Published online Jul 7, 2017. doi: 10.3748/wjg.v23.i25.4538
Peer-review started: January 14, 2017
First decision: February 9, 2017
Revised: February 24, 2017
Accepted: March 4, 2017
Article in press: March 6, 2017
Published online: July 7, 2017
Processing time: 175 Days and 17.1 Hours
To determine the number of mutations in the NS5A region of the hepatitis C virus (HCV) and its relationship to the response to antiviral therapy in patients with chronic hepatitis C genotype 1 who are non-responders to two or more treatments.
Sequences within HCV NS5A [PKR binding domain (PKRBD) and the interferon-sensitivity-determining region (ISDR)] were analysed via direct sequencing in a selected cohort of 72 patients, with a total of 201 treatments [interferon-alpha (IFN-α), n = 49; IFN-α + ribavirin (RBV), n = 75; pegylated (peg) IFN-α + RBV, n = 47; first-generation direct-acting antivirals (DAAs), n = 13; and second-generation DAAs, n = 17]. Of these, 48/201 achieved a sustained virological response (SVR) and 153/201 achieved no virological response (NVR).
For both regions, treatments resulting in SVR were associated with more baseline mutations than were treatments resulting in NVR (SVR vs NVR; PKRBD: 5.82 ± 3 vs 4.86 ± 2 mutations, P = 0.045; ISDR: 2.65 ± 2 vs 1.51 ± 1.7 mutations, P = 0.005). A decrease or no change in the number of mutations over time between treatments in the PKRBD or ISDR, as shown by sequencing, was associated with patients who usually failed to respond to treatment (PKRBD, P = 0.02; ISDR, P = 0.001). Moreover, patients showing a post-treatment baseline viral load > 600000 IU/mL and increased ISDR mutations with respect to the previous treatment were 9.21 times more likely to achieve SVR (P = 0.001).
The obtained results show that among patients who have shown no response to two or more antiviral treatments, the likelihood of achieving SVR increases with the genetic variability in the ISDR region (≥ 2 mutations or number of substitutions from the HCV-J and HCV-1 prototype), especially when the viral load is greater than 600000 IU/mL.
Core tip: To the best of our knowledge, this study is the first to evaluate the change in the number of mutations of the hepatitis C virus (HCV) NS5A region in patients with chronic hepatitis C genotype 1 and who were non-responders to two or more treatments over a period of two decades. The number of mutations in the HCV NS5A region was identified as an independent predictor of a sustained virological response regardless of the interferon-containing regimen or direct-acting antiviral-containing interferon-free protocol applied.
- Citation: Muñoz de Rueda P, Fuentes Rodríguez JM, Quiles Pérez R, Gila Medina A, Martín Álvarez AB, Casado Ruíz J, Ruíz Extremera A, Salmerón J. Hepatitis C virus NS5A region mutation in chronic hepatitis C genotype 1 patients who are non-responders to two or more treatments and its relationship with response to a new treatment. World J Gastroenterol 2017; 23(25): 4538-4547
- URL: https://www.wjgnet.com/1007-9327/full/v23/i25/4538.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i25.4538
The hepatitis C virus (HCV) is a globally prevalent human pathogen. This infection currently affects more than 170 million people worldwide, corresponding to approximately 3% of the world’s population[1], and the prevalence has increased by 2.3% since 1990[2]. HCV leads to chronic liver conditions, such as hepatitis, cirrhosis and hepatocellular carcinoma[3-6]. HCV encodes its own RNA-dependent RNA polymerase, which lacks 3’-to-5’ exonuclease proofreading activity; as a result, the HCV genome is constantly mutating. The combination of a high HCV replication rate, the low fidelity of its RNA polymerase and selective pressures from the immune system and drug treatments have led to the development of highly diverse viral sequences[7,8]. These high mutation rates, together with a restricted genome dimension and large population size, provide the perfect conditions for rapid evolution to occur.
This genetic diversity plays an important role in the virus’s ability to evade the selective pressures exerted by immune responses and antiviral therapies, which is the main cause of the therapy-related problems[9]. In addition, the different HCV genotypes show different treatment responses, and these determine the treatment strategy and patient outcome. Until 2011, the standard treatment for chronic hepatitis C (CHC) was based on interferon-alpha (IFN-α), but in recent years, new direct-acting antivirals (DAAs) that specifically target essential viral functions have been introduced into treatment regimens for HCV genotype 1 infections, in combination with IFN-based therapies and, subsequently, with IFN-free regimens.
The sustained virological response (SVR) rates to the different treatments for patients with CHC genotype 1 (CHC-1) range from 5%-20%, 40%-45%, 50% and > 90% for IFN-α, IFN-α + ribavirin (RBV), pegylated (peg)-IFN + RBV and DAAs, respectively[10-17]. The high degree of genetic diversity of HCV and the various mutations at different positions in the genes for the NS3 protease, NS5B polymerase and NS5A protein may be associated with viral sensitivity or resistance to IFN-based therapy and IFN-free therapy. Several studies have highlighted the influence of sequence heterogeneity within particular regions of the HCV genome, such as the core, E2, NS3 and NS5A regions, on the outcome of IFN-based therapy.
Of these regions, the one coding for NS5A has been the most extensively studied, due to its relationship with IFN responsiveness. This multifunctional phosphoprotein modulates HCV replication, which is mediated through interactions with other viral proteins and certain host proteins to form the HCV replication complex and regulate host cell functions, including responses to IFN and viral pathogenesis[18]. Several studies have associated an increase in the number of mutations in this region compared to the number in the HCV-J subtype 1b prototype with a better response to antiviral therapy[19-25].
In this study, we analysed HCV genetic diversity in the NS5A region in a cohort of 72 CHC-1 patients who were non-responders to their first treatment and the relationship between this diversity and subsequent treatment outcomes (2 to 8 consecutive treatments). These patients were treated with several different therapies over the course of more than two decades, and the analysis was conducted on archived serum samples that were obtained before each retreatment.
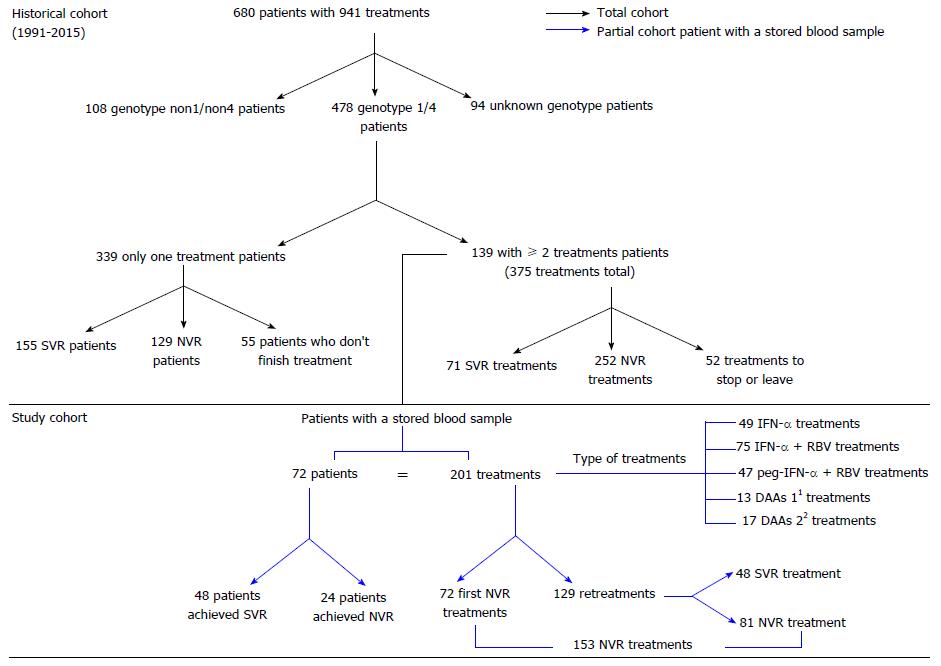
A retrospective study of a prospective cohort was conducted at San Cecilio University Hospital (Granada, Spain) from 1991 until 2015 (Figure 1). A total of 680 patients with CHC were treated with 941 anti-HCV treatments (historical cohort). Of these patients, 478 had genotype 1/4 HCV, of whom 139 received two or more anti-HCV treatments. A total of 375 treatments were applied. The study cohort was selected by applying the following inclusion criteria: patients with genotype 1/4 HCV, two or more anti-HCV treatments, and blood samples stored at -80 °C. A total of 72 patients with stored blood samples, having received a total of 201 treatments, were included in the study (Study cohort; Figure 1).
A total of 48/72 patients (66.7%) achieved SVR after several treatments. Of the 201 treatments, 153 resulted in no virological response (NVR), and 48 led to SVR. The outcomes of treatments resulting in NVR were defined as follows: relapse (R), treatment with serum HCV RNA undetectable in patients at 12 wk but detectable subsequently; partial virological response (pVR), treatment with a low viral load > 2log in patients at 12 wk; null response (NR), treatment with a low viral load < 2log in patients at 12 wk; and, unknown, i.e., treatments for patients whose reason for NR was not stated in their medical history.
The study protocol conformed to the ethical guidelines of the 1975 Helsinki Declaration, and all patients involved in the study were informed verbally and in writing of the characteristics of the study and provided their consent to participate by signing an informed consent form. This study was approved by the Ethics Committee of the San Cecilio University Hospital.
HCV genomes were analysed in serum samples obtained before the patients were subjected to treatments involving different antiviral therapies. The sequences of viruses from baseline serum samples associated with 201 treatments were amplified; samples that failed long RT-PCR amplification were excluded (n = 16). The amplified HCV genome region corresponded to a fragment from gene NS5A (nucleotides 6966 to 7164 in the reference HCV genome sequence, GenBank accession number AF009606), including the PKR binding domain (PKRBD) and the interferon-sensitivity-determining region (ISDR). These regions include structural motifs related to antiviral treatment.
Viral RNA was extracted from 300 μL of serum using the PrepitoViral DNA/RNA 300 Kit (Chemagen Technologie, PerkinElmer, Madrid, Spain). Two microlitres of the RNA solution were subjected to reverse transcription using a qScript Flex cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD, United States). PKRBD sequences were amplified via RT-PCR using SYBR Green Supermix Low ROX (Quanta Biosciences). The primers used for reverse transcription and first and second round amplifications of the NS5A region (nt 6966-7164) are shown in Table 1.
| Application | Direction | Sequence |
| First PCR | Sense | 5´-ACAACTCCCATGCGAGCCCGAGCCGGA-3´ |
| RT and first PCR | Antisense | 5´-GAGGGGGAGCCGGGGGATCCCGATCTC-3´ |
| Second PCR | Sense | 5´-TGCTCACCGACCCATCCCACATTACAGCAGA-3´ |
| Antisense | 5´-ATGCCCCCTCTCGAGGGGGAGCCGGG-3´ |
RT was performed at 45 °C for 30 min and terminated at 85 °C for 5 min, followed by the first-round PCR over 35 cycles, with each cycle consisting of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 70 s. The second-round PCR was performed under the same conditions. The sequences of the amplified fragments were determined via direct sequencing. The second-round PCR product was purified using the Wizard SV Gel and the PCR Clean-Up System (Promega, Promega Biotech Ibérica, Madrid, Spain) and resuspended in 20 μL of water. Next, 2-5 μL of purified PCR product was sequenced in a 3130 XL Genetic Analyzer automatic sequencer (Applied Biosystems, Foster City, CA, United States).
All of the nucleotide sequences obtained in the present study were aligned. To detect amino acid substitutions, sequences were aligned with reference strands, HCV-J (GenBank accession number D90208) for genotype 1b and HCV-1 (GenBank accession number M62321) for genotype 1a, and translated into amino acid sequences with Mega 5.0 software (Molecular Evolutionary Genetics Analysis)[26].
Quantitative variables are expressed as the means ± SD, and qualitative variables are expressed as absolute values with percentages. The χ2 test was used for qualitative variables. Comparisons of continuous variables with a non-normal distribution were performed using the Mann-Whitney U test or Kruskal-Wallis test. The degree of association between dependent and independent variables was determined by calculating the crude odds ratio and its 95%CI (bivariate analysis). Interaction studies were performed using logistic regression. Synergy studies were conducted in accordance with the method of Andersson et al[27]. The criterion for statistical significance was P ≤ 0.05. All statistical analyses were performed using SPSS 15.0 for Windows software (SPSS Inc, Chicago, IL, United States).
The characteristics of the historical cohort (n = 680 patients) are shown in Table S1. After application of the inclusion criteria, 72 patients were selected [45/72 (63.5%) were men], with a total of 201 treatments (72 first treatments and 129 retreatments; Figure 1). The number of treatments received was distributed as follows: 2 treatments (n = 35, 48.6%), 3 treatments (n = 24, 33.3%), 4 treatments (n = 9, 12.5%), 5 treatments (n = 3, 4.2%) or 8 treatments (n = 1, 1.4%). The following anti-HCV therapies were provided: IFN-α alone (24%, n = 49), IFN-α + RBV (37%, n = 75), peg-IFN-α + RBV (23%, n = 47) and DAAs (16%, n = 30) (including first-generation DAAs: peg-IFN-α + RBV + telaprevir 24% and peg-IFN-α + RBV + boceprevir, 20%; and second-generation DAAs: peg-IFN-α + RBV + simeprevir, 7%; simeprevir + sofosbuvir + RBV, 7%; simeprevir + sofosbuvir, 3%; sofosbuvir + daclatasvir, 3%; ledipasvir + sofosbuvir + RBV, 13%; ledipasvir + sofosbuvir, 13% and ombitasvir/paritaprevir/ritonavir + dasabuvir, 10%).
One patient was treated with eight different therapies (one treatment with IFN-α (Wellferon; the Wellcome Foundation Ltd, part of GlaxoSmithKline PLC, Brentford, United Kingdom), three with IFN-α + RBV, two with peg-IFN-α + RBV, one with peg-IFN-α + RBV + telaprevir and one with simeprevir + sofosbuvir + RBV) and finally achieved SVR. Of the 72 patients, 66.7% (n = 48) achieved SVR, while 33.3% (n = 24) did not respond to any of the treatments received. In the latter scenario, the clinical decision in almost all cases was not to perform a retreatment until a new treatment became available. Regarding the 201 total treatments in our cohort, 24% (48 treatments) achieved SVR, while 76% (153 treatments) showed NVR. The types of NVR in the 153 treatments were as follows: R, 21% (n = 32); pVR, 8% (n = 13); NR, 34% (n = 52); and unknown 37% (n = 56).
We then analysed how the prior type of SVR or NVR affected subsequent treatment responses (Table 2), and we found that previously treated patients showing R were 6.09 times more likely to respond to a new treatment than were patients with a previous NR (P = 0.006). In our study cohort, we also analysed the response achieved according to the type of therapy employed (Table 3). As expected, retreatments based on therapies introduced before the use of DAAs were significantly less likely to achieve SVR than were those based on DAAs.
| Previous response | NVR, n = 46 | SVR, n = 32 | OR (95%CI) | P value |
| Relapse | 7 (30) | 16 (70) | 6.09 (2.0-18.4) | 0.006 |
| Partial response viral | 7 (64) | 4 (36) | 1.5 (0.3-6.1) | n.s. |
| Null response | 32 (73) | 12 (27) | 1 |
| Total, n = 129 | NVR, n = 81 | SVR, n = 48 | OR (95%CI) | P value | |
| IFN | 16 (12) | 14 (88) | 2 (12) | 0.05 (0.01-0.3) | 0.001 |
| IFN + RBV | 40 (31) | 27 (68) | 13 (32) | 0.2 (0.06-0.5) | 0.001 |
| pegIFN + RBV | 43 (34) | 32 (74) | 11 (26) | 0.12 (0.04-0.4) | 0.00 |
| DAAs | 30 (23) | 8 (27) | 22 (73) | 1 |
When we distinguished between first- and second-generation DAA treatments, as expected, logistic regression analysis showed that first-generation DAA treatments were significantly less likely to achieve SVR than second-generation DAA treatments (OR = 0.16, 95%CI: 0.026-0.97, P = 0.04). Regarding this analysis, it should be noted that the patients recruited from the historical cohort who responded to the initial treatment (treatment-naïve patients) were not included in the study cohort, which accounts for the low rates of SVR observed in the study. To avoid bias in the analysis, the first 72 treatments were removed from the analysis as these all involved NVR according to the application of the inclusion criteria, regardless of the treatment received.
The change from one type of therapy to another in retreatments increased the probability of achieving SVR. In our cohort, of the total of 129 retreatments, 82 were based on a change to a more effective therapy, while 47 involved no change in therapy. In neither case was the difference in responses statistically significant (data not shown). It should be noted that the 2 patients who did not respond to treatment with second-generation DAAs (relapsers) previously showed NVR to treatment with first-generation DAAs.
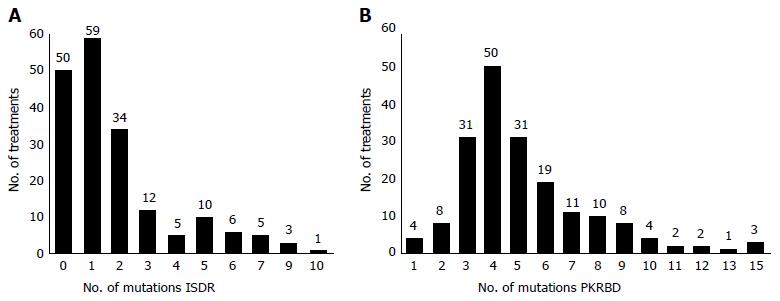
The genetic evolution of the HCV NS5A-PKRBD and NS5A-ISDR regions in the course of the different treatments received was investigated using 201 baseline samples from each treatment, of which 16 were subsequently excluded due to non-amplification (total sample, n = 185). Figure 2 shows the number of mutations found in each region for the 185 amplified samples. The mean number of mutations in the PKRBD region (min-max = 1-15) compared to the number in the reference sequence HCV-J and HCV-1 was 5.19 ± 2.6; in the ISDR (min-max = 0-10), this number was 1.85 ± 2.1.
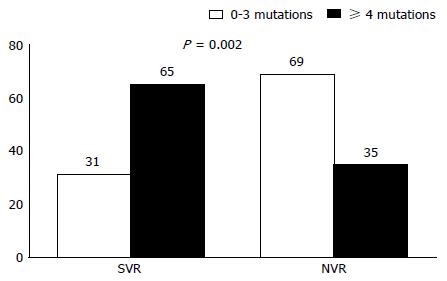
With respect to the association between the number of mutations and the response to antiviral treatment, the treatments that resulted in SVR were associated with significantly more baseline mutations in both regions than were the treatments resulting in NVR (SVR vs NVR; PKRBD: 5.82 ± 3 vs 4.86 ± 2 mutations, P = 0.045; ISDR: 2.65 ± 2 vs 1.51 ± 1.7 mutations, P = 0.005). When we distinguished patient outcomes by type of NVR (R, pVR, NR), significant differences were found in the ISDR region (P = 0.029; Table 4), with R patients presenting fewer mutations than SVR patients (P = 0.025). In the ISDR region, and applying the criteria recommended by Enomoto et al[19], we found that treatments associated with 4 or more mutations in the baseline sample showed an SVR rate of 65%, compared to 31% for those associated with 0-3 baseline mutations (P = 0.002; Figure 3).
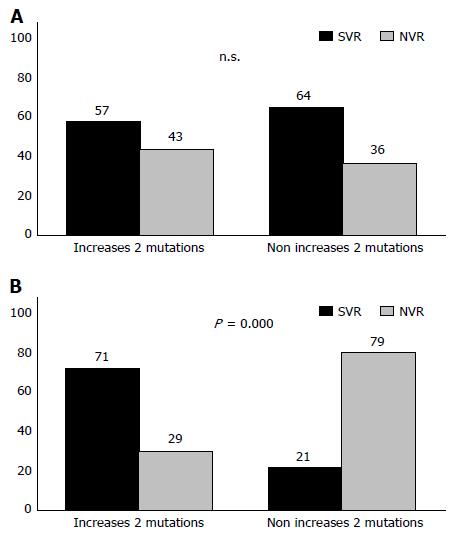
With respect to changes in the viral population during the time between one treatment and another and its relationship with the response to antiviral therapy, our analysis shows that when there was no change in the number of mutations both in the PKRBD and in the ISDR, or when this number decreased, between one treatment and another, the patient tended to be unresponsive to antiviral therapy (PKRBD, P = 0.02; ISDR, P = 0.001) (Table 5). From a clinical standpoint, we studied the viral variability of the ISDR region of the HCV between one treatment and another to determine how this variability was associated with the response or non-response to antiviral therapies (i.e., its use as a diagnostic test), and the following diagnostic values were obtained: 33% sensitivity, 90% specificity, 68% positive predictive value and 70% negative predictive value.
Viral load is another variable that can be considered a predictor of the treatment response. In our cohort study, logistic regression analysis showed that treatments for patients with a baseline viral load ≤ 600000 IU/mL were 3.3 times more likely to result in SVR (OR = 3.3, 95%CI: 1.4-7.7, P = 0.005). However, the analysis also showed that the variation in response according to baseline viral load was influenced by the change in the number of mutations between one treatment and another (Figure 4). Thus, when the treatments were associated with a low baseline viral load (≤ 600000 IU/mL), a response was usually achieved, whereas for treatments associated with a baseline viral load > 600000 IU/mL, depending on the genetic variability of the virus, a response may or may not be achieved; when the number of mutations had increased since the previous treatment, there was a 71% probability of SVR (P ≤ 0.0001).
Using logistic regression, we then sought to corroborate these results for the interaction between the two variables (P = 0.023) (Table 6). Treatments in which the patient had a baseline viral load of > 600000 IU/mL were 9.21 times more likely to achieve SVR if there had been an increase in the number of mutations in the ISDR region since the earlier treatment (P = 0.001). However, in those treatments in which the patient’s baseline viral load was < 600000 IU/mL, the increase in the number of mutations did not affect the response (P = 0.75) (Table 6). No synergy was detected between the two variables [relative excess risk due to interaction (RERI) = -6.052; proportion attributable to the interaction (PA) = -0.74; synergy index, S = 0.54].
| Viral > 600000 IU/mL | Viral ≤ 600000 IU/mL | |
| Decrease or no increase in mutations | 1 | 1 |
| ≥ 2 increase in mutations | OR = 9.21 (2.5-33.3) | OR = 0.76 (0.13-4.3) |
| P = 0.001 | P = 0.75 |
IFN-based therapies have been used for the treatment of CHC for more than two decades. Since 2011, the incorporation of first-generation DAAs (boceprevir and telaprevir) into the standard treatment with pegIFN-α + RBV and, currently, the IFN-free therapies using various combinations of second-generation DAAs (simeprevir, sofosbuvir, daclatasvir, ledipasvir, ombitasvir/paritaprevir/ritonavir and dasabuvir) has proved to be a breakthrough in the treatment of HCV-1, achieving SVR rates of 90%-95% in naïve and retreated patients[16,17]. In addition to the incorporation of new antiviral molecules, a greater understanding of the viral and host factors involved in treatment outcomes has contributed to therapeutic advances. With respect to viral factors, numerous studies have reported a correlation between heterogeneity of the NS5A region of HCV-1 and treatment outcomes[19-25].
In 1995, Enomoto et al[19] demonstrated the clinical influence of NS5A sequence heterogeneity on responses to IFN treatment. These authors identified what they termed the IFN sensitivity-determining region (ISDR), located within the PKRBD region of the NS5A of HCV-1b, and found that Japanese patients presenting four or more mutations in the ISDR (ISDR ≥ 4) relative to the sequence of the HCV-1b prototype (HCV-J) successfully responded to IFN therapy, whereas patients with three or fewer mutations (ISDR ≤ 3) were non-responders. Subsequently, Pascu et al[21] confirmed the importance of the ISDR in determining IFN treatment outcome in European and Japanese patients infected with HCV-1b and treated with IFN monotherapy. For peg-IFN + RBV therapy, the predictive value of the ISDR remained significant, but the criterion of ISDR ≥ 4 to predict SVR was replaced by ISDR ≥ 2[28-30] due to the selective impact of IFN monotherapy, whereby the prevalence of sensitive isolates with ISDR ≥ 4 decreased and that of HCV isolates of ISDR ≤ 3 increased. Therefore, the isolates with ISDR ≥ 2 were selected as sensitive isolates and those with ISDR ≤ 1 as resistant isolates for peg-IFN plus RBV therapy[22,23,30]. On the basis of these criteria, we assumed an increased genetic variability in the majority HCV sequence when the number of ISDR mutations was ≥ 2 throughout this study.
This paper describes a retrospective study of a selected cohort of 72 patients with CHC-1 who received two or more antiviral treatments (specifically, 2-8 treatments). Thus, it represents a study of HCV evolution in response to retreatments conducted over the course of two decades (1991-2015) for a group of patients for whom clinical treatment was problematic and who had achieved very low response rates. No previous studies with these particular characteristics have been performed. As expected, our results show that patients who relapsed following a previous treatment were more likely to respond to retreatment than were null responders[31]. Moreover, treatments with second-generation DAAs achieved a much higher response rate than previous IFN-based treatments (including first-generation DAAs), both in naïve patients and in difficult-to-treat patients who were retreated[16,17].
The results of our analysis of the number of mutations in the NS5A region corroborate previous findings that a higher number of mutations in the PKRBD and ISDR in the majority HCV sequence is associated with a greater probability that SVR will be achieved[19-25]. However, when we differentiated NVR treatment outcomes into R, NR and pVR, the number of mutations in the PKRBD lost statistical significance (although this was not the case in the ISDR). The R patients showed fewer mutations than the SVR patients, as is to be expected in NR patients[32,33]. Previous studies of the number of mutations also analysed the influence of quasispecies. Sakuma et al[33] demonstrated that during treatment with IFN-α, NR patients show greater numbers of minority quasispecies that present the smallest number of mutations in the ISDR region (non-mutant or wild-type quasispecies), while SVR patients have increased numbers of quasispecies with ≥ 3 ISDR mutations prior to treatment. These data are corroborated by other studies, which show that R patients have viruses with a greater number of mutations in the ISDR than do NR patients, both before and after antiviral treatment[32]. By contrast, no studies have reported how the genetic variability of the HCV evolves in patients who undergo various treatments over a period of two decades, as in the present study.
It should be noted that we did not perform a study of quasispecies but rather of the evolution of the genetic variation of the majority ISDR sequence in patients subjected to two or more treatments over time. Taking into account the findings of previous studies[22,23,30], we assumed that viral variability would increase when the PKRBD and ISDR regions presented ≥ 2 mutations. Thus, our results show that patients who have undergone different treatments and who do not present viruses with genetic variability (either no increase or a return to the wild-type sequence) tend not to respond to a new treatment, regardless of its overall effectiveness.
In a similar study, Watanabe et al[34] analysed a 44-year-old patient with CHC-1 and a high baseline viral load who received two IFN-α treatments and was NR in each case. Between the first and the second treatment, the patient’s baseline viral load decreased, and the number of ISDR mutations increased from one to six. At this stage, a new treatment was performed, and a rapid virological response was achieved. In view of these results, the authors concluded that retreatment should be considered for NR and R patients when the viral load falls and when the number of mutations in ISDR increases, as the chances of achieving SVR would then be higher.
In our cohort study, we found similar results in a patient who had undergone eight different treatments over a period of 22 years; on five occasions, he showed NR, on one a R, on one a breakthrough and then showed SVR after the final treatment. During the time elapsed between the first and second treatments, the number of mutations in the PKRBD region decreased by four, while in the ISDR region, the number remained unchanged. In both cases, the number of mutations remained unchanged for 19 years, at which point they increased in both regions, and SVR then was achieved.
Conversely, Ibarrola et al[35] analysed 62 patients, 42 of whom showed NR and 20 a R in response to previous treatments with IFN-α, who were then retreated with IFN-α + RBV. In 38 of these patients, an analysis was performed on the relationship between the mutations in the ISDR region before treatment and the SVR achieved. There was no correlation between the presence of mutations in and around the ISDR region and the response to the new treatment, although the number of patients was rather low for a reliable evaluation of these results. In addition to the predictive response value of the genetic variability in the PKRBD and ISDR regions, which has been amply demonstrated[19-25], many other factors also influence the response, one of which is the baseline viral load[36].
According to previous research, ISDR sequences with ≥ 4 mutations are correlated with a low viral load[20,37,38], although Nakagawa et al[30] found that more patients had ≥ 2 ISDR mutations and higher virus loads. Logistic regression analysis showed that both factors were independent variables to predict SVR, although the number of mutations within the ISDR was associated with a higher OR and was a predictive factor associated with SVR by multivariate analysis[30]. Our results indicate that patients with CHC-1, subjected to retreatment and with a high baseline viral load (≥ 600000 IU/mL) and who are classified as difficult to treat because of the low response rate obtained, will respond to new treatment depending on the evolution of viral genetic variability. Thus, if there is no change in the majority sequence of the viral population despite the application of different types of HCV treatment or if it tends to revert to the original status or to a wild-type sequence, the patient will not respond. Conversely, if there is an increase in the genetic variability of the ISDR region, despite the presence of a high viral load, the patient will have a greater likelihood of achieving SVR with a new treatment.
In conclusion, when the evolution of genetic variability in the ISDR region of HCV increases in patients with CHC-1 who have received two or more antiviral treatments and who have a high viral load, there is a greater probability of these patients achieving SVR with a new treatment. The present study shows that the genetic diversity of the HCV genome during decades of infection continues to be an independent predictor of SVR regardless of the treatment applied.
When antiviral treatments are used against hepatitis C virus (HCV), some patients are non-responders (NR). Our aim in this study was to analyse the change of mutations in the NS5A-interferon-sensitivity-determining region and PKR binding domain regions in patients with chronic hepatitis C genotype 1 and who showed NR to one or more treatment cycles over time. We also analysed the relationship between this change and sustained virologic response (SVR) to a new treatment.
To our knowledge, this is the first study providing data about how the genetic variability of HCV evolves in patients who undergo various treatments over a period of two decades.
This study represents a significant contribution to knowledge of the genetic diversity of the NS5A protein from HCV and its relationship to the treatment response in patients for whom clinical treatment is problematic and who achieve very low response rates.
This report shows that the genetic diversity of the HCV genome, during decades of infection, continues to be an independent predictor of SVR, regardless of the treatment applied. No previous studies with these particular characteristics have been performed.
This manuscript presents a single-centre, retrospective, observational study of the genetic evolution of HCV genotype 1 in patients who received antiviral therapies but were NR to prior therapies. The major strength of this manuscript is its relevance to contemporary practice, and the results could be applied to clinical decisions regarding therapeutic strategies to treat non-responders who receive novel DAAs.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Gigi E, Lu K, Zhao HT S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Li D
| 1. | Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 939] [Article Influence: 58.7] [Reference Citation Analysis (1)] |
| 2. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1770] [Cited by in RCA: 1847] [Article Influence: 153.9] [Reference Citation Analysis (3)] |
| 3. | Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:15S-20S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 481] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 4. | Di Bisceglie AM. Hepatitis C. Lancet. 1998;351:351-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 267] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 5. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1764] [Cited by in RCA: 1843] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 6. | Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 633] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 7. | Zeuzem S. Clinical implications of hepatitis C viral kinetics. J Hepatol. 1999;31 Suppl 1:61-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Palanisamy N, Danielsson A, Kokkula C, Yin H, Bondeson K, Wesslén L, Duberg AS, Lennerstrand J. Implications of baseline polymorphisms for potential resistance to NS3 protease inhibitors in Hepatitis C virus genotypes 1a, 2b and 3a. Antiviral Res. 2013;99:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Figlerowicz M, Alejska M, Kurzyńska-Kokorniak A, Figlerowicz M. Genetic variability: the key problem in the prevention and therapy of RNA-based virus infections. Med Res Rev. 2003;23:488-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Carithers RL, Emerson SS. Therapy of hepatitis C: meta-analysis of interferon alfa-2b trials. Hepatology. 1997;26:83S-88S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 166] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 11. | Zeuzem S, Feinman SV, Rasenack J, Heathcote EJ, Lai MY, Gane E, O’Grady J, Reichen J, Diago M, Lin A. Peginterferon alfa-2a in patients with chronic hepatitis C. N Engl J Med. 2000;343:1666-1672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 851] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 12. | Poynard T, Marcellin P, Lee SS, Niederau C, Minuk GS, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT). Lancet. 1998;352:1426-1432. [PubMed] |
| 13. | McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2509] [Cited by in RCA: 2434] [Article Influence: 90.1] [Reference Citation Analysis (0)] |
| 14. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4847] [Cited by in RCA: 4748] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 15. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [PubMed] |
| 16. | Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med. 2014;370:211-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 911] [Article Influence: 82.8] [Reference Citation Analysis (0)] |
| 17. | Lawitz E, Poordad FF, Pang PS, Hyland RH, Ding X, Mo H, Symonds WT, McHutchison JG, Membreno FE. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383:515-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 443] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 18. | Polyak SJ, Paschal DM, McArdle S, Gale MJ, Moradpour D, Gretch DR. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 113] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 452] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 20. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 734] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 21. | Pascu M, Martus P, Höhne M, Wiedenmann B, Hopf U, Schreier E, Berg T. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut. 2004;53:1345-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 109] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | El-Shamy A, Shoji I, Saito T, Watanabe H, Ide YH, Deng L, Kawata S, Hotta H. Sequence heterogeneity of NS5A and core proteins of hepatitis C virus and virological responses to pegylated-interferon/ribavirin combination therapy. Microbiol Immunol. 2011;55:418-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 23. | Hayashi K, Katano Y, Ishigami M, Itoh A, Hirooka Y, Nakano I, Urano F, Yoshioka K, Toyoda H, Kumada T. Mutations in the core and NS5A region of hepatitis C virus genotype 1b and correlation with response to pegylated-interferon-alpha 2b and ribavirin combination therapy. J Viral Hepat. 2011;18:280-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Shirakawa H, Matsumoto A, Joshita S, Komatsu M, Tanaka N, Umemura T, Ichijo T, Yoshizawa K, Kiyosawa K, Tanaka E. Pretreatment prediction of virological response to peginterferon plus ribavirin therapy in chronic hepatitis C patients using viral and host factors. Hepatology. 2008;48:1753-1760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 25. | Muñoz de Rueda P, Casado J, Patón R, Quintero D, Palacios A, Gila A, Quiles R, León J, Ruiz-Extremera A, Salmerón J. Mutations in E2-PePHD, NS5A-PKRBD, NS5A-ISDR, and NS5A-V3 of hepatitis C virus genotype 1 and their relationships to pegylated interferon-ribavirin treatment responses. J Virol. 2008;82:6644-6653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-2739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31130] [Cited by in RCA: 28337] [Article Influence: 2024.1] [Reference Citation Analysis (0)] |
| 27. | Andersson T, Alfredsson L, Källberg H, Zdravkovic S, Ahlbom A. Calculating measures of biological interaction. Eur J Epidemiol. 2005;20:575-579. [PubMed] |
| 28. | Murayama M, Katano Y, Nakano I, Ishigami M, Hayashi K, Honda T, Hirooka Y, Itoh A, Goto H. A mutation in the interferon sensitivity-determining region is associated with responsiveness to interferon-ribavirin combination therapy in chronic hepatitis patients infected with a Japan-specific subtype of hepatitis C virus genotype 1B. J Med Virol. 2007;79:35-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 29. | Murphy MD, Rosen HR, Marousek GI, Chou S. Analysis of sequence configurations of the ISDR, PKR-binding domain, and V3 region as predictors of response to induction interferon-alpha and ribavirin therapy in chronic hepatitis C infection. Dig Dis Sci. 2002;47:1195-1205. [PubMed] |
| 30. | Nakagawa M, Sakamoto N, Ueyama M, Mogushi K, Nagaie S, Itsui Y, Azuma S, Kakinuma S, Tanaka H, Enomoto N. Mutations in the interferon sensitivity determining region and virological response to combination therapy with pegylated-interferon alpha 2b plus ribavirin in patients with chronic hepatitis C-1b infection. J Gastroenterol. 2010;45:656-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Spadaro A, Freni MA, Ajello A, Alessi N, Barbaro E, Resta ML, Ferraü O. Interferon retreatment of patients with chronic hepatitis C. A long-term follow-up. Hepatogastroenterology. 1999;46:3229-3233. [PubMed] |
| 32. | Durante Mangoni E, Forton DM, Ruggiero G, Karayiannis P. Hepatitis C virus E2 and NS5A region variability during sequential treatment with two interferon-alpha preparations. J Med Virol. 2003;70:62-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Sakuma I, Enomoto N, Kurosaki M, Izumi N, Marumo F, Sato C. Differential effect of interferon on hepatitis C virus 1b quasispecies in the nonstructural protein 5A gene. J Infect Dis. 1999;180:1001-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 34. | Watanabe K, Yoshioka K, Terazawa Y, Kobayashi M, Ishigami M, Yano M, Fuji A, Hattori M, Kakumu S. A patient with chronic hepatitis C who obtained sustained response by retreatment of interferon after decrease of viral load and mutation in interferon sensitivity determining region. Intern Med. 2001;40:489-492. [PubMed] |
| 35. | Ibarrola N, Moreno-Monteagudo JA, Sáiz M, García-Monzón C, Sobrino F, García-Buey L, Lo Iacono O, Moreno-Otero R, Martínez-Salas E. Response to retreatment with interferon-alpha plus ribavirin in chronic hepatitis C patients is independent of the NS5A gene nucleotide sequence. Am J Gastroenterol. 1999;94:2487-2495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 36. | Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Watahiki S, Sato J. Predictive factors of virological non-response to interferon-ribavirin combination therapy for patients infected with hepatitis C virus of genotype 1b and high viral load. J Med Virol. 2006;78:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 37. | Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 152] [Article Influence: 5.4] [Reference Citation Analysis (0)] |