Published online Oct 28, 2017. doi: 10.3748/wjg.v23.i40.7265
Peer-review started: August 5, 2017
First decision: August 14, 2017
Revised: August 25, 2017
Accepted: September 13, 2017
Article in press: September 13, 2017
Published online: October 28, 2017
Processing time: 85 Days and 4.5 Hours
To study the type and frequency of adverse events associated with anti-tumor necrosis factor (TNF) therapy and evaluate for any serologic and genetic associations.
This study was a retrospective review of patients attending the inflammatory bowel disease (IBD) centers at Cedars-Sinai IBD Center from 2005-2016. Adverse events were identified via chart review. IBD serologies were measured by ELISA. DNA samples were genotyped at Cedars-Sinai using Illumina Infinium Immunochipv1 array per manufacturer’s protocol. SNPs underwent methodological review and were evaluated using several SNP statistic parameters to ensure optimal allele-calling. Standard and rigorous QC criteria were applied to the genetic data, which was generated using immunochip. Genetic association was assessed by logistic regression after correcting for population structure.
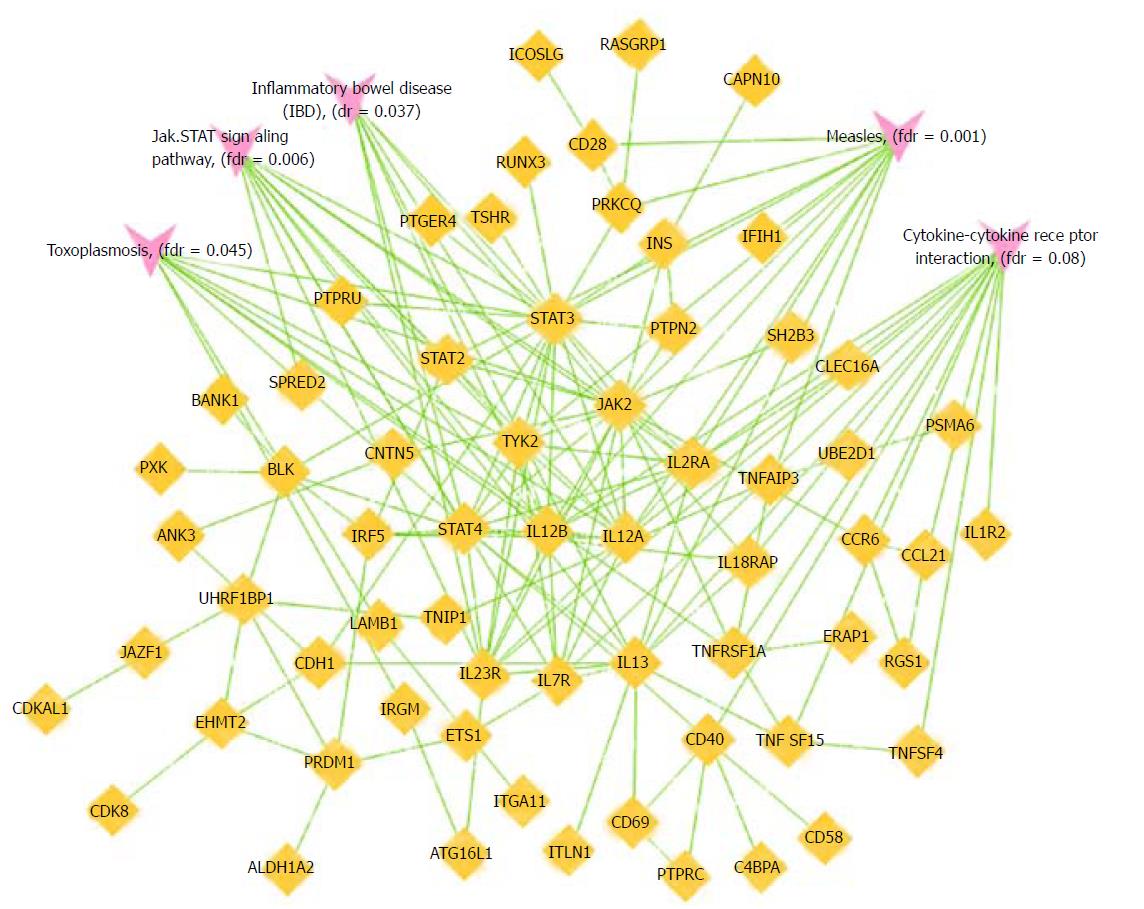
Altogether we identified 1258 IBD subjects exposed to anti-TNF agents in whom Immunochip data were available. 269/1258 patients (21%) were found to have adverse events to an anti-TNF-α agent that required the therapy to be discontinued. 25% of women compared to 17% of men experienced an adverse event. All adverse events resolved after discontinuing the anti-TNF agent. In total: n = 66 (5%) infusion reactions; n = 49 (4%) allergic/serum sickness reactions; n = 19 (1.5%) lupus-like reactions, n = 52 (4%) rash, n = 18 (1.4%) infections. In Crohn’s disease, IgA ASCA (P = 0.04) and IgG-ASCA (P = 0.02) levels were also lower in patients with any adverse events, and anti-I2 level in ulcerative colitis was significantly associated with infusion reactions (P = 0.008). The logistic regression/human annotation and network analyses performed on the Immunochip data implicated the following five signaling pathways: JAK-STAT (Janus Kinase-signal transducer and activator of transcription), measles, IBD, cytokine-cytokine receptor interaction, and toxoplasmosis for any adverse event.
Our study shows 1 in 5 IBD patients experience an adverse event to anti-TNF therapy with novel serologic, genetic , and pathways associations.
Core tip: Tumor necrosis factor-α (TNF-α) plays a key role in the development and progression of inflammatory bowel disease (IBD). Anti-TNF therapy is highly efficacious in treating IBD patients, but many experience adverse events. Few studies have evaluated factors associated with adverse events to anti-TNF therapy. In this study, we found some genetic associations and pathways that are enriched for genes associated with the development of adverse events. Future studies will need to confirm these findings as the ability to identify subjects at high risk may help clinicians anticipate and therefore prevent or avoid these adverse events in the future.
- Citation: Lew D, Yoon SM, Yan X, Robbins L, Haritunians T, Liu Z, Li D, McGovern DP. Genetic associations with adverse events from anti-tumor necrosis factor therapy in inflammatory bowel disease patients. World J Gastroenterol 2017; 23(40): 7265-7273
- URL: https://www.wjgnet.com/1007-9327/full/v23/i40/7265.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i40.7265
Tumor necrosis factor-α (TNF-α) plays a key role in the development and progression of inflammatory bowel disease (IBD)[1-3]. While anti-TNF therapy is an effective therapeutic option for IBD patients[1,4,5], response to these agents is highly heterogeneous and a high proportion of patients either fail initial induction therapy or lose response during maintenance therapy[6-8]. Predicting response to these agents has been studied extensively by evaluating a multitude of factors including potential genetic components[7], but an important addition would be the ability to predict the development of adverse events associated with these agents including: infusion reactions; infections; rash; allergic reactions; serum sickness like reactions; and lupus-like reactions[9-11]. Minimizing the risks is important to increase patient compliance and improve response to therapy, and will also become more important as physicians struggle with decisions around where to position biologic therapies as therapeutics targeting novel mechanisms become available[12,13].
Currently, there are few studies designed to determine factors associated with adverse events to anti-TNF-α therapy. The objectives of this study were to describe the type and frequency of adverse events associated with anti-TNF-α therapy in a large cohort and evaluate for any serologic and genetic associations.
This study was a retrospective review of patients attending the IBD centers at Cedars-Sinai IBD Center from 2005-2016. Patients included in the study were those that had given consent, had available genotype data, carried a diagnosis of IBD (Crohn’s disease, ulcerative colitis, or IBDU), and who had been treated with anti-TNF-α agents (infliximab, adalimumab, certolizumab pegol). All research-related activities were approved by the Cedars-Sinai Medical Center Institutional Review Board (IRB #3358).
Detailed clinical information for each patient was obtained via chart review. Clinical information included age at disease diagnosis, type of IBD (CD, UC, or IBDU), gender, and type of anti-TNF-α agent used. All patients were seen by gastroenterologists, experienced in managing patients with IBD treated with anti-TNF agents, at the IBD centers at Cedars-Sinai Medical Center, Los Angeles.
Adverse events were identified via chart review by evaluating the “Allergies” section and the progress notes written by the gastroenterologist. Potential adverse events include infusion reactions, serum sickness-like reactions, drug-induced lupus, rash, infections, and non-specific symptoms (arthralgias, shortness of breath, rash, etc). An infusion reaction was defined as any significant adverse experience that occurred during or within two hours of infusion[14]. An allergic or a serum sickness-like reaction was defined clinically as the occurrence of myalgias, arthralgias, fever, or rash within 1-14 d after reinfusion of infliximab[15].
The likelihood of a causal relationship for each adverse event was determined based on the assessment of the gastroenterologist as documented in the progress note and as evidenced by the following: time elapsed between a dose and adverse event, resolution of the adverse event when the therapy was discontinued, and return of the adverse event if the therapy was resumed[16].
IBD serologies (ANCA, anti-nuclear cytoplasmic antibodies; anti-CBir1, anti-flagellin; anti-I2, anti-Pseudomonas fluorescens-associated sequence I2; anti-OmpC, anti-outer membrane porin C; ASCA, anti-Saccharomyces cerevisiae antibodies) were measured by enzyme-linked immunosorbent assay (ELISA) as previously described[17]. Results were expressed as ELISA units (EU/mL) relative to Cedars-Sinai Medical Center laboratory or a Prometheus laboratory standard derived from a pool of patient sera with well-characterized disease found to have reactivity to these antigens. All assays were performed in a blinded fashion.
DNA samples were genotyped at Cedars-Sinai using Illumina Infinium Immunochipv1 array per manufacturer’s protocol (Illumina, San Diego, CA, United States). Average genotyping call rate for samples that passed quality control was 99.8%; average replicate concordance and average heritability rates were > 99.99% and 99.94%, respectively. Single-nucleotide polymorphisms (SNPs) underwent methodological review and were evaluated using several SNP statistic parameters to ensure optimal allele-calling[18].
χ2 test and logistic regression were performed to identify demographic and clinical characteristics associated with development of adverse events. For continuous variables with skewed distribution (e.g., serology levels), Wilcox test signed rank test was performed. SNPs association with adverse events was evaluated using PLINK. Principal components (PCs) from population stratification analysis were included in the PLINK analysis to control for potential confounding[19]. Two-sided P-value of 0.05 was considered statistically significant.
For any reaction, infusion reactions, and allergic reactions, the logistic regression/human annotation and network analysis was performed with statistically significant SNPs (P < 0.001). These SNPs were first annotated into corresponding genes, and the genes were further analyzed with multiple biological functional databases including human protein reference databases (http://www.hprd.org), Reactome, NCI/Nature pathway interaction database and others. The final networks were then constructed form the known interactions from any of these databases. Pathways and gene set enrichment analysis was performed with STRING (http://string-db.org/) and cytoscape (http://www.cytoscape.org).
1258 IBD (954 CD patients, 260 UC, 44 IBDU) patients qualified for this study. The average age of onset was 25.7 years and, and the overwhelming majority were of European ancestry.
A total of 269/1258 patients (21%) were found to have experienced an adverse event. The different types of adverse events were similar among women and men, except for lupus-like reactions and rashes, which were both more commonly seen in women (Table 1).
| Clinical traits | All adverse events | Infusion reactions | Allergic reactions | Lupus-like reactions | Rash | Other |
| Type of IBD | ||||||
| Crohn's disease (n = 954) | 220 (23) | 52 (5) | 45 (5) | 14 (1) | 40 (4) | 69 (7) |
| Ulcerative colitis (n = 260) | 42 (16) | 14 (5) | 4 (2) | 4 (2) | 10 (4) | 10 (4) |
| IBDU (n = 44) | 7 (16) | 0 (0) | 0 (0) | 1 (2) | 2 (5) | 4 (9) |
| Total | 269 (21) | 66 (5) | 49 (4) | 19 (1.5) | 52 (4) | 83 (7) |
| Gender | ||||||
| Male (n = 624) | 108 (17) | 28 (4) | 24 (4) | 3 (0.5) | 17 (3) | 36 (6) |
| Female (n = 634) | 161 (25) | 38 (6) | 25 (4) | 16 (3) | 35 (6) | 47 (7) |
In CD patients we observed that IgA ASCA+/- was associated with a lower risk of developing any adverse event (OR = 0.68, P = 0.047), while IgG ASCA+/- and total ASCA+/- show a similar trend and borderline association respectively (OR = 0.72 and 0.70, P = 0.09 and 0.05, respectively). IgA ASCA (P = 0.04) and IgG ASCA (P = 0.02) levels were also lower in patients with any adverse events (Table 2). Anti-I2 level in UC was significantly associated with infusion reactions (P = 0.008) (Table 2). No other associations were seen with IBD-associated serologies (data not shown).
| IBD type | Adverse event | Serological marker | Serology levels in positive (U/mL) | Serology levels in negative (U/mL) | P value |
| CD | Any | IgA ASCA | 7 | 10 | 0.040 |
| CD | Any | IgG ASCA | 18 | 26.5 | 0.020 |
| UC | Infusion | Anti-I2 | 0 | 7 | 0.008 |
IBD-associated SNPs that achieved a nominal level of significance with adverse events are shown in Table 3. For any reaction, infusion reactions, and allergic reactions, the logistic regression/human annotation and network analyses performed on the Immunochip data implicated the following five signaling pathways: JAK-STAT (Janus Kinase-signal transducer and activator of transcription), Measles, IBD, Cytokine-cytokine receptor interaction, and Toxoplasmosis (Table 4 and Figure 1). After False Discovery Rate correction, all associations remained statistically significant except for the Cytokine-cytokine receptor interaction for infusion and allergic reactions.
| Adverse event | Risk allele | SNP | Gene (s) of interest | P value | OR (95%CI) |
| Infusion | C | rs6740462 | SPRED2 | 0.003 | 0.4 (0.2-0.7) |
| Infusion | G | rs1182188 | GNA12 | 0.007 | 1.8 (1.2-2.6) |
| Infusion | G | rs10061469 | TMEM174, FOXD1 | 0.010 | 0.5 (0.3-0.9) |
| Infusion | A | rs477515 | HLA-DRB1 | 0.010 | 1.7 (1.1-2.5) |
| Allergic | A | rs4692386 | SMIM20, RBPJ | 0.010 | 0.5 (0.3-0.9) |
| Allergic | A | rs10761659 | ZNF365 | 0.010 | 0.6 (0.3-0.9) |
| Lupus-like | G | rs13407913 | ADCY3 | 0.003 | 3.5 (1.6-7.9) |
| Lupus-like | C | rs10051722 | CHSY2, HINT1 | 0.010 | 2.7 (1.2-5.7) |
| Rash | A | rs1363907 | ERAP2 | 0.003 | 3.0 (1.5-6.2) |
| Rash | G | rs11010067 | PARD3 | 0.003 | 0.2 (0.1-0.6) |
| Rash | C | rs7746082 | PREP | 0.005 | 2.7 (1.3-5.3) |
| Any | C | rs6740462 | SPRED2 | 0.0007 | 0.6 (0.5-0.8) |
| Any | C | rs10774482 | ERC1 | 0.003 | 1.4 (1.1-1.8) |
| Type of adverse event | Pathway | Number of genes | P value | P value (Bonferroni) |
| Any | JAK-STAT signaling pathway | 13 | 3.98 × 10-6 | 7.96 × 10-4 |
| Any | Measles | 12 | 8.87 × 10-6 | 0.0018 |
| Any | IBD | 8 | 3.05 × 10-5 | 0.0061 |
| Any | Cytokine-cytokine receptor interaction | 16 | 5.03 × 10-5 | 0.0100 |
| Any | Toxoplasmosis | 10 | 5.49 × 10-5 | 0.0110 |
| Infusion | JAK-STAT signaling pathway | 12 | 3.68 × 10-5 | 0.0074 |
| Infusion | Measles | 12 | 8.24 × 10-6 | 0.0016 |
| Infusion | IBD | 7 | 2.11 × 10-4 | 0.0420 |
| Infusion | Cytokine-cytokine receptor interaction | 14 | 4.98 × 10-4 | 0.0990 |
| Infusion | Toxoplasmosis | 11 | 9.02 × 10-6 | 0.0018 |
| Allergic | JAK-STAT signaling pathway | 12 | 2.96 ×10-5 | 0.0060 |
| Allergic | Measles | 12 | 6.6 × 10-6 | 0.0010 |
| Allergic | IBD | 7 | 1.84 × 10-4 | 0.0370 |
| Allergic | Cytokine-cytokine receptor interaction | 14 | 4.02 ×10-4 | 0.0800 |
| Allergic | Toxoplasmosis | 9 | 2.25 ×10-4 | 0.0450 |
In our cohort approximately 1 in 5 IBD patients experienced adverse events to anti-TNFs that eventually led to the discontinuation of the therapy in keeping with the current literature, although most studies only report the presence or not of adverse events and do not comment on whether patients had to discontinue[20]. The incidence of serious lupus-like reactions requiring the discontinuation of anti-TNFs was found to be 1.1%[21], which was comparable to that seen in our population of 1.5%. To our knowledge, this is the largest study examining adverse events with anti-TNF agents.
We identified a number of genetic associations with known IBD loci including two (HLA-DRB1 and ERAP2 [endoplasmic reticulum aminopeptidase 2]) that are associated with a number of immune-mediated diseases as well as IBD and also, in the case of HLA-DRB1, with the development of extra-intestinal manifestations in IBD[22-24]. Furthermore, both are involved in peptide presentation by HLA molecules[25,26]. We also observed associations at other IBD genes including ZNF365, a transcription factor in maintaining genomic stability during DNA replication in the brain, heart, lung, pancreas, small intestine and colon[27,28]. Our genetic findings also implicated genes that maintain colonic wall permeability including SPRED (Sprouty-related EVH1 domain-containing protein) and PARD3 (Partitioning defective 3 homolog)[29,30]. In addition, GNA12 (Guanine nucleotide-binding protein alpha-12), a modulator of different transmembrane signaling systems, has also been implicated in the loss of barrier integrity[31]. Our pathway analyses strongly implicated five signaling pathways including JAK-STAT signaling pathway, Cytokine-cytokine receptor interaction pathway, Measles signaling pathway, Toxoplasmosis signaling pathway, and the IBD signaling pathway. The network analyses for allergic reactions (Figure 1) show a number of key nodes including TYK2, BLK and IL13, which have previously been shown to be associated with allergic susceptibility[32-34].
IBD serologies (ANCA, anti-CBir1, anti-I2, anti-OmpC, and ASCA) can distinguish CD from UC, risk stratify IBD patients, and also predict postoperative complications and occur as a result of an aberrant or exaggerated response to commensal flora[35]. The association with ASCA and I2 are interesting. Perhaps these markers identify patients with a predilection towards small bowel involvement. Patients with colonic disease tend to respond less to anti-TNFs or require higher doses[8,36] and, perhaps therefore, these patients are more likely to develop antibodies or reactions to anti-TNFs. Further studies will be needed to confirm these borderline associations.
There are several potential limitations of this study including, the relatively small sample size and the retrospective nature of the study (despite it being the largest of its kind to date). Additionally, we did not have information on anti-drug antibody formation as the majority of these patients developed adverse events prior to the widespread use of these parameters in clinical practice. It is also important to note that our study population was predominantly of European ancestry. While IBD is rising in non-Europeans, the highest prevalence is still seen in European ancestry populations. For this reason, and the location of Cedars-Sinai Medical Center in west Los Angeles, the majority of our patients are “European”. Previous work have shown ethnic differences in genetic associations with adverse events[37], and a study similar to this one should be performed for other ethnic groups.
In conclusion, our study revealed that approximately 1 in 5 IBD patients experienced an adverse event to anti-TNF therapies that required cessation of therapy. The majority of these were infusion/allergic reactions but approximately 1 in 30 women will develop a lupus-like reaction and we also observed other serious adverse events including pancreatitis and vascultitis but these were rare. We have demonstrated some genetic associations and pathways that are enriched for genes associated with development adverse events. Future studies will need to confirm these findings as the ability to identify subjects at high risk may help clinicians anticipate and therefore prevent or avoid these adverse events in the future.
Tumor necrosis factor (TNF) inhibitors are highly efficacious in treating inflammatory bowel disease (IBD). Response to these agents is highly heterogenous, and there have been a multitude of studies aimed at predicting the response to these agents. An important addition is the ability to predict the development of adverse events associated with these agents such as infusion reactions, infections, or rash. Minimizing the risk is important to increase patient compliance and improve response to therapy.
Recognizing the type and frequency of adverse events to anti-TNF therapy, and the potential genetic and serologic associations can help identify subjects at high risk, and may help clinicians anticipate and therefore prevent or avoid these adverse events in the future.
The objectives of this study were to describe the type and frequency of adverse events associated with anti-TNF-α therapy in a large cohort and evaluate for any serologic and genetic associations. The significance of realizing these objectives is that it can identify subjects at high risk for developing adverse events and can help clinicians anticipate and therefore prevent or avoid these adverse events in the future.
This study was a retrospective review, and detailed clinical information was collected via manual chart review. χ2 test and logistic regression were performed to identify demographic and clinical characteristics associated with development of adverse events.
The serological data was measured by ELISA assay at Cedars-Sinai, which was performed in a blinded fashion, and analyzed with Wilcox test signed rank test.
DNA samples were genotyped at Cedars-Sinai using Illumina Infinium Immunochipv1 array per manufacturer’s protocol (Illumina, San Diego, CA, United States). Average genotyping call rate for samples that passed quality control was 99.8%; average replicate concordance and average heritability rates were > 99.99% and 99.94%, respectively. Single-nucleotide polymorphisms (SNPs) underwent methodological review and were evaluated using several SNP statistic parameters to ensure optimal allele-calling. SNPs association with adverse events was evaluated using PLINK, with two-sided p-value of 0.05 was considered statistically significant.
For any reaction, infusion reactions, and allergic reactions, the logistic regression/human annotation and network analysis was performed with statistically significant SNPs (P < 0.001). These SNPs were first annotated into corresponding genes, and the genes were further analyzed with multiple biological functional databases. The final networks were then constructed form the known interactions from any of these databases.
The research methods described above are standard for a retrospective review analyzing genetic and serologic data.
About 1 in 5 patients were found to have adverse events to an anti-TNF-α agent that required the therapy to be discontinued. All adverse events resolved after discontinuing the anti-TNF agent. The majority of patients developed infusion reactions. In CD patients we observed that IgA ASCA +/- was associated with a lower risk of developing any adverse event. IgA ASCA and IgG ASCA levels were also lower in patients with any adverse events. Anti-I2 level in UC was significantly associated with infusion reactions. The authors identified a number of genetic associations with known IBD loci including HLA-DRB1, ERAP2, ZNF365. Their pathway analyses strongly implicated JAK-STAT signaling pathway, Cytokine-cytokine receptor interaction pathway, Measles signaling pathway, Toxoplasmosis signaling pathway, and the IBD signaling pathway. The network analyses for allergic reactions showed a number of key nodes including TYK2, BLK and IL13, which have previously been shown to be associated with allergic susceptibility.
They have demonstrated some novel genetic associations and pathways that are enriched for genes associated with development adverse events. Future studies should be performed to confirm our results, and incorporate other ethnic groups besides European ancestry, and include data on anti-drug antibody formation.
The genetic and serologic associations found in concordance with adverse events to anti-TNF therapy. This is the first study to evaluate and describe these associations. There are potentially genetic and serologic associations with adverse events to anti-TNF therapy that can help clinicians anticipate and therefore prevent or avoid these adverse events in the future.
Current studies describe in great detail the efficacy of anti-TNF therapy and the ability to predict response to therapy. However, current studies are lacking in evaluating the ability to predict the development of adverse events. The results from this study reveal that there indeed are genetic and serologic associations with anti-TNF therapy that can potentially be targeted to prevent or avoid these adverse events in the future. The new hypothesis proposed by this study is that there serologic and genetic associations with anti-TNF therapy.
The methods used in this study were similar to other retrospective studies analyzing genetic and serologic data. Manual chart review was performed to generate detailed clinical information; ELISA assay was performed to gather serologic data; and genetic data was generated using Illumina Infinium Immunochipv1 array. χ2 test and logistic regression were performed to identify demographic and clinical characteristics associated with development of adverse events; serological data were analyzed with Wilcox test signed rank test; and SNPs association with adverse events were evaluated using PLINK, with two-sided P-value of 0.05 considered statistically significant.
The genetic and serologic associations found in concordance with adverse events to anti-TNF therapy are novel and have not been described elsewhere. This study confirmed that there are genetic and serologic associations with adverse events from anti-TNF therapy. Identifying the potential factors associated with adverse events from anti-TNF therapy can help clinicians anticipate and therefore prevent or avoid these adverse events in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Gassler N, Sebastian S, Sergi CM, Zhulina Y
S- Editor: Gong ZM L- Editor: A E- Editor: Ma YJ
| 1. | Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1030] [Cited by in RCA: 1055] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 2. | Neurath MF, Fuss I, Pasparakis M, Alexopoulou L, Haralambous S, Meyer zum Büschenfelde KH, Strober W, Kollias G. Predominant pathogenic role of tumor necrosis factor in experimental colitis in mice. Eur J Immunol. 1997;27:1743-1750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 309] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 3. | Targan SR, Hanauer SB, van Deventer SJ, Mayer L, Present DH, Braakman T, DeWoody KL, Schaible TF, Rutgeerts PJ. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn’s disease. Crohn’s Disease cA2 Study Group. N Engl J Med. 1997;337:1029-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2328] [Cited by in RCA: 2268] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 4. | Dignass A, Lindsay JO, Sturm A, Windsor A, Colombel JF, Allez M, D’Haens G, D’Hoore A, Mantzaris G, Novacek G. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 2: current management. J Crohns Colitis. 2012;6:991-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 702] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 5. | Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, Travers S, Rachmilewitz D, Hanauer SB, Lichtenstein GR. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2005;353:2462-2476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2744] [Cited by in RCA: 2884] [Article Influence: 144.2] [Reference Citation Analysis (2)] |
| 6. | Ordás I, Feagan BG, Sandborn WJ. Therapeutic drug monitoring of tumor necrosis factor antagonists in inflammatory bowel disease. Clin Gastroenterol Hepatol. 2012;10:1079-1087; quiz e85-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 7. | Siegel CA, Melmed GY. Predicting response to Anti-TNF Agents for the treatment of crohn’s disease. Therap Adv Gastroenterol. 2009;2:245-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Yoon SM, Haritunians T, Chhina S, Liu Z, Yang S, Landers C, Li D, Ye BD, Shih D, Vasiliauskas EA. Colonic Phenotypes Are Associated with Poorer Response to Anti-TNF Therapies in Patients with IBD. Inflamm Bowel Dis. 2017;23:1382-1393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 9. | Colombel JF, Loftus EV Jr, Tremaine WJ, Egan LJ, Harmsen WS, Schleck CD, Zinsmeister AR, Sandborn WJ. The safety profile of infliximab in patients with Crohn’s disease: the Mayo clinic experience in 500 patients. Gastroenterology. 2004;126:19-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 621] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 10. | Antoni C, Braun J. Side effects of anti-TNF therapy: current knowledge. Clin Exp Rheumatol. 2002;20:S152-S157. [PubMed] |
| 11. | Scheinfeld N. Adalimumab: a review of side effects. Expert Opin Drug Saf. 2005;4:637-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 109] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 12. | Siegel CA, Hur C, Korzenik JR, Gazelle GS, Sands BE. Risks and benefits of infliximab for the treatment of Crohn’s disease. Clin Gastroenterol Hepatol. 2006;4:1017-1024; quiz 976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 13. | Baert F, Noman M, Vermeire S, Van Assche G, D’ Haens G, Carbonez A, Rutgeerts P. Influence of immunogenicity on the long-term efficacy of infliximab in Crohn’s disease. N Engl J Med. 2003;348:601-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1523] [Cited by in RCA: 1519] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 14. | Remicade (infliximab) for IV injection. Package insert. Malvern, PA: Centocor Inv 2002; . |
| 15. | Kugathasan S, Levy MB, Saeian K, Vasilopoulos S, Kim JP, Prajapati D, Emmons J, Martinez A, Kelly KJ, Binion DG. Infliximab retreatment in adults and children with Crohn’s disease: risk factors for the development of delayed severe systemic reaction. Am J Gastroenterol. 2002;97:1408-1414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 99] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Bégaud B, Evreux JC, Jouglard J, Lagier G. [Imputation of the unexpected or toxic effects of drugs. Actualization of the method used in France]. Therapie. 1985;40:111-118. [PubMed] |
| 17. | Mow WS, Vasiliauskas EA, Lin YC, Fleshner PR, Papadakis KA, Taylor KD, Landers CJ, Abreu-Martin MT, Rotter JI, Yang H. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 372] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 18. | Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8:e68095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 207] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 19. | Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24221] [Cited by in RCA: 24202] [Article Influence: 1344.6] [Reference Citation Analysis (0)] |
| 20. | Lichtenstein L, Ron Y, Kivity S, Ben-Horin S, Israeli E, Fraser GM, Dotan I, Chowers Y, Confino-Cohen R, Weiss B. Infliximab-Related Infusion Reactions: Systematic Review. J Crohns Colitis. 2015;9:806-815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 21. | Beigel F, Schnitzler F, Paul Laubender R, Pfennig S, Weidinger M, Göke B, Seiderer J, Ochsenkühn T, Brand S. Formation of antinuclear and double-strand DNA antibodies and frequency of lupus-like syndrome in anti-TNF-α antibody-treated patients with inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Roussomoustakaki M, Satsangi J, Welsh K, Louis E, Fanning G, Targan S, Landers C, Jewell DP. Genetic markers may predict disease behavior in patients with ulcerative colitis. Gastroenterology. 1997;112:1845-1853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 176] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 23. | Ahmad T, Marshall SE, Jewell D. Genetics of inflammatory bowel disease: the role of the HLA complex. World J Gastroenterol. 2006;12:3628-3635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 97] [Cited by in RCA: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ. HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999;45:395-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 166] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Hulur I, Gamazon ER, Skol AD, Xicola RM, Llor X, Onel K, Ellis NA, Kupfer SS. Enrichment of inflammatory bowel disease and colorectal cancer risk variants in colon expression quantitative trait loci. BMC Genomics. 2015;16:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 375] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 27. | Zhang Y, Park E, Kim CS, Paik JH. ZNF365 promotes stalled replication forks recovery to maintain genome stability. Cell Cycle. 2013;12:2817-2828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Haritunians T, Jones MR, McGovern DP, Shih DQ, Barrett RJ, Derkowski C, Dubinsky MC, Dutridge D, Fleshner PR, Ippoliti A. Variants in ZNF365 isoform D are associated with Crohn’s disease. Gut. 2011;60:1060-1067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Takahashi S, Yoshimura T, Ohkura T, Fujisawa M, Fushimi S, Ito T, Itakura J, Hiraoka S, Okada H, Yamamoto K. A Novel Role of Spred2 in the Colonic Epithelial Cell Homeostasis and Inflammation. Sci Rep. 2016;6:37531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, Howdle P, Holmes G, Mulder CJ, Dijkstra G. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57:463-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 127] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 31. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2019] [Cited by in RCA: 1882] [Article Influence: 134.4] [Reference Citation Analysis (2)] |
| 32. | Seto Y, Nakajima H, Suto A, Shimoda K, Saito Y, Nakayama KI, Iwamoto I. Enhanced Th2 cell-mediated allergic inflammation in Tyk2-deficient mice. J Immunol. 2003;170:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Ashley SE, Tan HT, Peters R, Allen KJ, Vuillermin P, Dharmage SC, Tang MLK, Koplin J, Lowe A, Ponsonby AL. Genetic variation at the Th2 immune gene IL13 is associated with IgE-mediated paediatric food allergy. Clin Exp Allergy. 2017;47:1032-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 34. | Liu Y, Ke X, Kang HY, Wang XQ, Shen Y, Hong SL. Genetic risk of TNFSF4 and FAM167A-BLK polymorphisms in children with asthma and allergic rhinitis in a Han Chinese population. J Asthma. 2016;53:567-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 35. | Kuna AT. Serological markers of inflammatory bowel disease. Biochem Med (Zagreb). 2013;23:28-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 36. | Cohen RD, Lewis JR, Turner H, Harrell LE, Hanauer SB, Rubin DT. Predictors of adalimumab dose escalation in patients with Crohn’s disease at a tertiary referral center. Inflamm Bowel Dis. 2012;18:10-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 37. | Yang SK, Hong M, Baek J, Choi H, Zhao W, Jung Y, Haritunians T, Ye BD, Kim KJ, Park SH. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet. 2014;46:1017-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 421] [Article Influence: 38.3] [Reference Citation Analysis (0)] |