Published online Sep 28, 2017. doi: 10.3748/wjg.v23.i36.6665
Peer-review started: June 12, 2017
First decision: July 27, 2017
Revised: August 27, 2017
Accepted: September 5, 2017
Article in press: September 5, 2017
Published online: September 28, 2017
Processing time: 106 Days and 13.2 Hours
To investigate the intestinal luminal microbiota (LM) and mucosa-associated microbiota (MAM) in Chinese patients with functional gastrointestinal disorders (FGIDs) and examine the association between these communities and the expression of toll-like receptor (TLR) 2 and TLR4.
Thirty-two Chinese subjects who suffered from symptoms of FGIDs, as confirmed by gastroenterologists, were enrolled in this study. Fresh faecal samples and descending colonic mucosal biopsies were collected from the subjects before (faecal) and during (mucosal) flexible colonoscopy. For analysis of the samples, we performed high-throughput sequencing of the V3-V4 region of the 16S rRNA gene and reverse transcription (RT)-PCR to detect the expression of colonic TLR2 and TLR4. Differences in the stool and mucosal microbiota were examined and a correlation network analysis was performed.
The microbiota of faecal samples was significantly more diverse and richer than that of the mucosal samples, and the LM and MAM populations differed significantly. TLR2 expression showed a significant positive correlation with TLR4 expression. In the MAM samples, the genera Faecalibacterium and Ruminococcus, which belong to the family Ruminococcaceae, were inversely correlated with TLR4 expression (r = -0.45817, P = 0.0083 and r = -0.5306, P = 0.0018, respectively). Granulicatella, which belongs to Carnobacteriaceae, and Streptococcus, which belongs to Streptococcaceae, were inversely correlated with TLR2 expression (r = -0.5573, P = 0.0010 and r = -0.5435, P = 0.0013, respectively). In the LM samples, examination at phylum, class, or order level revealed no correlation with TLR4 expression. Faecalibacterium, which belongs to Ruminococcaceae, and Streptococcus, which belongs to Streptococcaceae, were inversely correlated with TLR2 expression (r = -0.5743, P = 0.0058 and r = -0.3905, P = 0.0271, respectively).
Microbial compositions of LM and MAM in Chinese patients with FGIDs are different. Expression of TLRs may be affected by the type of bacteria that are present in the gut.
Core tip: To explore which bacteria regulate the expression of toll-like receptors (TLRs) and thereby affect intestinal functions, we performed high-throughput pyrosequencing of the bacterial 16S rRNA gene, compared the microbial communities in the faeces and mucosa of Chinese patients with functional gastrointestinal disorders, and studied their association with the expression of colonic mucosal TLR2 and TLR4. Samples of luminal microbiota were different from those of mucosa-associated microbiota (MAM), and MAM samples were closely associated with TLR2/4 expression. The abundance of Faecallibacterium and Ruminococcus was lower in patients with gut disease, while the expression of TLRs is higher than in healthy controls. The presence of Faecallibacterium and Ruminococcus was significantly and negatively correlated with TLR4 expression, suggesting that these two bacteria, which colonize on the colonic mucosa, play a key role in gut diseases by regulating mucosal TLR4 expression. Granulicatella, which belongs to Carnobacteriaceae, and Streptococcus, which belongs to Streptococcaceae, were inversely correlated with TLR2 expression. Because the two genera contained not only pathogenic species but also probiotic species, it will be important to establish a better understanding of the relationship between TLRs and bacterial strains in future studies.
- Citation: Dong LN, Wang JP, Liu P, Yang YF, Feng J, Han Y. Faecal and mucosal microbiota in patients with functional gastrointestinal disorders: Correlation with toll-like receptor 2/toll-like receptor 4 expression. World J Gastroenterol 2017; 23(36): 6665-6673
- URL: https://www.wjgnet.com/1007-9327/full/v23/i36/6665.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i36.6665
The intestinal microbiota is a complex community of bacteria, archaea, viruses, and eukarya. A wide variety of bacterial species in the gastrointestinal tract exerts numerous effects on the host and influences a variety of gastrointestinal functions[1]. Faecal samples (representing the luminal niche) are examined in most studies of the intestinal microbiota because their collection is simple. However, recent studies have shown that the microbial compositions of the luminal microbiota (LM) and the mucosa-associated microbiota (MAM) differ, suggesting that these two distinct microbial populations play different roles within the intestinal microbiota ecosystem[2]. LM is present in the whole intestine, whereas MAM represents a special niche. Because MAM is in close contact with the host, it may play a more prominent role in the intestine, whereas LM may play a key role in metabolic activities and nutrient harvest[3]. Toll-like receptors (TLRs) are pattern recognition receptors expressed by various cells in the gastrointestinal tract. TLRs detect conserved microbial products and play a central role in the activation of innate and adaptive immune pathways. TLR2 and TLR4 are two of the best characterized TLRs that respond to microbial membrane components. A number of studies have examined the role of TLR signalling in microbiota-induced chronic inflammation and immunopathology[4]. The microbiota may directly interact with the TLRs and regulate gut immune responses[5].
In some intestinal diseases, such as inflammatory bowel disease (IBD) or irritable bowel syndrome (IBS), the expression of TLR2 and TLR4 is increased and the dysbiosis is observed. However, limited data are available on the correlation between the intestinal microbiota and TLRs in humans. To explore which bacteria regulate the expression of TLRs and thereby affect intestinal functions, we performed high-throughput pyrosequencing of the bacterial 16S rRNA gene, compared the microbial communities in the faeces and mucosa of Chinese patients with functional gastrointestinal disorders (FGIDs), and studied their association with the expression of colonic mucosal TLR2 and TLR4.
Thirty-two Chinese subjects aged between 18 and 65 years, who were diagnosed with FGIDs, were recruited at the Department of Gastroenterology, Shanxi Provincial People’s Hospital from 2013 to 2014. None of the subjects enrolled in the study had taken corticosteroids, opioids, probiotic and prebiotic supplements or antibiotics in the 6 mo preceding the study, no one had systemic comorbidity, and no one had a history of excessive alcohol intake (> 20 alcoholic drinks per week). Patients with a prior history of gastrointestinal surgery or intestinal organic disease were excluded. All subjects provided signed informed consent before participation. The study was performed in accordance with the principles of the Declaration of Helsinki and the study protocol was approved by the Ethics Committee of Shanxi Provincial People’s Hospital, China.
Faecal samples were collected at home before bowel preparation, frozen immediately at -20 °C, and transported within 6 h to the study centre, where they were stored at -80 °C until analysis. Bacterial DNA was extracted from the faecal samples with the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. The DNA concentrations were quantified with an Eppendorf BioSpectrometer (Eppendorf, Hamburg, Germany).
Colonic mucosal biopsies were collected from each subject during an unsedated flexible colonoscopy. Colonic mucosal biopsies were taken from the descending colon. After removal from the colon, the biopsies were immediately frozen at -20 °C and transported within 6 h to the study centre, where they were stored at -80 °C until analysis. Total genomic DNA was extracted from the colon samples with the QIAamp DNA Mini Kit (Qiagen), according to the manufacturer’s instructions. The concentration and purity of the genomic DNA were measured with an Eppendorf BioSpectrometer.
The bacterial genomic DNA was used as the template for amplification of the V3-V4 hypervariable region of the 16S rRNA gene with the forward primer 5′-GACTACHVGGGTATCTAATCC-3′ and the reverse primer 5′-CCTACGGGNGGCWGCAG-3′.
Pairs of reads from the original DNA fragments were merged using FLASH, which was designed in case the original DNA fragments were shorter than twice the read length. The sequencing reads of each sample were given a unique barcode and analysed with the Quantitative Insights Into Microbial Ecology (QIIME) software and the UPARSE pipeline. In brief, the reads were filtered with the QIIME quality filters using the default settings for Illumina processing and the operational taxonomic units (OTUs) were selected using the UPARSE pipeline. The samples were sequenced on an Illumina MiSeq Benchtop Sequencer and the bioinformatic analysis was performed by Genesky Biotechnologies Inc. (Shanghai, China).
The size of the bacterial groups is expressed as percentage of the total bacteria.
Total mucosal RNA was extracted from the colonic biopsies using the TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan), according to the manufacturer’s instructions. After reverse transcription with PrimeScript Reverse Transcriptase Mix (TaKaRa), the expression of TLR2 and TLR4 genes was determined by quantitative real-time polymerase chain reaction (qPCR) and SYBR Green technology on a Bio-Rad CFX96 Q-PCR instrument (Bio-Rad, United States) in duplicate. The specific primers for TLR2 were: 5’-TGATGCTGCCATTCTCATTC-3’ (forward) and 5’-CGCAGCTCTCAGATTTACCC-3’ (reverse); and for TLR4: 5’-CAGGGCTTTTCTGAGTCGTC-3’ (forward) and 5’-TGAGCAGTCGTGCTGGTATC-3’ (reverse). Each amplification reaction was run in duplicate in a final volume of 20 μL. All qPCR amplifications were optimized and performed in 0.2 mL 96-well plates with the following cycling program: initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Negative controls lacking the template DNA were included in triplicate. The relative amount of each mRNA was normalized to GAPDH (forward: 5’-CCATCAATGACCCCTTCATTG-3’, reverse: 5’-CTTGACGGTGCCATGGAATT-3’) and data were analysed using the 2-ΔΔCT method.
All statistical analyses were performed with SPSS 22.0 for Windows (SPSS Inc., United States). To determine the statistical differences between the two groups, we used the independent t test and the Mann-Whitney test. Correlations were determined with Spearman’s correlation test. Heat maps were generated by R-packages for TLR2 and TLR4 as environmental factors, to reveal the relationship between the diversity and environmental factors based on the Pearson’s correlation analyses.
We investigated 64 samples from 32 subjects with FGIDs [14 with diarrhoea-predominant IBS, 14 with functional dyspepsia (FD), and 4 with other diseases]. All subjects provided both a faecal sample and a colonic mucosal sample. The study population consisted of 50% females, and had a mean age of 49 years (range, 20-65 years) and a mean body mass index of 23.24 kg/m2 (range, 20-26 kg/m2).
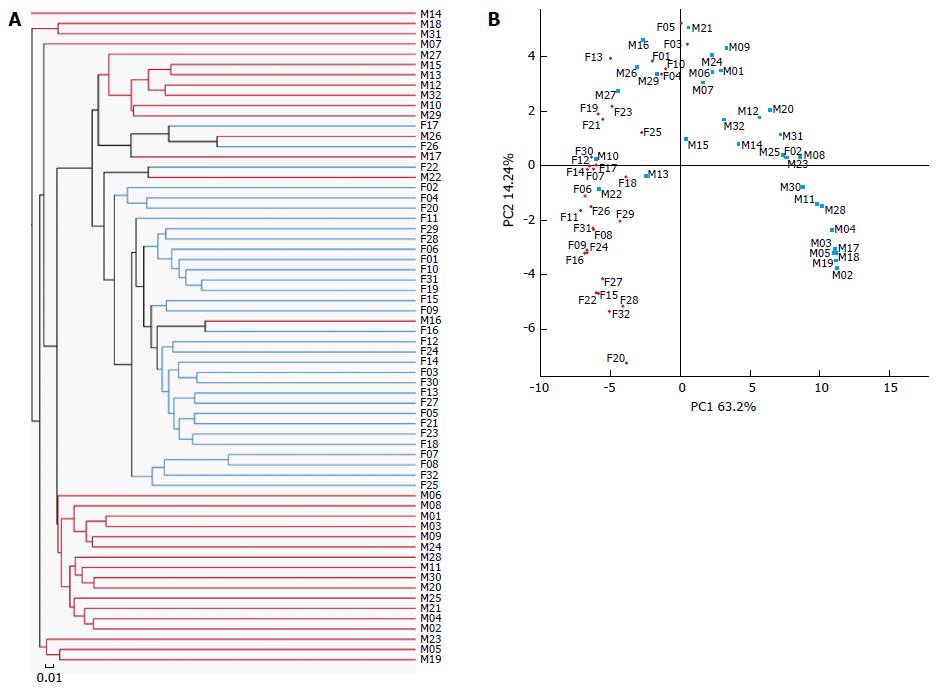
We obtained a total of 4351929 raw reads and 3545053 reads remained after filtering. The sequencing analysis of the 64 samples identified 1026 OTUs. The rarefaction curves had a tendency to approach a saturation plateau, indicating that the number of samples in this study was sufficient. The same tendency was found in the Shannon-Wiener curves, indicating that the number of 16S rRNA gene sequences in the database was very abundant and reflected the vast majority of the microbiota. The gut faecal samples showed significantly more diversity and richness of the microbiota than the mucosal samples (Table 1) and samples of LM and MAM were significantly different (Figure 1A and B).
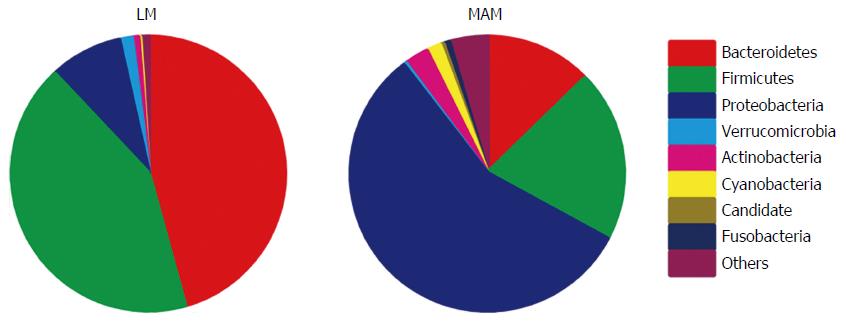
Significant differences between samples of LM and MAM were identified for almost all populated phyla. Bacteroidetes (44.7%) and Firmicutes (42.2%) were the most represented phyla in LM samples, followed by Proteobacteria (8.5%), whereas Proteobacteria (56.6%) was the most represented phylum in MAM samples, followed by Firmicutes (20.2%) and Bacteroidetes (12.7%) (P < 0.05; Figure 2).
Examination at the genus level showed that the relative abundance of Escherichia-Shigella, Streptococcus, Clostridium sensu stricto1, Sphingomonas, Acinetobacter, Brevundimonas, and Enhydrobacter was significantly greater in samples of MAM than in LM samples, whereas abundance of Bacteroides, Faecalibacterium, Incertae sedis, Subdoligranulum, Pseudobutyrivibrio, Megasphaera, Parasutterella, Akkermansia, Alistipes, and Lachnospira was significantly lower in samples of MAM than in LM samples (P < 0.05).
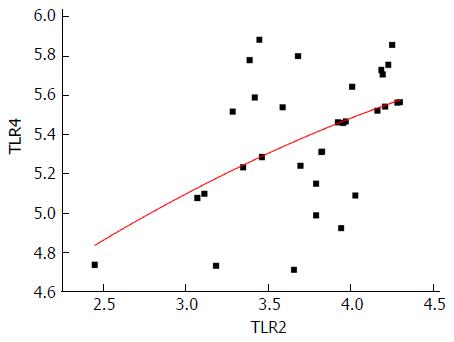
We measured the mRNA expression of TLR2 (3.7394 ± 0.43514) and TLR4 (5.3866 ± 0.33556) in all samples and expression of TLR2 was significantly and positively correlated with that of TLR4 (r = 0.492, P = 0.004) (Figure 3).
Regardless of phylum, class or order level, no bacterium was correlated with expression of TLR4 in LM samples.
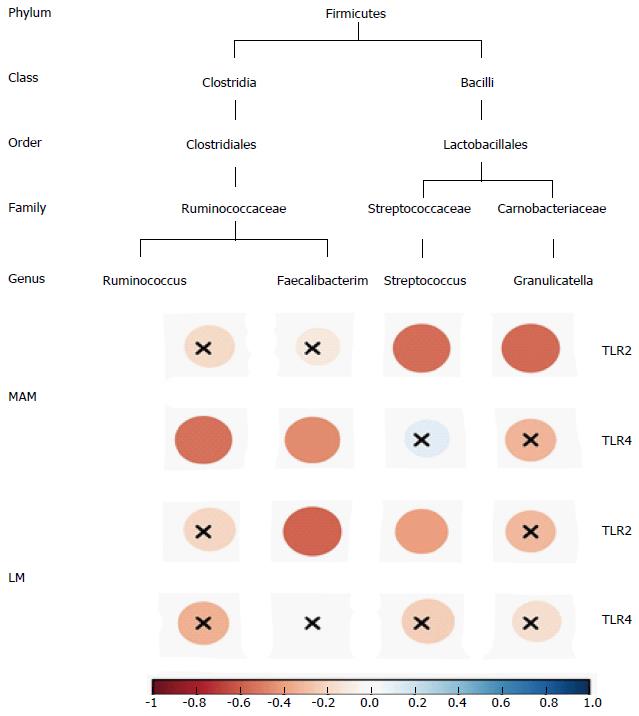
At the phylum level, Firmicutes was inversely and significantly correlated with TLR4 expression in MAM samples (r = -0.4676, P = 0.0070). At the class level, Clostridia, which belongs to Firmicutes, was inversely and significantly correlated with TLR4 expression in MAM samples (r = -0.3913, P = 0.0268). At the order level among the class Clostridia, the order Clostridia was inversely and significantly correlated with TLR4 expression (r = -0.3906, P = 0.0271), as well as the families Defluviitaleaceae (r = -0.4227, P = 0.0159), Peptostreptococcaceae (r = -0.3611, P = 0.0422), and Ruminococcaceae (r = -0.4740, P = 0.0061), which belong to the order Clostridia. The genera Faecalibacterium and Ruminococcus, which belong to the family Ruminococcaceae, were also inversely and significantly correlated with TLR4 expression (r = -0.45817, P = 0.0083 and r = -0.5306, P = 0.0018, respectively) (Figure 4).
The presence of bacteria in both LM and MAM samples was correlated with TLR2 expression. However, the taxa of microbiota was very different in these two sample populations.
In LM samples, Clostridia and Bacilli at the class level were inversely and significantly correlated with TLR2 expression (r = -0.4238, P = 0.0156 and r = -0.4482, P = 0.0101, respectively), as well as Clostridiales at the order level, which belongs to Clostridia, and Lactobacillales, which belongs to Bacilli (r = -0.4482, P = 0.0156 and r = -0.4207, P = 0.0165, respectively). At the family level, Ruminococcaceae, which belongs to Clostridiales, and Streptococcaceae, which belongs to Lactobacillales, were inversely and significantly correlated with TLR2 expression (r = -0.4437, P = 0.0110 and r = -0.3839, P = 0.0300, respectively). And at the genus level, Faecalibacterium, which belongs to Ruminococcaceae, and Streptococcus, which belongs to Streptococcaceae, were inversely and significantly correlated with TLR2 expression (r = -0.5743, P = 0.0058 and r = -0.3905, P = 0.0271, respectively) (Figure 4).
In MAM samples, Bacilli at the class level was inversely and significantly correlated with TLR2 expression (r = -0.4676, P = 0.0070), as well as Lactobacillales at the order level, which belongs to Bacilli (r = -0.4574, P = 0.0085). At the family level, Carnobacteriaceae and Streptococcaceae, which belong to Lactobacillales, were inversely and significantly correlated with TLR2 expression (r = -0.5554, P = 0.0010 and r = -0.5445, P = 0.0013, respectively). And at the genus level, Granulicatella, which belongs to Carnobacteriaceae, and Streptococcus, which belongs to Streptococcaceae, were correlated with TLR2 expression (r = -0.5573, P = 0.0010 and r = -0.5435, P = 0.0013, respectively) (Figure 4).
Deep sequencing showed that more than 95% of the sequences in all stool and mucosal samples belonged to the three most popular bacterial phyla, consisting of Firmicutes, Bacteroidetes, and Proteobacteria. This is consistent with the findings of previous studies[6], which showed that these phyla account for the majority of the gut microbiota in both stool and mucosal samples. The faecal samples displayed significantly greater bacterial diversity and richness than the mucosal samples, as in the study of Ringel et al[2]. Comparing the proportions of the dominant bacterial taxa in the faecal and mucosal samples revealed significant differences. In this study, Proteobacteria was the predominant phylum in MAM samples. This is different from other reports and might reflect geographical differences[7]. It is well known that the Chinese diet and genetics are very different from those in Western countries and these factors markedly influence the gut microbiota. Furthermore, MAM samples were taken from a distinct location in the colon, whereas LM samples displayed the whole intestinal microbiota. Therefore, MAM samples may reflect a stronger relationship with the host.
A correlation analysis was performed between the two bacterial populations at the phylum, class, order, family, and genus levels and the expression of TLR2 and TLR4. The results showed that although the Proteobacteria was the predominant phylum in MAM samples, the bacteria that correlated strongest with expression of TLRs were part of the phylum Firmicutes. The taxa that showed a significant correlation was selected and studied from the phylum to genus level. Finally, Faecalibacterium, Ruminococccaceae, Streptococcus and Granulicatella genera were the bacteria that correlated strongest with TLRs expression. Although a positive correlation existed between the expression of TLR2 and TLR4, the types of bacteria associated with TLR4 expression were very different from those related to the expression of TLR2. However, this was in accordance with their distinct microbial compositions. The bacteria that correlated with TLR4 expression were only found in MAM samples. Bacteria that correlated with TLR2 expression originated from both LM and MAM. Both TLR4 and TLR2 expression were negatively correlated with MAM, however, TLR2 expression was mainly associated with bacteria belonging to the Bacilli class, whereas TLR4 expression was mainly associated with bacteria belonging to the Clostridia class.
The enteric commensal bacteria of the genus Faecallibacterium, which belongs to the group Clostridium, exert an anti-inflammatory effect[8]. In the present study, Faecallibacterium from LM samples was negatively correlated with the expression of TLR2 and Faecallibacterium from MAM samples was negatively correlated with the expression of TLR4. Faecallibacterium richness was reduced in patients with IBS and IBD[9,10]. TLR signalling, activated by pathogens, is involved in the pathogenesis of several infectious and inflammatory diseases. Imbalanced relationships among the environment, genetics, and host immunity may promote aberrant TLR signalling, thereby contributing on a large scale to acute and chronic intestinal inflammatory processes, such as IBD, colitis, and colorectal cancer[11]. In our previous study, the expression of TLR2 and TLR4 was increased in diarrhoea-predominant IBS patients and decrease of Clostridia was strongly associated with higher TLR2 and TLR4 expression[12]. In an animal study, Faecallibacterium prausnitzii and its supernatant (SN) had beneficial effects on intestinal epithelial barrier impairment in a chronic low-grade inflammation model[13]. Round et al[14] showed that unlike pathogens whose TLR ligands trigger inflammation, some commensal bacteria exploit the TLR pathway to actively suppress immune reactions. The anti-inflammatory effect of Faecallibacterium may be involved in the upregulation of the TLRs expression. Ruminococcus is another common bacterium that is decreased in patients with IBD or IBS. In addition, a reduction in the relative abundance of potentially immunomodulatory gut bacteria including Ruminococcus is associated with exaggerated inflammatory cytokine responses to TLR ligands and subsequent development of IgE-associated eczema[15]. In the present study, the decrease of Faecallibacterium and Ruminococcus in MAM samples was strongly associated with higher TLR4 expression, suggesting that the decrease of mucosal Faecallibacterium and Ruminococcus induced an increase in expression of TLR4 mRNA, which plays an important role in the causation of immune related gut diseases including IBD and IBS.
Streptococcus is a genus of (spherical) coccus Gram-positive bacteria belonging to the phylum Firmicutes phylum and the order Lactobacillales. Some streptococcal species, e.g., S. pneumoniae, are pathogenic. Tomlinson et al[16] suggested that leukocyte responses to bacterial lipoproteins are required for TLR2- and IL-1R-associated kinase-4-mediated inflammatory responses to S. pneumoniae. However, many other streptococcal species, e.g., S. thermophilus, are not pathogenic[17] and are part of the commensal human microbiota of the mouth, skin, intestine, and upper respiratory tract. S. thermophilus is considered a probiotic and is a key strain of VSL#3, which is considered a therapeutic immunomodulatory tool[18]. Streptococcaceae was negatively correlated with TLR2 expression in samples of both MAM and LM in our study. Although there is no direct evidence of inhibition of TLRs by Streptococcaceae, some beneficial streptococcal species may affect disease processes via the TLR2 pathway. Future research will explore which species belonging to Streptococcaceae can influence the expression of TLR2. Granulicatella is part of the normal body flora and is often difficult to culture in traditional media. A few reports mention that it can induce an infection similar to Streptococci[19].
The composition of LM samples was different than that of MAM; MAM samples showed a stronger correlation with the expression of TLR2 and TLR4. The abundance of Faecallibacterium and Ruminococcus was lower in IBD and IBS, while TLRs expression was higher in patients than in healthy controls. Faecallibacterium and Ruminococcus were significantly and negatively correlated with TLR4 expression, suggesting that these two bacteria colonizing on the colonic mucosa play a key role in the pathogenesis of gut diseases by regulating the expression of mucosal TLR4. Granulicatella, which belongs to Carnobacteriaceae, and Streptococcus, which belongs to Streptococcaceae, were inversely correlated with TLR2 expression.
One limitation of our study was the small biopsy from the colonic endoscopy and multiple sampling might cause bleeding, therefore we were not able to measure the expression of proteins. The other limitation of our study was the scattered distribution of the correlation between species levels, which might be explained by the restriction of pyrosequencing of species levels, therefore we did not report the results. It will be important to establish a better understanding of the relationship between TLRs and bacteria strains in future studies. Probiotics play an important role in the treatment and prevention of diseases. Further studies will have to verify the relationship between TLRs and bacteria[20,21].
Toll-like receptors (TLRs) detect conserved microbial products and play a central role in the activation of innate and adaptive immune pathways. TLR2 and TLR4 are two of the best characterized TLRs that respond to microbial membrane components. Faecal samples (representing the luminal niche) were used in most studies on intestinal microbiota because their collection is easy. However, more recent studies revealed distinct microbial composition between luminal microbiota (LM) and mucosa-associated microbiota (MAM). To explore which bacteria may regulate the expression of TLRs and thereby affect intestinal functions, we performed high-throughput pyrosequencing of the bacterial 16S rRNA gene, compared the microbial communities in the faeces and mucosa of Chinese patients with functional gastrointestinal disorders, and studied their association with the expression of colonic mucosal TLR2 and TLR4. LM differed from MAM; MAM showed a stronger association with the expression of TLR2 and TLR4.
Recent advances in the understanding of intestinal immunology suggest that functional gastrointestinal disorders (FGIDs) may not all be ‘functional’, as considered for decades. This view was largely developed by the absence of active inflammation. Faecal microbiota and MAM play a vital role in gut immunity. It will be important to establish a better understanding of the relationship between TLRs and bacteria strains in FGIDs.
In some intestinal diseases, such as inflammatory bowel disease or irritable bowel syndrome, the expression of TLR2 and TLR4 is increased and dysbiosis is observed. However, limited data are available on the correlation between the intestinal microbiota and TLRs in humans. In this study, the abundance of Faecallibacterium and Ruminococcus was lower in many gut diseases, while the TLRs expression was higher than in healthy controls. Faecallibacterium and Ruminococcus were significantly and negatively correlated with TLR4 expression, suggesting that these two bacteria colonizing on the colonic mucosa play a key role in gut diseases by regulating the expression of mucosal TLR4. Granulicatella, which belongs to Carnobacteriaceae, and Streptococcus, which belongs to Streptococcaceae, were inversely correlated with TLR2.
Probiotics play an important role in the treatment and prevention of diseases. Further studies will have to verify the relationship between inflammatory proteins and bacteria. The bacteria may be used as probiotics in the treatment of diseases by regulating immunity.
According to the literature, they described the microbiota from faecal samples as LM.
This is an interesting paper that investigated the intestinal LM and the MAM in Chinese patients with FGIDs and examined the association between these communities and the expression of TLR2 and TLR4. The manuscript is well written.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Biondi A, Kucherlapati MH, Teramoto-Matsubara OT S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Zhang FF
| 1. | Kataoka K. The intestinal microbiota and its role in human health and disease. J Med Invest. 2016;63:27-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 120] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 2. | Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes. 2015;6:173-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 156] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 3. | Sundin J, Rangel I, Fuentes S, Heikamp-de Jong I, Hultgren-Hörnquist E, de Vos WM, Brummer RJ. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther. 2015;41:342-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 4. | Kramer CD, Genco CA. Microbiota, Immune Subversion, and Chronic Inflammation. Front Immunol. 2017;8:255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 5. | de Kivit S, Tobin MC, Forsyth CB, Keshavarzian A, Landay AL. Regulation of Intestinal Immune Responses through TLR Activation: Implications for Pro- and Prebiotics. Front Immunol. 2014;5:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 6. | Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474:1823-1836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1710] [Cited by in RCA: 2052] [Article Influence: 256.5] [Reference Citation Analysis (7)] |
| 7. | Escobar JS, Klotz B, Valdes BE, Agudelo GM. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014;14:311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 162] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 8. | Hornef MW, Pabst O. Real friends: Faecalibacterium prausnitzii supports mucosal immune homeostasis. Gut. 2016;65:365-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 9. | Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1036] [Cited by in RCA: 1325] [Article Influence: 120.5] [Reference Citation Analysis (3)] |
| 10. | Lopez-Siles M, Martinez-Medina M, Abellà C, Busquets D, Sabat-Mir M, Duncan SH, Aldeguer X, Flint HJ, Garcia-Gil LJ. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol. 2015;81:7582-7592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 11. | Frosali S, Pagliari D, Gambassi G, Landolfi R, Pandolfi F, Cianci R. How the Intricate Interaction among Toll-Like Receptors, Microbiota, and Intestinal Immunity Can Influence Gastrointestinal Pathology. J Immunol Res. 2015;2015:489821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 174] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 12. | Guo WT, Liu P, Dong LN, Wang JP. [The correlation study between the changes of intestinal mucosa predominant bacteria and Toll-like receptor 2, Toll-like receptor 4 gene expressions in diarrhea-predominant irritable bowel syndrome patients]. Zhonghua Neike Zazhi. 2016;55:541-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 13. | Martín R, Miquel S, Chain F, Natividad JM, Jury J, Lu J, Sokol H, Theodorou V, Bercik P, Verdu EF. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 2015;15:67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 197] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 14. | Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974-977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1311] [Cited by in RCA: 1209] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 15. | West CE, Rydén P, Lundin D, Engstrand L, Tulic MK, Prescott SL. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy. 2015;45:1419-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 16. | Tomlinson G, Chimalapati S, Pollard T, Lapp T, Cohen J, Camberlein E, Stafford S, Periselneris J, Aldridge C, Vollmer W. TLR-mediated inflammatory responses to Streptococcus pneumoniae are highly dependent on surface expression of bacterial lipoproteins. J Immunol. 2014;193:3736-3745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 17. | Marcial G, Villena J, Faller G, Hensel A, de Valdéz GF. Exopolysaccharide-producing Streptococcus thermophilus CRL1190 reduces the inflammatory response caused by Helicobacter pylori. Benef Microbes. 2017;8:451-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Ekmekciu I, von Klitzing E, Fiebiger U, Neumann C, Bacher P, Scheffold A, Bereswill S, Heimesaat MM. The Probiotic Compound VSL#3 Modulates Mucosal, Peripheral, and Systemic Immunity Following Murine Broad-Spectrum Antibiotic Treatment. Front Cell Infect Microbiol. 2017;7:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 19. | York J, Fisahn C, Chapman J. Vertebral Osteomyelitis Due to Granulicatella Adiacens, a Nutritionally Variant Streptococci. Cureus. 2016;8:e808. [PubMed] |
| 20. | Uccello M, Malaguarnera G, Basile F, D’agata V, Malaguarnera M, Bertino G, Vacante M, Drago F, Biondi A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012;12 Suppl 1:S35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 21. | Sánchez B, Delgado S, Blanco-Míguez A, Lourenço A, Gueimonde M, Margolles A. Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res. 2017;61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 647] [Article Influence: 71.9] [Reference Citation Analysis (0)] |