor-1 receptor in human carcinomas. Hum Pathol. 2003;34:803-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 172] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 80. | Chang CK, Ulrich CM. Hyperinsulinaemia and hyperglycaemia: possible risk factors of colorectal cancer among diabetic patients. Diabetologia. 2003;46:595-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 81. | Tran TT, Medline A, Bruce WR. Insulin promotion of colon tumors in rats. Cancer Epidemiol Biomarkers Prev. 1996;5:1013-1015. [PubMed] |
| 82. | Chu E. The IGF-1R pathway as a therapeutic target. Oncology (Williston Park). 2011;25:538-539, 543. [PubMed] |
| 83. | Pandini G, Vigneri R, Costantino A, Frasca F, Ippolito A, Fujita-Yamaguchi Y, Siddle K, Goldfine ID, Belfiore A. Insulin and insulin-like growth factor-I (IGF-I) receptor overexpression in breast cancers leads to insulin/IGF-I hybrid receptor overexpression: evidence for a second mechanism of IGF-I signaling. Clin Cancer Res. 1999;5:1935-1944. [PubMed] |
| 84. | Wu W, Ma J, Shao N, Shi Y, Liu R, Li W, Lin Y, Wang S. Co-Targeting IGF-1R and Autophagy Enhances the Effects of Cell Growth Suppression and Apoptosis Induced by the IGF-1R Inhibitor NVP-AEW541 in Triple-Negative Breast Cancer Cells. PLoS One. 2017;12:e0169229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 85. | Sanderson MP, Hofmann MH, Garin-Chesa P, Schweifer N, Wernitznig A, Fischer S, Jeschko A, Meyer R, Moll J, Pecina T. The IGF1R/INSR Inhibitor BI 885578 Selectively Inhibits Growth of IGF2-Overexpressing Colorectal Cancer Tumors and Potentiates the Efficacy of Anti-VEGF Therapy. Mol Cancer Ther. 2017;16:2223-2233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 86. | Stanton MJ, Dutta S, Polavaram NS, Roy S, Muders MH, Datta K. Angiogenic growth factor axis in autophagy regulation. Autophagy. 2013;9:789-790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 87. | Shimizu M, Shirakami Y, Iwasa J, Shiraki M, Yasuda Y, Hata K, Hirose Y, Tsurumi H, Tanaka T, Moriwaki H. Supplementation with branched-chain amino acids inhibits azoxymethane-induced colonic preneoplastic lesions in male C57BL/KsJ-db/db mice. Clin Cancer Res. 2009;15:3068-3075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 88. | Iwasa J, Shimizu M, Shiraki M, Shirakami Y, Sakai H, Terakura Y, Takai K, Tsurumi H, Tanaka T, Moriwaki H. Dietary supplementation with branched-chain amino acids suppresses diethylnitrosamine-induced liver tumorigenesis in obese and diabetic C57BL/KsJ-db/db mice. Cancer Sci. 2010;101:460-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 89. | Yoshiji H, Noguchi R, Kitade M, Kaji K, Ikenaka Y, Namisaki T, Yoshii J, Yanase K, Yamazaki M, Tsujimoto T. Branched-chain amino acids suppress insulin-resistance-based hepatocarcinogenesis in obese diabetic rats. J Gastroenterol. 2009;44:483-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 90. | Wubetu GY, Utsunomiya T, Ishikawa D, Ikemoto T, Yamada S, Morine Y, Iwahashi S, Saito Y, Arakawa Y, Imura S. Branched chain amino acid suppressed insulin-initiated proliferation of human cancer cells through induction of autophagy. Anticancer Res. 2014;34:4789-4796. [PubMed] |
| 91. | Ekyalongo RC, Mukohara T, Kataoka Y, Funakoshi Y, Tomioka H, Kiyota N, Fujiwara Y, Minami H. Mechanisms of acquired resistance to insulin-like growth factor 1 receptor inhibitor in MCF-7 breast cancer cell line. Invest New Drugs. 2013;31:293-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 92. | Feng Y, Gao S, Gao Y, Wang X, Chen Z. Anti-EGFR antibody sensitizes colorectal cancer stem-like cells to Fluorouracil-induced apoptosis by affecting autophagy. Oncotarget. 2016;7:81402-81409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 93. | Koustas E, Karamouzis MV, Mihailidou C, Schizas D, Papavassiliou AG. Co-targeting of EGFR and autophagy signaling is an emerging treatment strategy in metastatic colorectal cancer. Cancer Lett. 2017;396:94-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 94. | Chen Z, Gao S, Wang D, Song D, Feng Y. Colorectal cancer cells are resistant to anti-EGFR monoclonal antibody through adapted autophagy. Am J Transl Res. 2016;8:1190-1196. [PubMed] |
| 95. | Oberthür R, Seemann H, Gehrig J, Rave-Fränk M, Bremmer F, Halpape R, Conradi LC, Scharf JG, Burfeind P, Kaulfuß S. Simultaneous inhibition of IGF1R and EGFR enhances the efficacy of standard treatment for colorectal cancer by the impairment of DNA repair and the induction of cell death. Cancer Lett. 2017;407:93-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 96. | Renna M, Bento CF, Fleming A, Menzies FM, Siddiqi FH, Ravikumar B, Puri C, Garcia-Arencibia M, Sadiq O, Corrochano S. IGF-1 receptor antagonism inhibits autophagy. Hum Mol Genet. 2013;22:4528-4544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 97. | Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1195] [Cited by in RCA: 1195] [Article Influence: 91.9] [Reference Citation Analysis (0)] |
| 98. | Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 599] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 99. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 1913] [Article Influence: 159.4] [Reference Citation Analysis (0)] |
| 100. | Eskelinen EL, Prescott AR, Cooper J, Brachmann SM, Wang L, Tang X, Backer JM, Lucocq JM. Inhibition of autophagy in mitotic animal cells. Traffic. 2002;3:878-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 132] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 101. | Liu JJ, Nilsson A, Oredsson S, Badmaev V, Zhao WZ, Duan RD. Boswellic acids trigger apoptosis via a pathway dependent on caspase-8 activation but independent on Fas/Fas ligand interaction in colon cancer HT-29 cells. Carcinogenesis. 2002;23:2087-2093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 127] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 102. | Liu JJ, Huang B, Hooi SC. Acetyl-keto-beta-boswellic acid inhibits cellular proliferation through a p21-dependent pathway in colon cancer cells. Br J Pharmacol. 2006;148:1099-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 103. | Takahashi M, Sung B, Shen Y, Hur K, Link A, Boland CR, Aggarwal BB, Goel A. Boswellic acid exerts antitumor effects in colorectal cancer cells by modulating expression of the let-7 and miR-200 microRNA family. Carcinogenesis. 2012;33:2441-2449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 104. | Pathania AS, Guru SK, Kumar S, Kumar A, Ahmad M, Bhushan S, Sharma PR, Mahajan P, Shah BA, Sharma S. Interplay between cell cycle and autophagy induced by boswellic acid analog. Sci Rep. 2016;6:33146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |