Published online Sep 7, 2017. doi: 10.3748/wjg.v23.i33.6194
Peer-review started: May 3, 2017
First decision: June 5, 2017
Revised: June 27, 2017
Accepted: July 22, 2017
Article in press: July 24, 2017
Published online: September 7, 2017
Processing time: 129 Days and 8.3 Hours
Attaran et al[1] have recently shown that decreased susceptibility of established Helicobacter pylori (H. pylori) biofilms to specific antibiotics, was associated with the overtly enhanced transcription of two efflux pump genes, hp1165 and hefA, involved in specific resistance to tetracycline and multiple antibiotics, respectively. Apart from antibiotic exposure, secretion of multiple antimicrobial peptides, such as human β-defensins (hβDs), by the gastric epithelium upon Hp challenge, may act as early triggering events that positively impact biofilm formation and thus, antibiotic resistance. In this regard, we undertook genomic transcriptional studies using Hp 26695 strain following exposure to sublethal, similar to those present in the gastric niche, concentrations of hβDs in an attempt to provide preliminary data regarding possible mechanisms of immune evasion and selective sensitivity of Hp. Our preliminary results indicate that hβD exposure ignites a rapid response that is largely due to the activation of several, possibly interconnected transcriptional regulatory networks – origons - that ultimately coordinate cellular processes needed to maintain homeostasis and successful adaptation of the bacterium in the gastric environment. In addition, we have shown that both antibiotic and hβD resistance are mediated by dedicated periplasmic transporters, including the aforementioned efflux pump genes hp1165 and hefA, involved in active export of antibiotics from the cell membrane and/or, as recently suggested, substrate sensing and signalling. Furthermore, it appears that sublethal doses of hβDs may enhance biofilm formation by the sustained expression of, mainly, quorum sensing-related genes. In conclusion, we provide additional data regarding the role of specific innate immune molecules in antibiotic cross-resistance mechanisms that may deepen our understanding in the context of the development of novel eradication regimens.
Core tip: In the course of Helicobacter pylori infection, epithelium-derived human β-defensins may act as early triggering signals that induce biofilm formation and enhanced expression of antibiotic resistance genes, regardless of prior antibiotic exposure.
- Citation: Kazakos EI, Dorrell N, Polyzos SA, Deretzi G, Kountouras J. Comment on “Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics”. World J Gastroenterol 2017; 23(33): 6194-6196
- URL: https://www.wjgnet.com/1007-9327/full/v23/i33/6194.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i33.6194
Attaran et al[1] concluded that, in biofilm-forming populations, overexpression of two efflux pump genes, hp1165 and hefA, conferring resistance to tetracycline and multiple antibiotics respectively, may favor reduced antibiotic susceptibility of Helicobacter pylori (H. pylori) in vivo.
Further to antibiotic exposure, additional, epithelial-derived molecules may function as triggering signals during the dynamic H. pylori interaction with the gastric mucosa, provoking overexpression of efflux pumps that in turn, regulate the bacterium’s biofilm-producing capacity and promote its virulence. Several studies have unraveled the role of constitutive and/or induced expression of human β-defensins (hβDs)1 - 4 in the bacterium’s adaptation in the human stomach and H. pylori -related pathologies [2,3].
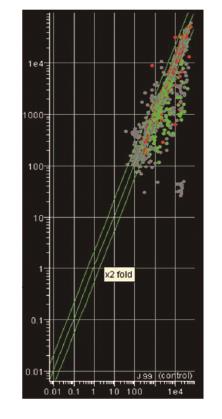
In this respect, we performed whole genome transcriptome analyses (competitive genomic RNA/ RNA hybridisations) using H. pylori -specific microarrays based on the Hp 26695 and J99 genome sequences and annotation available at the time. Briefly, H. pylori 26695 strain was exposed to sublethal, similar to those encountered at the gastric epithelium concentrations of hβDs, in an attempt to identify possible mechanisms of H. pylori immune escape and clarify their role in biofilm development in vitro. Our preliminary results have identified profound changes in the transcriptional profile of H. pylori 26695 demonstrated by the induction or suppression of multiple gene components of distinct regulatory and signaling cascades activated as a result of environmental stress (Figure 1, unpublished data). Overall, the vast majority of genes affected, encoded components of the cell wall stimulon, possibly as means to prevent hβD-specific binding and proper immune recognition, or could be further assigned to certain origons, essential for colonisation of the gastric niche and long-term adaptation, intracellular metal homeostasis and urease activation that largely determine H. pylori pathogenicity. Apart from the marked induction of hp1165 and hefA, also reported by the authors[1], several other genes coding for transmembrane ABC transporters (glnP, dppF, hp1458, hp1486), efflux proteins (hp0656, hp0946), multidrug and toxic extrusion proteins were found to be significantly up-regulated, thereby indicating their prominent role in the cellular response to hβDs challenge, membrane detoxification and maintenance of osmotic balance.
Interestingly, enhanced biofilm production by Hp 26695, observed in our studies upon exposure to sublethal concentrations of hβD1 and hβD3, was primarily attributed to the down-regulation of metK and luxS genes, involved in synthesis of quorum-sensing autoinducer-2, in accordance to previously published data[4,5].
Collectively, our results indicate that sublethal doses of epithelial-secreted antimicrobial peptides such as hβDs, may select co-resistance to antibiotics commonly used in Hp eradication therapies and vice versa, considering that they provoke the activation of shared, contact-dependent signaling networks, including efflux pumps. Furthermore, it appears that hβDs may independently act as triggering stimuli promoting biofilm formation in vivo which in turn, accounts, at least partly, for the observed failure of eradication regimens and the establishment of H. pylori -related chronic inflammation.
Given the complexity of H. pylori -host epithelial crosstalk aforementioned data warrant further investigation to achieve the development of successful anti-biofilm strategies that will ultimately re-enforce our therapeutic options mainly towards eradication of H. pylori -related resistance. Furthermore, future research focus on the polymorphic variability of the human genome that directly affects epithelial dynamics of hβDs expression may reveal important correlation patterns between H. pylori pathogenesis, including biofilm formation, and individual disease susceptibility.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Greece
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Ahmed Said ZN, Gonzalez-Reimers E, Slomiany BL, Zamani M S- Editor: Qi Y L- Editor: A E- Editor: Huang Y
| 1. | Attaran B, Falsafi T, Ghorbanmehr N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J Gastroenterol. 2017;23:1163-1170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 2. | Kountouras J, Deretzi G, Gavalas E, Zavos C, Polyzos SA, Kazakos E, Giartza-Taxidou E, Vardaka E, Kountouras C, Katsinelos P. A proposed role of human defensins in Helicobacter pylori-related neurodegenerative disorders. Med Hypotheses. 2014;82:368-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 3. | Pero R, Coretti L, Nigro E, Lembo F, Laneri S, Lombardo B, Daniele A, Scudiero O. β-Defensins in the Fight against Helicobacter pylori. Molecules. 2017;22:pii: E424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 4. | Bessa LJ, Grande R, Di Iorio D, Di Giulio M, Di Campli E, Cellini L. Helicobacter pylori free-living and biofilm modes of growth: behavior in response to different culture media. APMIS. 2013;121:549-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. MBio. 2015;6:e00379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |