Published online Sep 14, 2017. doi: 10.3748/wjg.v23.i34.6330
Peer-review started: April 10, 2017
First decision: May 12, 2017
Revised: June 7, 2017
Accepted: July 12, 2017
Article in press: July 12, 2017
Published online: September 14, 2017
Processing time: 159 Days and 11.2 Hours
To determine whether circular RNAs (circRNAs) are involved in pathological processes of gastric cancer (GC).
Three circRNAs with differential expression in GC and colorectal cancer were randomly selected for validation by quantitative reverse transcription-polymerase chain reaction (qRT-PCR), using 20 pairs of gastric tissues and normal tissues. Based on the predicted circRNA-miRNA network, we then focused on hsa_circ_0000745, which was found to be down-regulated in 20 GC tissues compared with normal tissues. The hsa_circ_0000745 levels were further analyzed by qRT-PCR in 60 GC tissues and paired adjacent non-tumor tissues, as well as 60 plasma samples from GC patients and 60 plasma samples from healthy controls. The associations between the levels of hsa_circ_0000745 and the clinicopathological features of GC patients were statistically assessed. A receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value of hsa_circ_0000745 in GC.
Hsa_circ_0000745 was down-regulated in GC tissues vs non-tumorous tissues (P < 0.001) and in plasma samples from patients with GC vs healthy controls (P < 0.001). The expression level of hsa_circ_0000745 in GC tissues correlated with tumor differentiation, while the expression level in plasma correlated with tumor-node-metastasis stage. The area under the ROC curve (AUC) of hsa_circ_0000745 in plasma was 0.683, suggesting good diagnostic value. Plasma hsa_circ_0000745 level combined with carcinoembryogenic antigen (CEA) level increased the AUC to 0.775.
Hsa_circ_0000745 plays an important role in GC and its expression level in plasma in combination with CEA level is a promising diagnostic marker for this malignancy.
Core tip: This study demonstrates that expression of hsa_circ_0000745 is down-regulated in human gastric cancer (GC), with significantly lower levels detected in human GC tissues compared to paired adjacent non-tumor tissues, and in plasma of GC patients compared to plasma from normal healthy controls. Detection of this stable circular RNA in plasma may represent a clinically useful and convenient marker for GC diagnosis, with higher sensitivity and specificity when analyzed in combination with carcinoembryogenic antigen.
- Citation: Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol 2017; 23(34): 6330-6338
- URL: https://www.wjgnet.com/1007-9327/full/v23/i34/6330.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i34.6330
Gastric cancer (GC) is one of the most common malignancies worldwide, and is the most frequently diagnosed cancer among East Asian populations[1-3]. Despite extensive research attention, the mortality rate of GC remains high due to the fact that it is usually diagnosed at late stages. To improve early diagnosis and reduce the high mortality rate of GC[4-6], it is critical to continue elucidating the molecular mechanisms of GC, which may help identify markers for earlier diagnosis and novel therapeutic targets.
Endogenous noncoding RNAs, including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and the recently identified circular RNAs (circRNAs), play key regulatory roles in various cellular physiological processes and may contribute to pathological processes such as cancer formation and progression[7,8]. Unlike linear RNAs which terminate with 5' caps and 3' tails, circRNAs form covalently closed loop structures with neither 5' to 3' polarity nor a polyadenylated tail. They are not simply by-products of mis-spliced RNAs or splicing errors, but rather they are the products of regulated back-spliced RNAs with distinct sets of cis-elements and/or trans-factors[9]. Moreover, the formation of circRNAs involves the occurrence of back-splicing by the canonical spliceosome[10].
Recent research has demonstrated that circRNAs are widely involved in myriad physiological and pathological processes, including neuronal differentiation, synaptogenesis, neurological disorders, angiogenesis, prion disease, and cancer[11-16]. Likewise, circRNAs have been described as a class of biomarkers related to aging in Drosophila[17] and as putative disease biomarkers in human saliva[18]. Li et al[19] reported that hsa_circ_002059, a typical circRNA, was significantly down-regulated in GC tissues compared with paired adjacent non-tumorous tissues, suggesting its potential as a novel and stable biomarker for the diagnosis of GC.
The collective findings from the research on circRNAs indicate that they may play crucial roles in the molecular pathways of cancer. However, only a few publications have reported on the relationship between circRNAs and cancer. Whether and how circRNAs expression is dysregulated in GC remains unknown.
In this study, we randomly selected three differentially expressed circRNAs (hsa_circ_0000745, hsa_circ_0085616, and chr16:30740286-30740893|+) from two circRNA databases: CircBase (http://circbase.org/) and circ2Traits (http://gyanxet-beta.com/circdb/), which had been characterized using RNA-Seq analysis of ribosomal RNA-depleted total RNA from three GC or colorectal cancerous tissues and paired normal tissues[20,21] (Table 1). We then focused our attention on circRNA hsa_circ_0000745, which is encoded by the sperm antigen with calponin homology and coiled-coil domains 1 gene (SPECC1) that is located at chr17:20107654-20109225. We selected hsa_circ_0000745 for investigation based on preliminary broad sequencing analysis and prediction of its binding to various miRNAs with known relations to GC. By expanding the numbers of human GC and control samples for investigation, we were able to identify a statistically significant down-regulation of hsa_circ_0000745 expression in both GC tissues and plasma samples from patients with GC. Moreover, we were able to determine that the down-regulated expression correlated with tumor differentiation. Overall, the data identifies hsa_circ_0000745 as a novel potential biomarker for diagnosing GC.
| CircRNA | Chrom | TxStart | TxEnd | circBaseID | Best_transcript | Gene name | Catalog |
| Chr16: 30740287-30740893|+ | 16 | 30740286 | 30740893 | 266 | NM_006662 | SRCAP | Exonic |
| Hsa_circ_0000745 | 17 | 20107645 | 20109225 | 790 | NM_152904 | SPECC1 | Exonic |
| Hsa_circ_0085616 | 8 | 131370262 | 131374017 | 127 | NM_001247996 | ASAP1 | Exonic |
This study was approved by the Institutional Ethics Review Board of Anhui Provincial Hospital Affiliated to Anhui Medical University (China) and was conducted according to the Ethical Guidelines for Human Genome/Gene Research issued by the Chinese Government. Human samples of GC tissues and paired adjacent non-tumor tissues were prospectively collected from 60 patients at the Anhui Provincial Hospital from January 2016 to January 2017. Upon removal, the respective tissues were immediately submerged in RNAlater RNA stabilization reagent (Qiagen, Hilden, Germany) and stored at -80 °C until analysis. None of the patients had undergone chemotherapy or radiotherapy prior to the operation (resection).
The GC diagnosis was confirmed by histopathology. Tumor stages were determined according to the American Joint Committee on Cancer tumor-node-metastasis (TNM) staging criteria.
Before any treatment was applied, each study participant’s peripheral blood (2-3 mL) was collected in an ethylenediaminetetraacetic acid tube, centrifuged at 3000 × g for 10 min to obtain plasma, and stored at -80 °C. Following the age- and sex-matching criteria of the study design, fresh normal plasma samples were collected from 60 healthy volunteers.
All research subjects provided written informed consent prior to study enrollment.
Total RNA was isolated from three samples of gastric tumor and paired adjacent non-tumor (normal) tissues, as well as from plasma samples, using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. RNA integrity was assessed using standard denaturing agarose gel electrophoresis. Total RNA from each specimen was quantified and quality assurance was provided using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, United States).
Transcriptome high-throughput sequencing and subsequent bioinformatics analysis were performed by Cloud-Seq Biotech (Shanghai, China). The paired-end reads were harvested on a HiSeq 4000 System (Illumina, San Diego, CA, United States). The high-quality reads were aligned to the reference genome/transcriptome with STAR software[22], and circRNAs were detected and identified with DCC software[23]. EdgeR software[24] was used to normalize the data and perform analysis to determine differential expression among the circRNAs.
Total RNA samples were reverse-transcribed into cDNA with a random primer using SuperScript™ III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions.
The expression of circRNAs was measured using quantitative polymerase chain reaction (qPCR) SYBR Green Master Mix (Takara, Tokyo, Japan) in a ViiA 7 Real-time PCR System (Applied Biosystems Inc, Foster City, CA, United States). The sequences of the divergent primers for the detection of hsa_circ_0000745 by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) were 5’-GTTGAAAGTAGCCCGAGCAG-3’ and 5’-ACGTGGCACAGACCTCTCTC-3’. The primer sequences for glyceraldehyde-3-phosphate dehydrogenase (GAPDH; used as a control) were 5’-CTCGCTTCGGCAGCACA-3’ and 5’-AACGCTTCACGAATTTGCGT-3’. These primers were synthesized by Invitrogen (Shanghai, China). The reaction conditions were as follows: 95 °C for 10 min, and 40 cycles of 95 °C for 10 s and 60 °C for 60 s. The RNA levels were normalized to human GAPDH. The expression levels were analyzed by the 2-ΔΔCt method.
All of the quantitative PCR reactions were conducted in triplicate. The appearance of a single-peak in the melt-curve suggested the specificity of the PCR products.
All experimental data were analyzed using SPSS software (version 22.0; IBM, Armonk, NY, United States) and GraghPad Prism 5.0 (GraphPad Software, La Jolla, CA, United States). The expression level of each circRNA was represented as fold-change using the 2-ΔΔCt method. Differences of hsa_circ_0000745 levels between GC tissues and adjacent non-tumor tissues, and between plasma samples from patients with GC and healthy controls were assessed using t-test. The correlation between hsa_circ_0000745 levels and clinicopathological factors was further analyzed by one-way analysis of variance. The receiver operating characteristic (ROC) curve was established to evaluate the diagnostic value. The cut-off value of hsa_circ_0000745 was analyzed with SPSS software. A P value < 0.05 was considered statistically significant.
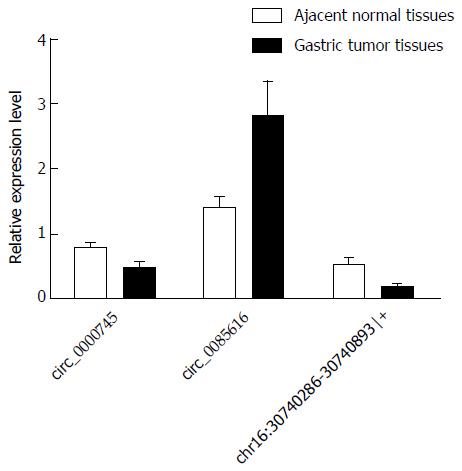
Among the three differentially expressed circRNAs examined (hsa_circ_0000745, hsa_circ_0085616, and chr16:30740286-30740893|+), hsa_circ_0085616 was reported to be up-regulated in colorectal tissues compared to that in normal tissues[20], while hsa_circ_0000745 and chr16:30740286-30740893|+ were found to be down-regulated in GC tissues compared to paired adjacent non-tumor tissues[21]. The observed changes in expression levels were validated by qRT-PCR analysis using 20 sets of gastric tissues and paired adjacent non-tumor tissues.
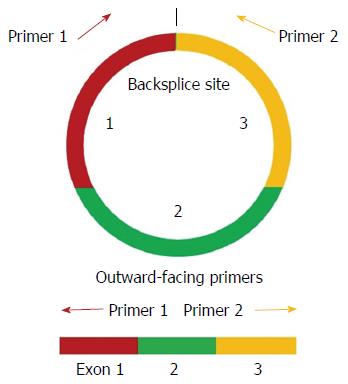
Divergent primers, rather than the more commonly used convergent primers, were designed for quantitative amplification of the circRNAs (Figure 1 and Table 2). The qRT-PCR results showed that circ_0085616 was up-regulated, while circ_0000745 and chr16:30740286-30740893|+ were down-regulated in the GC tissues compared to the paired adjacent non-tumor tissues (Figure 2), suggesting that the general expression patterns of the three circRNAs in GC were consistent with those in colorectal cancer.
| Target ID | Primer sequence, 5’-3’ | Product size in bp |
| Circ_0000745 | F: GTTGAAAGTAGCCCGAGCAG | 204 |
| R: ACGTGGCACAGACCTCTCTC | ||
| Circ_0085616 | F: GACTACAACTCGCCCACCAC | 200 |
| R: TCCATTTCTGGGCCATAATC | ||
| Chr16:30740286-30740893|+ | F: CCTTTGCACCGTATTGTGTG | 199 |
| R: GGCAGGGTACAGAAATCCAG | ||
| GAPDH | F: CTCGCTTCGGCAGCACA | 122 |
| R: AACGCTTCACGAATTTGCGT |
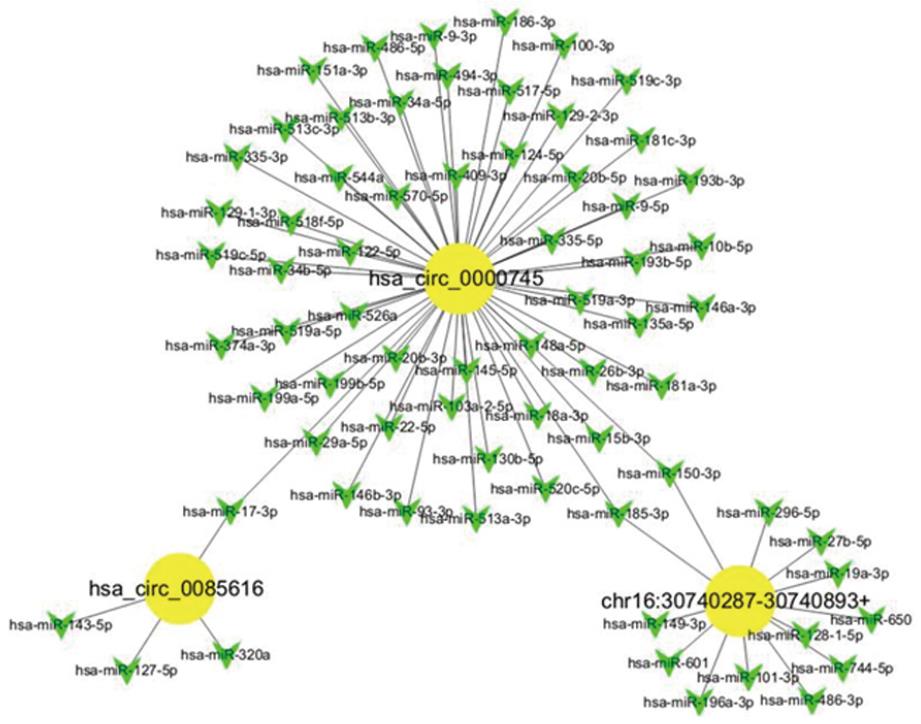
Given that miRNAs play important roles in the progression of GC, some circRNAs could be involved in GC, likely via interacting with miRNAs. We analyzed the potential binding miRNAs for the three circRNAs using sequence analysis. The association of miRNAs with GC indicated that circRNAs may have a regulatory role in GC. A tree diagram of circRNAs and their potential binding miRNAs is shown in Figure 3. Based on these potential circRNA/miRNA interactions, hsa_circ_0000745 was predicted to be able to bind a spectrum of miRNAs, with the known functions suggesting its potential role in the regulation of GC growth.
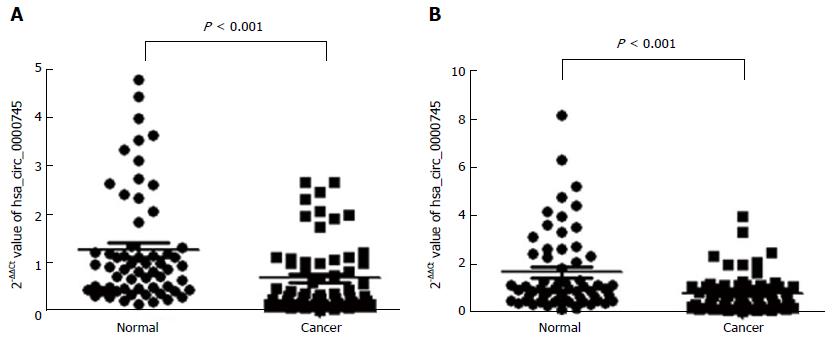
We tested hsa_circ_0000745 levels in GC tissues and plasma samples from patients with GC. As shown in Figure 4A, hsa_circ_0000745 expression was found to be down-regulated in the GC tissues (P < 0.001). Furthermore, hsa_circ_0000745 levels in the plasma samples of patients with GC were lower than those in plasma samples from healthy controls (P < 0.001; Figure 4B).
Since we found that hsa_circ_0000745 expression levels were lower in GC tissues and patients’ plasma samples, we further analyzed their association with clinicopathological features of patients with GC. As shown in Table 3, hsa_circ_0000745 levels in the GC tissues were significantly related to tumor differentiation (P = 0.012). However, we found no association between its levels with other clinicopathological features, such as age, sex, tumor diameters, lymphatic metastasis, TNM stage and carcinoembryogenic antigen (CEA) levels.
| Clinicopathological factor | Tissue hsa_circ_0000745 | Plasma hsa_circ_0000745 | |||
| n | Mean ± SD | P value | mean ± SD | P value | |
| Age (yr) | 0.757 | 0.195 | |||
| < 60 | 33 | 0.77 ± 0.83 | 0.90 ± 0.96 | ||
| ≥ 60 | 27 | 0.58 ± 0.60 | 0.69 ± 0.53 | ||
| Sex | 0.203 | 0.252 | |||
| Male | 43 | 0.59 ± 0.67 | 0.68 ± 0.60 | ||
| Female | 17 | 0.92 ± 0.85 | 1.13 ± 1.12 | ||
| Diameter in cm | 0.168 | 0.127 | |||
| ≥ 5 | 26 | 0.76 ± 0.77 | 0.87 ± 0.76 | ||
| < 5 | 34 | 0.63 ± 0.71 | 0.76 ± 0.84 | ||
| Differentiation | 0.012a | 0.087 | |||
| Well | 2 | 0.96 ± 1.10 | 0.89 ± 0.44 | ||
| Moderate | 25 | 0.65 ± 0.71 | 0.70 ± 0.66 | ||
| Poor | 33 | 0.70 ± 0.77 | 0.88 ± 0.92 | ||
| Lymphatic metastasis | 0.064 | 0.087 | |||
| Present | 44 | 0.71 ± 0.71 | 0.85 ± 0.78 | ||
| Absent | 16 | 0.61 ± 0.81 | 0.69 ± 0.88 | ||
| TNM stage | 0.056 | 0.046a | |||
| I-II | 14 | 0.63 ± 0.81 | 0.60 ± 0.53 | ||
| III-IV | 46 | 0.70 ± 0.72 | 0.87 ± 0.87 | ||
| CEA in ng/mL | 0.077 | 0.130 | |||
| ≥ 5 | 10 | 0.79 ± 0.72 | 0.65 ± 0.59 | ||
| < 5 | 50 | 0.66 ± 0.74 | 0.85 ± 0.78 | ||
Meanwhile, hsa_circ_0000745 levels in plasma from GC patients were significantly related to TNM stage (P = 0.046). Nevertheless, we did not find any association between hsa_circ_0000745 and the other clinicopathological features.
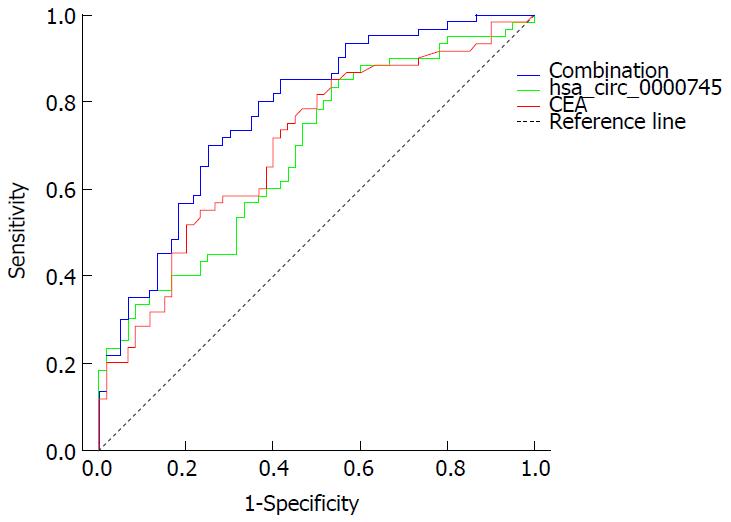
To evaluate the potential diagnostic value, a ROC curve was generated for hsa_circ_0000745 levels in plasma. We found that the area under the ROC curve (AUC) was 0.683 (Figure 5). The sensitivity and specificity were 0.855 and 0.45, respectively. When the expression level of plasma hsa_circ_0000745 was combined with CEA level, the AUC increased to 0.775, while the sensitivity and specificity changed to 0.800 and 0.633, respectively.
Gastric carcinogenesis is a complicated biological process that involves a wide array of molecular abnormalities. In recent decades, the molecular mechanisms of GC have been extensively investigated. Expression arrays for genes and noncoding RNAs, such as miRNAs and lncRNAs, have been applied and recognized as feasible and useful approaches to profiling the molecular signatures of GC[25,26].
Today, circRNAs are the newest type of noncoding RNAs, and their discovery expanded our knowledge of the complexity of noncoding RNAs. The circRNAs were originally thought to be by-products of splicing errors or the mRNA process[27]. They represent a stable, diverse and conserved class of RNA molecules[11,28]. Ongoing research is continuing to provide evidence that circRNAs are involved in the development and progression of diseases, especially cancer[29].
Hsa_circ_002059 was first found to be significantly down-regulated in GC and proposed as a potential novel and stable biomarker for the diagnosis of this particular cancer type[19]. According to other reports, expression of the circular RNA-ITCH is typically down-regulated in esophageal squamous cell carcinoma (ESCC) and colorectal carcinoma, and may exert an inhibitory effect on ESCC and colorectal cancer by suppressing the Wnt/β-catenin pathway[30,31]. Collectively, these findings have suggested that circRNAs play a crucial role in cancers, and that they may serve as the basis for developing new strategies for diagnosis and therapy. However, the tumorigenesis mechanisms of GC are far from being completely elucidated and this idea is still in its infancy.
In this study, we randomly selected three differently expressed circRNAs, which we characterized using RNA-Seq analysis of ribosomal RNA-depleted total RNA from three GC or colorectal cancer tumor tissues and paired normal tissues. Circ_0085616 was up-regulated in colorectal cancer tissues compared with non-cancerous tissues, while circ_0000745 and chr16:30740286-30740893|+ were down-regulated in GC tissues compared with normal tissues. We further confirmed the RNA-Seq results of three circRNAs in 20 paired normal and cancerous gastric tissues by qRT-PCR analysis; circ_0085616 up-regulation and circ_0000745 and chr16:30740286-30740893|+ down-regulation in the GC tissues suggested that the circRNA sequencing results of GC are consistent with the expression patterns observed in colorectal cancer.
Recent studies have demonstrated that circRNAs can function as miRNA sponges or potent competitive endogenous RNA molecules[27,32-35]. Given that miRNAs play important roles in the progression of GC, it is possible and even likely that some circRNAs are involved in GC via interacting with miRNAs. We analyzed the potential binding miRNAs of the three circRNAs by sequence analysis. The association of miRNAs with GC indicated that circRNAs may have a regulatory role in GC. Based on the potential circRNA/miRNA interactions identified, hsa_circ_0000745 is potentially able to bind various miRNAs that may play critical roles in regulating GC growth and migration. However, additional studies are required to clarify whether these circRNAs are truly involved in GC, and whether circRNAs acting as miRNA sponges are a general phenomenon.
In this study, we found that hsa_circ_0000745 was down-regulated in GC tissues as well as in the plasma samples from patients with GC. It was suggested that hsa_circ_0000745 may be involved in other diseases besides GC, including various cancers (i.e., breast cancer, colorectal cancer, hepatocellular carcinoma, lung cancer, ovarian cancer, and rhabdomyosarcoma) and cerebellar neurodegeneration, Waldenstrom macroglobulinemia, cardiac hypertrophy, cardiomyopathy, coronary artery disease, heart failure, myeloproliferative disorder, myocardial infarction, retinitis pigmentosa, and schizophrenia, according to the previous analysis (http://gyanxet-beta.com/circdb/). That is the main reason why we choose hsa_circ_0000745 for further study.
We found that expression levels of hsa_circ_0000745 in GC tissues were associated with tumor differentiation. However, its expression did not show any statistically significant relationship with tumor size, lymphatic metastasis or TNM stage (Table 3). Moreover, we determined its potential diagnostic value, reported herein for the first time. When compared with serum CEA levels, hsa_circ_0000745 had a higher sensitivity and specificity in the screening of GC (Figure 5). The combination of plasma hsa_circ_0000745 and serum CEA levels, however, increased the diagnostic value. In summary, our study revealed that the circular RNA hsa_circ_0000745 may serve as a diagnostic marker for GC.
We are grateful to the members of our laboratory for helpful discussions regarding this study. We thank Cloud-Seq Biotech Ltd. Co. (Shanghai, China) for the transcriptome sequencing service and the subsequent bioinformatics analysis.
Circular RNA (circRNA) is a recently identified type of noncoding RNA that plays an important role in the biological processes of tumor development and progression. The stability of circRNA makes it an attractive candidate as a serological diagnostic marker for cancer. However, the circRNA expression profiles in gastric cancer (GC) have not been elucidated.
Recent studies have shown that different tumors have distinctive differential expression profiles of various circRNAs, such as hsa_circ_002059 in GC and hsa_circ_0004277 in acute myeloid leukemia.
This is the first study to demonstrate that hsa_circ_0000745 is differentially expressed in plasma samples of GC patients and normal healthy controls, as well as in gastric tumor tissues and paired adjacent non-tumor tissues. Moreover, the data suggest the possibility that hsa_circ_0000745 may serve as a novel diagnostic marker for GC.
Expression of hsa_circ_0000745 was found to correlate with tumor differentiation and tumor-node-metastasis stage. When plasma hsa_circ_0000745 expression was combined with serum level of carcinoembryogenic antigen, the diagnostic ability (especially specificity) for GC was improved.
Circular RNA is a type of RNA which, unlike the better known linear RNA, forms a covalently closed continuous loop. GC is also known as stomach cancer, developing from the lining of the stomach. Diagnostic biomarkers are found in blood, urine or tissues and are abnormally expressed, in a trackable manner, in the presence of diseases.
This is the first study to show the promising diagnostic value of measuring circulating hsa_circ_0000745 levels in GC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Fujita T S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Huang Y
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21359] [Article Influence: 2135.9] [Reference Citation Analysis (3)] |
| 2. | Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441-5447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 142] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 3. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20496] [Article Influence: 2049.6] [Reference Citation Analysis (20)] |
| 4. | Ishihara R. Infrared endoscopy in the diagnosis and treatment of early gastric cancer. Endoscopy. 2010;42:672-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Tanizawa Y, Terashima M. Lymph node dissection in the resection of gastric cancer: review of existing evidence. Gastric Cancer. 2010;13:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the Role of Circulating Long Non-Coding RNA H19 as a Promising Novel Biomarker in Plasma of Patients with Gastric Cancer. J Clin Lab Anal. 2016;30:1100-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 7. | Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1266] [Cited by in RCA: 1470] [Article Influence: 113.1] [Reference Citation Analysis (0)] |
| 8. | Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 823] [Cited by in RCA: 953] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 9. | Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1045] [Cited by in RCA: 1505] [Article Influence: 167.2] [Reference Citation Analysis (0)] |
| 10. | Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 617] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 11. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6017] [Article Influence: 501.4] [Reference Citation Analysis (0)] |
| 12. | Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1370] [Cited by in RCA: 1806] [Article Influence: 180.6] [Reference Citation Analysis (0)] |
| 13. | You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603-610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 740] [Cited by in RCA: 911] [Article Influence: 91.1] [Reference Citation Analysis (0)] |
| 14. | Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-5612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 758] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 15. | Boeckel JN, Jaé N, Heumüller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res. 2015;117:884-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 297] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 16. | Satoh J, Yamamura T. Gene expression profile following stable expression of the cellular prion protein. Cell Mol Neurobiol. 2004;24:793-814. [PubMed] |
| 17. | Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966-1980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 774] [Cited by in RCA: 755] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 18. | Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 526] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 19. | Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 631] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 20. | Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 518] [Cited by in RCA: 608] [Article Influence: 60.8] [Reference Citation Analysis (0)] |
| 21. | Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 570] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 22. | Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22949] [Cited by in RCA: 32967] [Article Influence: 2535.9] [Reference Citation Analysis (0)] |
| 23. | Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 270] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 24. | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22632] [Cited by in RCA: 28986] [Article Influence: 1811.6] [Reference Citation Analysis (0)] |
| 25. | Huang T, Wang-Johanning F, Zhou F, Kallon H, Wei Y. MicroRNAs serve as a bridge between oxidative stress and gastric cancer (Review). Int J Oncol. 2016;49:1791-1800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N, Bin J, Liao Y, Liao W. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 165] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 27. | Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1350] [Article Influence: 135.0] [Reference Citation Analysis (0)] |
| 28. | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3481] [Cited by in RCA: 3412] [Article Influence: 284.3] [Reference Citation Analysis (0)] |
| 29. | Mirzaei H, Khataminfar S, Mohammadparast S, Sales SS, Maftouh M, Mohammadi M, Simonian M, Parizadeh SM, Hassanian SM, Avan A. Circulating microRNAs as Potential Diagnostic Biomarkers and Therapeutic Targets in Gastric Cancer: Current Status and Future Perspectives. Curr Med Chem. 2016;23:4135-4150. [PubMed] |
| 30. | Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001-6013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 449] [Cited by in RCA: 567] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 31. | Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One. 2015;10:e0131225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 247] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 32. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6041] [Article Influence: 503.4] [Reference Citation Analysis (0)] |
| 33. | Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919-3931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 276] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 34. | Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602-2611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 713] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 35. | Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1230] [Cited by in RCA: 1618] [Article Influence: 179.8] [Reference Citation Analysis (0)] |