Published online Jul 28, 2017. doi: 10.3748/wjg.v23.i28.5158
Peer-review started: April 18, 2017
First decision: May 16, 2017
Revised: May 24, 2017
Accepted: July 4, 2017
Article in press: July 4, 2017
Published online: July 28, 2017
Processing time: 105 Days and 21.6 Hours
To investigate the effect of a single amino acid mutation in human class B scavenger receptor I (SR-BI) on the infectivity of cell culture-derived hepatitis C virus (HCVcc) in SR-BI knock-down Huh7-siSR-BI cells.
Site-directed mutagenesis was used to construct the SR-BI S112F mutation, and the mutation was confirmed by nucleotide sequencing. SR-BI knock-down Huh7-siSR-BI cells were transfected with SR-BI S112F, SR-BI wild type (WT) and control plasmids, and then infected with HCVpp (HCV pseudoparticles) and hepatitis C virus derived from cell culture (HCVcc). A fluorescence assay was performed to analyze the effect of the S112F mutation on HCV entry; quantitative real-time PCR, immunofluorescence, and Western blot assays were used to analyze the effect of the S112F mutation on HCV infectivity. CHO cells expressing WT and SR-BI S112F were incubated with the HCV E2 protein expressed in HEK 293T cells, and flow cytometry was performed to examine the ability of SR-BI S112F to bind to the HCV E2 protein. Huh7-siSR-BI cells were transfected with SR-BI WT and the S112F mutant, and then DiI-HDL was added and images captured under the microscope to assess the ability of SR-BI S112F to take up HDL.
The SR-BI S112F mutation was successfully constructed. The S112F mutation decreased the expression of the SR-BI mRNA and protein. SR-BI S112F decreased HCV entry and HCVcc infectivity in Huh7-siSR-BI cells. The S112F mutation impaired the binding of SR-BI to HCV E2 protein and decreased the HDL uptake of SR-BI.
The S112F single amino acid mutation in SR-BI decreased the levels of the SR-BI mRNA and protein, as well as the ability of SR-BI to bind to the HCV E2 protein. Amino acid 112 in SR-BI plays important roles in HCV entry and the infectivity of HCVcc in vitro.
Core tip: Human class B scavenger receptor I (SR-BI) plays important roles in both host lipid metabolism and the entry of hepatitis C virus (HCV). Single nucleotide polymorphisms (SNPs) in the host genome that affect the virus-host interaction have received increasing attention in recent years. Several SR-BI SNPs have been reported to affect the high-density lipoprotein cholesterol levels in populations carrying SR-BI mutations; however, the impact of SR-BI SNPs on HCV infection has not been studied intensively. Based on our results, the S112F single amino acid mutation in SR-BI inhibited the infectivity of hepatitis C virus derived from cell culture in a cell culture model by downregulating the expression of the SR-BI protein.
- Citation: Gao R, Gao W, Xu G, Xu J, Ren H. Single amino acid mutation of SR-BI decreases infectivity of hepatitis C virus derived from cell culture in a cell culture model. World J Gastroenterol 2017; 23(28): 5158-5166
- URL: https://www.wjgnet.com/1007-9327/full/v23/i28/5158.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i28.5158
Hepatitis C virus (HCV) is an enveloped RNA virus that belongs to the Hepacivirus genus of the Flaviviridae family. The HCV genome encodes a large precursor polyprotein, which is cleaved by host and viral proteases to generate at least 10 functional viral proteins[1]. HCV infection is a global health problem, with an estimated 180 million persons infected worldwide, and HCV infection is the leading cause of cirrhosis and hepatocellular carcinoma[2]. The prevalence of chronic HCV infection in China was 3.2% in 1992 and 0.4% in 2006. Recent reports from the Chinese Ministry of Health have identified 70861 cases in 2006 and 201622 cases in 2012[3]. The most recent investigation showed a prevalence of HCV infection of 3.0% in northeastern China[4]. In recent years, the Chinese government has increased its investment in the prevention and control of viral hepatitis. However, an effective vaccine is not available and treatment with the combination of interferon and ribavirin therapy produces a response in approximately half of infected patients. More recently, a new therapy comprising novel direct-acting antivirals (DAAs), such as protease inhibitors (telaprevir, boceprevir and simeprevir) and an RNA polymerase inhibitor (sofosbuvir), increased the sustained virological response rate in HCV-infected patients[5-7]. However, the DAA therapy also produces significant side effects[8]. Therefore, novel anti-HCV methods, including host targets, are still needed.
HCV entry is a multi-step process that requires many host molecules, including the tetraspanin molecule CD81, human class B scavenger receptor I (SR-BI), and the tight-junction proteins claudin-1 (CLDN1) and occludin (OCLN)[9-12]. Among these proteins, SR-BI plays a crucial role since both SR-BI and its ligand lipoprotein are involved in the HCV entry process[13,14]. SR-BI has a number of common ligands, including high-density lipoprotein (HDL), low-density lipoprotein (LDL) and oxidized LDL[13]. As shown in a study by Dreux et al[15], HDL enhances the infectivity of HCVpp (HCV pseudoparticles) and hepatitis C virus derived from cell culture (HCVcc).
SR-BI was originally defined as a class B scavenger receptor in a family that includes CD36, LIMP II (lysosome membrane protein II), and SR-BII (a form of SR-BI with an alternate C-terminal cytoplasmic tail)[16]. SR-BI is a lipoprotein receptor composed of 509 amino acids (aa) in which the cytoplasmic C- and N-terminal domains are separated by a large extracellular domain. As an HDL receptor, SR-BI mediates selective uptake of HDL-derived cholesteryl ester (CE) in vitro and in vivo[17]. The conformation of the extracellular domain is important for the binding of SR-BI and HDL, and hence affects the function of SR-BI[18]. Eleven N-linked glycosylation sites (aa102, 108, 116, 173, 212, 227, 255, 288, 310, 330 and 383) have been identified in the extracellular domain, and two glycosylation sites (Asn108 and Asn173) were proven to be indispensable for the expression and function of SR-BI[19]. In addition to mediating selective CE transport, SR-BI has been shown to play important roles in many human diseases, including atherosclerosis, apoptosis, immune responses, HCV and dengue virus entry, and malaria parasite infection[20].
In recent years, the influence of single nucleotide polymorphisms (SNPs) in the host genome on the virus-host interaction has received increasing attention. For HCV, most studies have focused on SNPs in the IL28B gene and HCV prognosis[21]. Currently, few reports on virus entry and host genomic SNPs have been published[22]. For SR-BI, researchers have focused on its regulation of HDL-cholesterol and other metabolites, and a very recent report showed that polymorphisms in the SR-BI gene are associated with the virological response in HCV-infected patients[23,24]. According to the results of a GWAS (genome-wide association study) of 10000 individuals, SNPs in the SR-BI gene are associated with a small, but significant elevation in plasma HDL-cholesterol levels[25]. Recently, a single loss-of-function mutation (P297S) in SR-BI was identified, and the mutation increased HDL-cholesterol levels and reduced cholesterol efflux from macrophages[26]. Subsequently, two novel missense mutations, S112F (nucleotide C588T) and T175A (nucleotide A776T) were also shown to be associated with elevated HDL-cholesterol levels[27,28]. However, few studies have investigated how the mutations impact HCV infection and development. In this study, we studied the effects of the SR-BI S112F single amino acid mutation on the infectivity of HCVcc using a cell culture model.
The Huh7 cell line, human embryonic kidney (HEK) 293T cell line and Huh7-siSR-BI cell line are maintained in our laboratory. Briefly, SR-BI shRNA was designed, cloned into the pGP-Lenti3 vector (Biovector, Science Lab, Beijing), and the positive recombinant Lv-SR-BI-shRNA vector was verified. This vector and helper plasmids were co-transfected into HEK 293T cells. The recombinant lenti-SR-BI-shRNA virus was used to infect Huh7 cells. Puromycin was added for screening, and real-time PCR and Western blot were conducted to detect the levels of the SR-BI mRNA and protein, respectively; finally, the Huh7-siSR-BI cell line was obtained[29].
Cells were grown in complete Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco BRL, United States), 1 mmol/L -glutamine, 100 nmol/L nonessential amino acids, 100 U/mL penicillin, and 100 μg/mL streptomycin.
The SR-BI S112F mutation was introduced into pcDNA-SR-BI (maintained in our lab) using a Quick Change Lightning Site-Directed Mutagenesis Kit (Stratagene, CA, United States), according to the manufacturer’s instructions, with the primers (S112F-F: 5’-ACAACGACACCGTGTTCTTCCTCGAGTACCGCACCT-3’ and S112F-R: 5’-AGGTGCGGTACTCGAGGAAGAACACGGTGTCGTTGT-3’; the italic letters represent the mutant nucleotide). The presence of the desired mutation was confirmed using nucleotide sequencing (Invitrogen, CA, United States).
RNA was isolated from harvested cells using TRIzol reagent (Invitrogen), and RNA was prepared according to the manufacturer’s instructions. RNA obtained from 1 × 105 cell equivalents was analyzed using RT-PCR. RNA samples were transcribed into cDNA using random primers and then quantitatively analyzed with the specific primers SR-BI-F: 5’-GCTGCAGGAA GCAAAACTGT-3’ and SR-BI-R: 5’-CCAGTAGAAAAGGG TCACAGG-3’ using the quantitative RT-PCR kit (Applied Biosystems, United States). Genome copy numbers were normalized to GAPDH levels determined in parallel (GAPDH-F: 5’-TGACTTCAACAGCGACACCCA-3’; GAPDH-R: 5’-CACCCTGTTGCTGTAGCCAAA-3’) using the comparative cycle threshold values.
Huh7 and SR-BI knock-down Huh7-siSR-BI cells were cultured in collagen-coated 96-well plates at a density of 1 × 104 cells/well on the day before transfection. Cells were transfected with the SR-BI wild type (WT) and SR-BI S112F plasmids using LipofectamineTM (Invitrogen, CA, United States), according to the manufacturer’s instructions. Fresh cell medium was replaced 6 h after transfection and the cells were cultured for 48 h. Cells were washed with PBS, fixed with cold methanol, and then stained with an anti-SR-BI mouse monoclonal antibody (mAb) (1:1000 dilution, BD, United States) or serum from HCV-infected patients (1:100 dilution) for 2 h at room temperature (RT). After being washed with PBS, cells were reacted with an Alexa Fluor 488-conjugated anti-mouse antibody or human IgG antibodies (1:1000 dilution, BD, United States). Nuclei were stained with DAPI (1:2000 dilution, BD, United States). Images were captured and infected foci were counted under a fluorescence microscope (Olympus IX81, Japan).
Protein extracts from transfected cells were prepared in a modified RIPA buffer containing 0.5% SDS and a protease inhibitor cocktail (Complete mini; Roche) on ice. After centrifugation, protein concentrations were determined using the BCA method (Beyotime). The proteins were separated using 10% (w/v) SDS-polyacrylamide gel electrophoresis and transferred onto PVDF membranes (Millipore, Billerica, MA, United States) using a Trans-Blot apparatus (Bio-Rad). Membranes were blocked with 5% nonfat milk, incubated with a primary anti-SR-BI mouse mAb (1: 2000 dilution, BD), and detected using a horseradish peroxidase (HRP)-conjugated species-specific secondary antibody (Santa Cruz Biotechnology). Immunoreactivity was visualized using enhanced chemiluminescence (GE Healthcare).
The pJFH1 plasmid, which was kindly provided by Dr. Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan), was used as template to generate HCVcc as described[30]. Briefly, pJFH-1 was linearized to generate the template used in the in vitro transcription reaction to produce the viral RNA using the MEGAscript kit (Promega, Madison, WI, United States). Huh7 cells were then transfected with JFH-1 transcripts by electroporation. Cell culture supernatants were collected 5 d later and the viral titer was quantified. For the infection assay, Huh7-siSR-BI cells were seeded in a 96-well plate, cultured in 10% FBS-DMEM overnight, and then infected with HCVcc. The cells were cultured for 48 h before the infectivity was measured using the immunofluorescence assays.
HCVpp were generated as described[31]. Briefly, HEK 293T cells were transfected with plasmids encoding the HCV envelope proteins, Gag/Pol (pLP1) and Rev (pLP2) and the pLenti6 transfer vector (Invitrogen) expressing the luciferase gene. Vesicular stomatitis virus pseudoparticles were produced as controls. Cell culture supernatants were collected 2 d later and the viral titer was quantified using immunofluorescence assay (IFA). For the HCV entry assay, Huh7-siSR-BI cells were seeded on a 96-well plate, cultured overnight, and transfected with pCDNA-SR-BI WT and mutant plasmids, inoculated with HCVpp, and then cultured for 72 h before the entry of HCVpp was measured using a fluorescence assay.
The SR-BI and HCV E2 protein binding assay was performed using a FACS-based assay, as previously described[32]. Briefly, 4 × 105 CHO cells expressing SR-BI were incubated with equivalent amounts of HCV E2 protein (transient expression in HEK 293T cells) for 1 h at RT, washed twice with PBS, and incubated with an anti-E2 mAb (1 h at RT). After labeling with Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen), the mean fluorescence intensity was quantified by flow cytometry (Beckman Coulter, Fullerton, CA, United States).
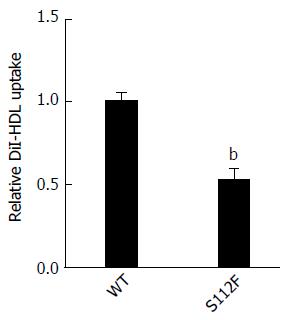
Huh7-siSR-BI cells were seeded in a 96-well plate at a density of 2 × 104 cells/well and cultured for 24 h. Cells were transfected with the SR-BI WT and mutant S112F plasmids using LipofectamineTM. Complete culture medium was replaced 6 h after transfection and the cells cultured for 48 h before DiI-HDL (200 μg/mL, Alfa Aesar, United States) was added and the cells were cultured for an additional 4 h. Nuclei were stained with Hoechst (Merk, NJ, United States) for 10 min, and cells were observed and images captured using a microscope.
The error bars represent the SD of means from at least three independent experiments. Statistical significance was analyzed using Student’s t test. P < 0.05 was considered statistically significant.
The pcDNA-SR-BI/S112F vector expressing the SR-BI single amino acid mutant was obtained by site-directed mutagenesis and confirmed by nucleotide sequencing. The Ser at site 112 was replaced with Phe in the mutant.
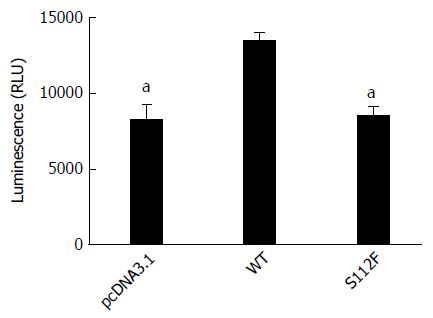
Huh7-siSR-BI cells were transfected with pcDNA3.1, pcDNA-SR-BI, or pcDNA-SR-BI/S112F first, and infected with HCVpp expressing the luciferase gene 3 d after transfection. Fluorescence was detected 3 d later. Compared with cells transfected with the pcDNA3.1 vector control and pcDNA-SR-BI WT control, SR-BI S112F decreased the entry of HCV in Huh7-siSR-BI cells (Figure 1).
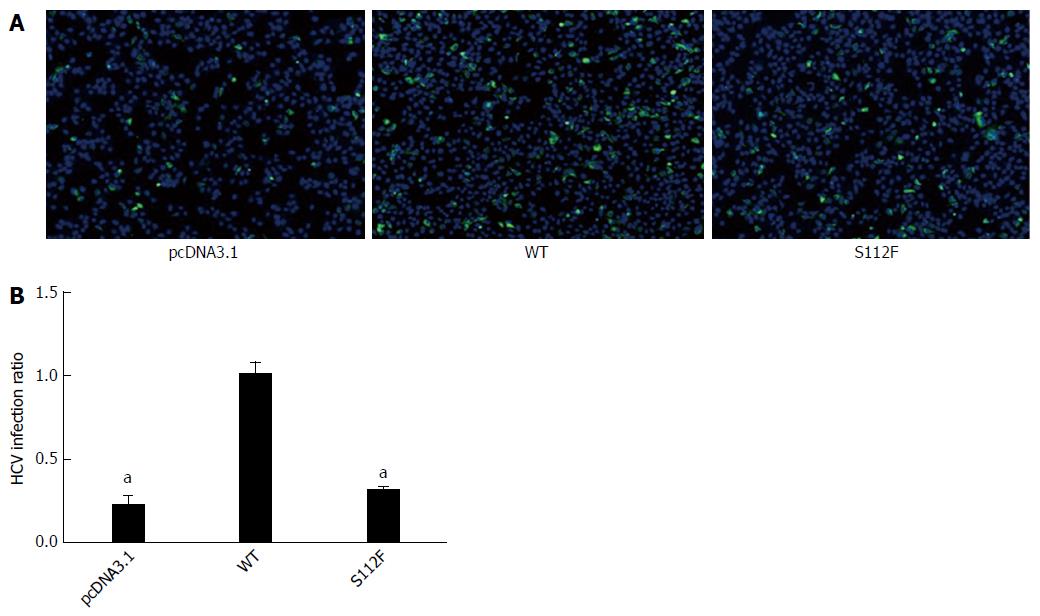
Huh7-siSR-BI cells were seeded in 96-well plates (for IFA) or 24-well plates (for qRT-PCR) and transfected with pcDNA3.1, pcDNA-SR-BI, or pcDNA-SR-BI/S112F; HCVcc (103FFU/mL) was added 24 h after transfection. Cells were harvested 72 h after infection and RNA was isolated for qRT-PCR. IFA was performed 48 h later. According to the IFA results, the expression of the HCV protein was decreased in the pcDNA-SR-BI/S112F group (Figure 2A). Based on the qRT-PCR results, the level of the HCV RNA was decreased in the pcDNA-SR-BI/S112F group (0.711 of the WT level), compared with pcDNA3.1 vector control and pcDNA-SR-BI WT control (Figure 2B).
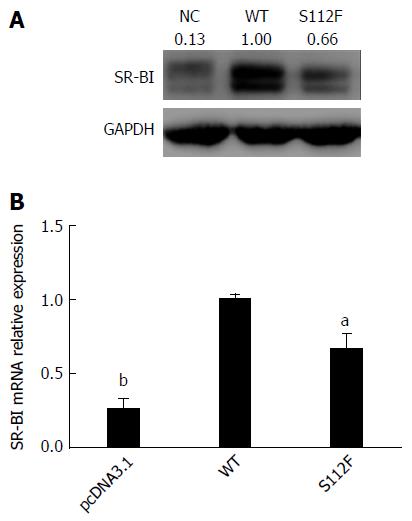
Huh7-siSR-BI cells were transfected with pcDNA3.1, pcDNA-SR-BI and pcDNA-SR-BI/S112F. Forty-eight hours after transfection, cells were harvested and analyzed by Western blot and qRT-PCR, and the results showed that the levels of the SR-BI S112F mRNA and protein were decreased compared with SR-BI WT cells (Figure 3).
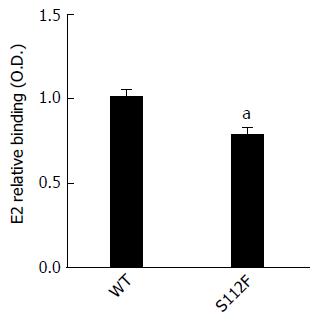
SR-BI has been reported to bind to HCV E2 protein. The ability of CHO cells expressing WT and mutant SR-BI to bind to the HCV E2 protein that had been expressed in HEK 293T cells was analyzed, and the results showed that the S112F mutation impaired the binding of SR-BI to the HCV E2 protein (Figure 4). We also performed the binding assay using HCVcc instead of the expressed HCV E2 protein and obtained similar results.
We performed the HDL uptake assay with DiI-HDL to determine whether the single amino acid mutation affects the HDL uptake ability of SR-BI. Huh7-siSR-BI cells were first transfected with SR-BI WT and mutant S112F plasmids, and then DiI-HDL was added. Based on the microscopic images, the S112F mutation decreased HDL uptake compared with SR-BI WT (Figure 5).
The SR-BI receptor is associated with lipid metabolism and participates in the bidirectional transport of cholesterol between cells and HDL. Increased clearance of HDL-CE from plasma and enhanced reverse cholesterol transport have been shown to significantly reduce atherosclerosis in animal models[23]. Previous studies have confirmed that the extracellular domain of SR-BI is critical for its receptor function[19].
Recently, the functions of SNPs in the SR-BI gene have been studied both in human and animal models. Acton and colleagues were the first to identify the associations between SNPs in the SR-BI gene and plasma lipid levels and body mass index in a Caucasian European population[33]. The nonsynonymous exon 1 SNP (rs4238001 [G2S]), which encodes a different amino acid, was significantly associated with higher HDL cholesterol (HDL-C) and lower LDL cholesterol (LDL-C) levels[33]. In 2011, a P297S missense mutation in SR-BI was reported, and people who carry the P297S mutation have increased HDL cholesterol levels[26]. Then, two novel missense mutations, S112F and T175A, in human SR-BI were identified in patients with atherosclerosis, which led to elevated HDL cholesterol levels[27]. A coding variant in SR-BI (I179N) significantly increased atherosclerosis, although the mutation did not dramatically affect the plasma lipid levels[34]. The hydrophobicity of N-terminal half of the extracellular domain of SR-BI was proven to be critical for the SR-BI-mediated cholesterol transport function, and this domain might function by interacting with other integral membrane proteins[35-37]. S112F, T175A and I179N point mutations occurred in the same region, but had different effects on the function of SR-BI and require further study.
HCV infection is a complicated process and is closely correlated with host lipid metabolism. In the attachment step, the HCV lipoviral particle (LVP) is recruited and binds to glycosaminoglycans on heparan sulfate and low-density lipoprotein receptor on host cells; then, HCV enters hepatocytes by interacting with several host entry factors, including CD81, SR-BI, the tight junction proteins CLDN1 and OCLN, and the cholesterol absorption receptor Niemann-Pick C1-like-1[38]. Lipids and lipid receptors play key roles in the early stage of HCV infection, and researchers have postulated that LVP was actually endocytosed into the hepatocytes as a regular lipoprotein[39].
During the entry step, the HCV E2 HVR1 region interacts with the extracellular loop of SR-BI in both the binding and post-binding steps[40,41]. Therefore, strategies targeting SR-BI have been reported to inhibit HCV infection. SR-BI binds to serum amyloid A (SAA), an acute-phase protein produced by the liver, promoting SAA internalization and inhibiting HCV entry[42]. A small-molecule antiviral compound, ITX5061, has been reported to impede the uptake of HDL by SR-BI and blocks the uptake of HCV viral particles by hepatocytes[43,44].
In addition to the entry step, the replication of HCV in the membrane web and release from hepatocytes are associated with the host lipid metabolism. Since SR-BI has a key role in the host lipid metabolism and SNPs in the SR-BI gene have been reported to modulate the function of SR-BI, we studied the effect of the S112F missense mutation in SR-BI on the infectivity of HCVcc in SR-BI knock-down Huh7 cells. The Huh7-siSR-BI cell line was established by screening Huh7 cells with puromycin after infection with Lv-SR-BI-shRNA[29]. We first constructed SR-BI S112F using site-directed mutagenesis, and then Huh7-siSR-BI cells were used to detect the effects of the single amino acid mutant in SR-BI on the entry and infectivity of HCV. The S112F single amino acid mutation decreased HCV entry and the infection of HCVcc compared with SR-BI WT. We further assessed the effects of S112F on the SR-BI mRNA and protein levels to determine how the S112F mutation affected SR-BI, and the results showed that the levels of both the SR-BI mRNA and protein decreased when Ser112 was replaced with Phe. Then, we detected the ability of SR-BI S112F to bind to HCV E2 and showed that the binding ability decreased, potentially due to the decreased level of the SR-BI protein. Finally, since the S112F mutation is associated with an abnormal HDL level and HCV replication is closely correlated with lipid metabolism, we measured the HDL uptake ability of SR-BI S112F and observed a decrease compared with SR-BI WT.
We performed a literature search to determine why the S112F mutant significantly decreased the expression of SR-BI and found that Ser112 is located in the extracellular domain of SR-BI. SR-BI and LIMP II belong to the same family and share 34% sequence identity and 56% sequence homology. The X-ray crystal structure of the extracellular domain of human LIMP II has been solved. Therefore, we used the LIMP II structure as a guide to generate a homology model of human SR-BI. Ser112 in SR-BI is located in a hydrophilic pocket, which is conserved in SR-BI. If the serine (hydrophilic amino acid) is mutated to phenylalanine (hydrophobic amino acid), this hydrophilic pocket will be destroyed. Thus, the protein will not fold correctly, which might be responsible for the downregulation of SR-BI expression in cells expressing the SR-BI S112F mutant.
In summary, we constructed the S112F single amino acid SR-BI mutant and analyzed the effects of this mutant on HCV entry and infectivity. The S112F single amino acid mutant decreased the levels of the SR-BI mRNA and protein and subsequently reduced the binding of SR-BI to HCV E2 protein, as well as the SR-BI-dependent HCV entry and infectivity of HCVcc. In this study, Huh-7-siSR-BI cells expressing SR-BI S112F also showed decreased HDL uptake, but the effects of this mutation on the release of progeny viruses require further investigation.
Human class B scavenger receptor I (SR-BI) is an important receptor associated with host lipid metabolism, and both SR-BI itself and its ligands also play crucial roles in the life cycle of hepatitis C virus (HCV). Single nucleotide polymorphisms (SNPs) in the host genome that affect the virus-host interaction have received increasing attention in recent years. SNPs in the IL28B gene are associated with HCV prognosis.
SNPs in the SR-BI gene are reported to be associated with the elevated plasma HDL-cholesterol levels and virological response of HCV-infected patients. However, the relation between SR-BI SNPs and HCV infectivity has not yet been clearly identified.
This study is the first to evaluate the effect of the single amino acid polymorphism SR-BI S112F on the infectivity of hepatitis C virus derived from cell culture (HCVcc) using a cell culture model. SR-BI S112F inhibited HCVcc infectivity by downregulating the expression of the SR-BI protein.
Based on the results from the present study, the SR-BI S112F single amino acid mutant affected both HCV entry and infectivity. Future studies of SR-BI SNPs will provide insights into the host-virus interaction and will help identify new treatments for HCV infection.
HCVpp, referred as HCV pseudoparticles, were prepared by inserting HCV envelope proteins into pLenti6 transfer vector, and transfected into HEK 293T cells together with helper plasmids Gag/Pol and Rev. HCVcc, referred as real HCV virus particles in cell culture, is prepared by transfection of whole genomic HCV RNA transcripts into Huh 7 or its derived cell lines.
The manuscript is well written. Authors demonstrated that single amino acid polymorphism of SR-BI S112F decreased the infectivity of HCVcc in SR-BI silenced Huh 7 cells. This is the first study evaluating the effect of SR-BI S112F on the infectivity of HCV even though in the in vitro cell culture system. The study showed that SNPs of SR-BI were critical for HCV infectivity.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Vaughan G, Waheed Y S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Zhang FF
| 1. | Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 221] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 2. | Cox AL. MEDICINE. Global control of hepatitis C virus. Science. 2015;349:790-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 3. | He Y, Zhang J, Zhong L, Chen X, Liu HM, Wan LK, Wang H, Li H, Tian L, Hu JL. Prevalence of and risk factors for hepatitis C virus infection among blood donors in Chengdu, China. J Med Virol. 2011;83:616-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Zhang Q, Qi W, Wang X, Zhang Y, Xu Y, Qin S, Zhao P, Guo H, Jiao J, Zhou C. Epidemiology of Hepatitis B and Hepatitis C Infections and Benefits of Programs for Hepatitis Prevention in Northeastern China: A Cross-Sectional Study. Clin Infect Dis. 2016;62:305-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 5. | Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405-2416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1905] [Cited by in RCA: 1861] [Article Influence: 132.9] [Reference Citation Analysis (0)] |
| 6. | Pelosi LA, Voss S, Liu M, Gao M, Lemm JA. Effect on hepatitis C virus replication of combinations of direct-acting antivirals, including NS5A inhibitor daclatasvir. Antimicrob Agents Chemother. 2012;56:5230-5239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 7. | Poordad F, McCone J Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP; SPRINT-2 Investigators. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1996] [Cited by in RCA: 1980] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 8. | Cheung MC, Walker AJ, Hudson BE, Verma S, McLauchlan J, Mutimer DJ, Brown A, Gelson WT, MacDonald DC, Agarwal K. Outcomes after successful direct-acting antiviral therapy for patients with chronic hepatitis C and decompensated cirrhosis. J Hepatol. 2016;65:741-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 316] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 9. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [PubMed] |
| 10. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [PubMed] |
| 11. | Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 991] [Cited by in RCA: 941] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 12. | Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 774] [Cited by in RCA: 729] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 13. | Van Eck M, Hoekstra M, Out R, Bos IS, Kruijt JK, Hildebrand RB, Van Berkel TJ. Scavenger receptor BI facilitates the metabolism of VLDL lipoproteins in vivo. J Lipid Res. 2008;49:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Rhainds D, Brissette L. The role of scavenger receptor class B type I (SR-BI) in lipid trafficking. defining the rules for lipid traders. Int J Biochem Cell Biol. 2004;36:39-77. [PubMed] |
| 15. | Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281:18285-18295. [PubMed] |
| 16. | Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518-520. [PubMed] |
| 17. | Murao K, Terpstra V, Green SR, Kondratenko N, Steinberg D, Quehenberger O. Characterization of CLA-1, a human homologue of rodent scavenger receptor BI, as a receptor for high density lipoprotein and apoptotic thymocytes. J Biol Chem. 1997;272:17551-17557. [PubMed] |
| 18. | Gu X, Trigatti B, Xu S, Acton S, Babitt J, Krieger M. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J Biol Chem. 1998;273:26338-26348. [PubMed] |
| 19. | Viñals M, Xu S, Vasile E, Krieger M. Identification of the N-linked glycosylation sites on the high density lipoprotein (HDL) receptor SR-BI and assessment of their effects on HDL binding and selective lipid uptake. J Biol Chem. 2003;278:5325-5332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Shen WJ, Hu J, Hu Z, Kraemer FB, Azhar S. Scavenger receptor class B type I (SR-BI): a versatile receptor with multiple functions and actions. Metabolism. 2014;63:875-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 21. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1706] [Cited by in RCA: 1686] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 22. | Bekker V, Chanock SJ, Yeager M, Hutchinson AA, von Hahn T, Chen S, Xiao N, Dotrang M, Brown M, Busch MP. Genetic variation in CLDN1 and susceptibility to hepatitis C virus infection. J Viral Hepat. 2010;17:192-200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 23. | Kent AP, Stylianou IM. Scavenger receptor class B member 1 protein: hepatic regulation and its effects on lipids, reverse cholesterol transport, and atherosclerosis. Hepat Med. 2011;3:29-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Hsu CS, Hsu SJ, Liu WL, Chen DS, Kao JH. Association of SCARB1 Gene Polymorphisms with Virological Response in Chronic Hepatitis C Patients Receiving Pegylated Interferon plus Ribavirin Therapy. Sci Rep. 2016;6:32303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707-713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3060] [Cited by in RCA: 2837] [Article Influence: 189.1] [Reference Citation Analysis (0)] |
| 26. | Vergeer M, Korporaal SJ, Franssen R, Meurs I, Out R, Hovingh GK, Hoekstra M, Sierts JA, Dallinga-Thie GM, Motazacker MM. Genetic variant of the scavenger receptor BI in humans. N Engl J Med. 2011;364:136-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 264] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 27. | Brunham LR, Tietjen I, Bochem AE, Singaraja RR, Franchini PL, Radomski C, Mattice M, Legendre A, Hovingh GK, Kastelein JJ. Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans. Clin Genet. 2011;79:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | Chadwick AC, Sahoo D. Functional characterization of newly-discovered mutations in human SR-BI. PLoS One. 2012;7:e45660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Gao W, Gao R, Ren H. Construction of recombinant lentivirus and screening of stable cell line with human SR-BI gene knock down. China Tropical Med. 2016;16:203-207. |
| 30. | Qian XJ, Zhang XL, Zhao P, Jin YS, Chen HS, Xu QQ, Ren H, Zhu SY, Tang HL, Zhu YZ. A Schisandra-Derived Compound Schizandronic Acid Inhibits Entry of Pan-HCV Genotypes into Human Hepatocytes. Sci Rep. 2016;6:27268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 31. | Guan M, Wang W, Liu X, Tong Y, Liu Y, Ren H, Zhu S, Dubuisson J, Baumert TF, Zhu Y. Three different functional microdomains in the hepatitis C virus hypervariable region 1 (HVR1) mediate entry and immune evasion. J Biol Chem. 2012;287:35631-35645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Tong Y, Zhu Y, Xia X, Liu Y, Feng Y, Hua X, Chen Z, Ding H, Gao L, Wang Y. Tupaia CD81, SR-BI, claudin-1, and occludin support hepatitis C virus infection. J Virol. 2011;85:2793-2802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 33. | Acton S, Osgood D, Donoghue M, Corella D, Pocovi M, Cenarro A, Mozas P, Keilty J, Squazzo S, Woolf EA. Association of polymorphisms at the SR-BI gene locus with plasma lipid levels and body mass index in a white population. Arterioscler Thromb Vasc Biol. 1999;19:1734-1743. [PubMed] |
| 34. | Picataggi A, Lim GF, Kent AP, Millar JS, Rader DJ, Stylianou IM. A coding variant in SR-BI (I179N) significantly increases atherosclerosis in mice. Mamm Genome. 2013;24:257-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 35. | Papale GA, Nicholson K, Hanson PJ, Pavlovic M, Drover VA, Sahoo D. Extracellular hydrophobic regions in scavenger receptor BI play a key role in mediating HDL-cholesterol transport. Arch Biochem Biophys. 2010;496:132-139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 36. | Wang Y, Maegawa S, Akiyama Y, Ha Y. The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J Mol Biol. 2007;374:1104-1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 37. | Hauser M, Kauffman S, Lee BK, Naider F, Becker JM. The first extracellular loop of the Saccharomyces cerevisiae G protein-coupled receptor Ste2p undergoes a conformational change upon ligand binding. J Biol Chem. 2007;282:10387-10397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 38. | Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 352] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 39. | Blaising J, Pécheur EI. Lipids: a key for hepatitis C virus entry and a potential target for antiviral strategies. Biochimie. 2013;95:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 40. | Lavie M, Voisset C, Vu-Dac N, Zurawski V, Duverlie G, Wychowski C, Dubuisson J. Serum amyloid A has antiviral activity against hepatitis C virus by inhibiting virus entry in a cell culture system. Hepatology. 2006;44:1626-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 41. | Prentoe J, Serre SB, Ramirez S, Nicosia A, Gottwein JM, Bukh J. Hypervariable region 1 deletion and required adaptive envelope mutations confer decreased dependency on scavenger receptor class B type I and low-density lipoprotein receptor for hepatitis C virus. J Virol. 2014;88:1725-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 42. | Catanese MT, Ansuini H, Graziani R, Huby T, Moreau M, Ball JK, Paonessa G, Rice CM, Cortese R, Vitelli A. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 121] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 43. | Syder AJ, Lee H, Zeisel MB, Grove J, Soulier E, Macdonald J, Chow S, Chang J, Baumert TF, McKeating JA. Small molecule scavenger receptor BI antagonists are potent HCV entry inhibitors. J Hepatol. 2011;54:48-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 44. | Zhu H, Wong-Staal F, Lee H, Syder A, McKelvy J, Schooley RT, Wyles DL. Evaluation of ITX 5061, a scavenger receptor B1 antagonist: resistance selection and activity in combination with other hepatitis C virus antivirals. J Infect Dis. 2012;205:656-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |