Published online Jan 14, 2017. doi: 10.3748/wjg.v23.i2.232
Peer-review started: August 23, 2016
First decision: September 5, 2016
Revised: October 12, 2016
Accepted: October 31, 2016
Article in press: October 31, 2016
Published online: January 14, 2017
Processing time: 91 Days and 14.1 Hours
To determine the relationship between five A3G gene single nucleotide polymorphisms and the incidence of hepatitis B virus (HBV) infection and hepatocellular carcinoma (HCC).
This association study was designed as a retrospective study, including 657 patients with chronic HBV infection (CHB) and 299 healthy controls. All subjects were ethnic Han Chinese. Chronic HBV-infected patients recruited between 2012 and 2015 at The First Hospital of Jilin University (Changchun) were further classified into HBV-related HCC patients (n = 287) and non-HCC patients (n = 370). Frequency matching by age and sex was performed for each group. Human genomic DNA was extracted from whole blood. Gene polymorphisms were identified using a mass spectroscopic method.
There were no significant differences between the genotype and allele frequencies of the rs7291971, rs5757465 and rs5757463 A3G gene polymorphisms, and risk of CHB and HBV-related HCC. The AG genotype and G allele for rs8177832 were significantly related to a decreased risk of CHB (OR = 0.67, 95%CI: 0.47-0.96; OR = 0.69, 95%CI: 0.50-0.95, respectively) and HCC (OR = 0.53, 95%CI: 0.34-0.84; OR = 0.58, 95%CI: 0.39-0.87, respectively). A significant relationship was found between rs2011861 computed tomography, TT genotypes and increased risk of HCC (OR = 1.69, 95%CI: 1.02-2.80; OR = 1.82, 95%CI: 1.08-3.06, respectively). Haplotype analyses showed three protective and four risk haplotypes for HCC. Also, one protective haplotype was found against CHB.
This study indicates that the A3G rs8177832 polymorphism is associated with a decreased risk of CHB infection and HCC, while the rs2011861 polymorphism is associated with an increased risk of HCC.
Core tip: A3G is a dominant cytidine deaminase that strongly inhibits synthesis and editing of hepatitis B virus (HBV) DNA. We studied the relationship between five A3G gene single nucleotide polymorphisms and the incidence of chronic HBV infection (CHB) and hepatocellular carcinoma (HCC), including 657 CHB patients (287 HCC and 370 non-HCC) and 299 healthy controls. The AG genotype and G allele for rs8177832 were potentially protective factors against CHB and HCC. Computed tomography and TT genotypes of rs2011861 were risk factors for HCC. Haplotype analyses showed three protective and four risk haplotypes for HCC. Also, one protective haplotype was found against CHB.
- Citation: He XT, Xu HQ, Wang XM, He XS, Niu JQ, Gao PJ. Association between polymorphisms of the APOBEC3G gene and chronic hepatitis B viral infection and hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2017; 23(2): 232-241
- URL: https://www.wjgnet.com/1007-9327/full/v23/i2/232.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i2.232
Hepatitis B virus (HBV) infection is one of the most common infectious diseases of global public health concern. There are more than 240 million chronic HBV carriers today, and about 620000 die per year from end-stage liver cirrhosis (LC) or hepatocellular carcinoma (HCC)[1]. Although HBV infection is a high-risk factor for liver disease, the clinical outcomes after exposure to HBV are highly variable. Genetic and environmental factors both critically modulate the susceptibility and progression of liver disease[2-4]. A number of epidemiologic studies have demonstrated that high alcohol consumption and cigarette smoking are associated with increased HCC risk[5,6].
APOBEC3s (A3s) are components of innate immunity that play an important role in defending against invading viruses, including HBV and human immunodeficiency virus (HIV). In humans, A3s are comprised of seven proteins: APOBEC-3A, -3B, -3C, -3DE, -3F, -3G and -3H[7]. A3s have one or two catalytic domains that have cytidine deaminase activity, which convert cytosine to uracil in DNA[8]. The presence of the enzyme in cells producing DNA viruses results in C to T transitions in negative stranded DNA and G to A transitions in positive stranded DNA during DNA replication without repair pathways[9]. A3s, especially A3G, can inhibit HBV through hypermutation-dependent and -independent mechanisms[10,11]. A3G is one of the most active deaminases, with a strong inhibitory effect on replication and editing of HBV DNA in vivo[12-16]. A3G is expressed widely in human tissues, and its mRNA levels broadly correlate with lymphoid cell content[17]. Levels of A3G were found to be the highest among A3s in human liver tissue[14].
Several studies identified genetic variants that are associated with risk of HIV infection and progression to acquired immune deficiency syndrome[18-20]. Since A3G is an important host factor that may inhibit HBV, we screened the A3G gene for both regulatory and coding region variants that could modify A3G transcription or amino acid sequence. The goal of this study was to evaluate the association of five A3G single nucleotide polymorphisms (SNPs) with the development of chronic HBV and HBV-related HCC in a Chinese Han population.
This association study was designed as a retrospective study, including 657 patients with chronic HBV infection (CHB) and 299 healthy controls. All subjects were ethnic Han Chinese. CHB patients recruited between 2012 and 2015 at The First Hospital of Jilin University (Changchun) were further classified into non-HCC (n = 370) and HBV-related HCC (n = 287) patients. Frequency matching by age and sex was performed for each group. CHB patients were defined by persistent or intermittent elevations in alanine transaminase level (≥ 2 times the upper limit of normal) and elevated HBV DNA levels for at least 6 mo. HBV-related HCC was diagnosed based on (1) positive results on computed tomography (CT), magnetic resonance imaging or ultrasonography; and (2) combined positive findings upon cytological or pathological examination. The non-HCC patients included CHB and LC patients, characterized by active necro-inflammatory liver disease without/with fibrosis on imaging examination without evidence of HCC, according to the guidelines for the prevention and treatment of CHB (2010 version), and the diagnostic criteria (10th National Conference on Viral Hepatitis and Hepatopathy 2000, China). All samples were HBV-positive, but hepatitis C virus (HCV)-, HIV-negative, according to serology tests and infection history. Exclusion criteria included the presence of autoimmune and other liver diseases, alcoholic liver disease, hemorrhagic liver disease, and intra- and extra-hepatic bile duct stones. The criteria for healthy participants included no previous diagnosis of cancer or liver-associated illness. Healthy individuals were recruited from The First Hospital of Jilin University during the same period. All patients were further confirmed as being negative for hepatitis B surface antigen, hepatitis B e antigen (HBeAg), hepatitis B e antibody (HBeAb), hepatitis B virus core antibody, and hepatitis C antibody, as measured by chemiluminescence methods (Roche E411, Basil, Switzerland). We also collected demographic data of each subject, such as smoking and drinking status. Individuals who smoked daily for at least 1 year were defined as smokers, and those who consumed alcoholic drinks more than once per wk for over 6 mo were considered drinkers. Written informed consent was obtained from all patients, and this study was approved by The First Hospital Ethical Committee of Jilin University.
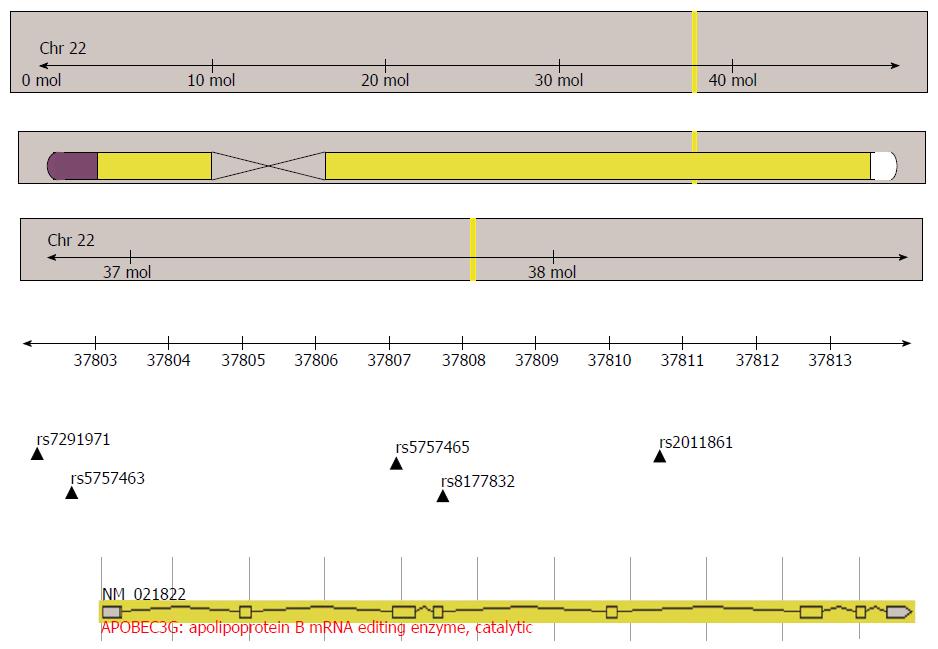
GeneView of NCBI and 1000 Genomes databases were used to select the SNPs in the functional region of A3G (minor allele frequency > 10% in CHB data), and the function was forecasted in the following website: http://snpinfo.niehs.nih.gov/.Rs7291971, located in the transcription factor binding site of the promoter region, was selected because it may play a role in genetic transcription. Previously reported SNPs (rs5757463, rs5757465, rs8177832) were chosen because they may change the expression and anti-virus function of A3G. Haploview software (http://www.broad.mit.edu/mpg/haploview) was used to perform linkage disequilibrium, and haplotype analysis of the SNPs, rs5757465 and rs2011861, was used to choose tag-SNPs for the subsequent studies. The location of the A3G gene and five selected SNPs are shown in Figure 1.
Human genomic DNA was extracted from whole blood. Genotyping of the rs7291971, rs5757465, rs5757463, rs8177832, and rs2011861 SNPs was performed using a mass spectroscopic method (SEQUENOM, BioMiao Biological Technology). Primers and the reaction conditions used for PCR are listed in Table 1.
| SNP | Sequences of the primers | Annealing temperature, °C |
| Rs7291971 | F: 5’-ACGTTGGATGGATCATCTGAGGTCAGTGTC-3’ | 59.6 |
| R: 5’-ACGTTGGATGCCATCTGGATGTATATGTGC-3’ | ||
| Rs5757465 | F: 5’-ACGTTGGATGTGTACAAGGGATATGGCCAC-3’ | 54.6 |
| R: 5’-ACGTTGGATGAATCTGGGTCCCAGAAGTAG-3’ | ||
| Rs5757463 | F: 5’-ACGTTGGATGTAATTTGTAGGTCACCACGC-3’ | 50.9 |
| R: 5’-ACGTTGGATGAGCCTGTCTGGAGCCTCCCT-3’ | ||
| Rs8177832 | F: 5’-ACGTTGGATGGAGCCTTGGAATAATCTGCC-3’ | 51.5 |
| R: 5’-ACGTTGGATGGAGACCCTCACCTGAGAATC-3’ | ||
| Rs2011861 | F: 5’-ACGTTGGATGTCTTTTCCCGCAGGATGAAG-3’ | 55.7 |
| R: 5’-ACGTTGGATGATTTGAGGATCAGGGCCTAC-3’ |
The Hardy-Weinberg equilibrium (H-WE) test was used to assess independent segregation of alleles. The rank-sum test or χ2 text was used to evaluate the differences in demographic and clinical data among the groups. Distributions of the allele and genotype frequencies were calculated by the χ2 test or Fisher’s exact test. Unphased3.1.4 software was used for haplotype analysis of polymorphisms. Logistic regression analysis was used to calculate the P value, ORs and 95%CIs after adjusting for age, sex and environmental factors. All two-sided P < 0.05 values were considered statistically significant. All data were analyzed by SPSS17.0 statistical software (SPSS, Chicago, IL, United States).
Detailed patient demographics for all groups, including sex, age, smoking, and alcohol intake, are listed in Table 2. Detailed clinical and virological characteristics of the HBV infection patients are shown in Supplementary Table 1. The genotype frequencies of each of the A3G gene polymorphisms were categorized into groups, as shown in Table 3. There were no differences in A3G polymorphisms between non-HCC and HCC in the HBV-infected patients (data not shown).
| Group | Healthy controls, | Chronic HBV infection patients | P value3 | |||
| n = 299 | Non-HCC | HBV-related HCC | ||||
| n = 370 | P value1 | n = 287 | P value2 | |||
| Male | 247 (82.6) | 295 (79.7) | 0.345 | 246 (85.7) | 0.304 | 0.921 |
| Age, M (P25, P75) | 50 (45, 55) | 49 (42, 55) | 0.078 | 50 (46, 56) | 0.589 | 0.408 |
| Smoking | 0.392 | 0.003 | 0.292 | |||
| Ever | 113 (37.8) | 128 (34.6) | 144 (50.2) | |||
| Never | 186 (62.2) | 242 (65.4) | 143 (49.8) | |||
| Drinking | 0.036 | 0.804 | 0.241 | |||
| Ever | 122 (40.8) | 122 (33.0) | 120 (41.8) | |||
| Never | 177 (59.2) | 248 (67.0) | 167 (58.2) | |||
| SNP | Healthy controls | Chronic HBV infection patients | Hepatitis B patients (n = 657) vs healthy controls (n = 299) | ||||||
| Non-HCC, n = 370 | HBV-related HCC, n = 287 | ||||||||
| n = 299 (%) | n (%) | OR (95%CI) | P value1 | n (%) | OR (95%CI) | P value2 | OR (95%CI) | P value3 | |
| Rs7291971 genotype and allele | |||||||||
| Detected number | n = 290 | n = 366 | n = 282 | n = 648 vs n = 290 | |||||
| CC | 28 (9.7) | 36 (9.8) | 1 | 26 (9.2) | 1 | 1 | |||
| CG | 113 (39.0) | 139 (38.0) | 0.92 (0.53-1.61) | 0.778 | 114 (40.4) | 1.14 (0.63-2.09) | 0.667 | 1.00 (0.61-1.66) | 0.987 |
| GG | 149 (51.4) | 191 (52.2) | 0.98 (0.57-1.68) | 0.935 | 142 (50.4) | 1.14 (0.63-2.06) | 0.664 | 1.04 (0.64-1.69) | 0.891 |
| CG + GG | 262 | 330 | 0.95 (0.57-1.61) | 0.867 | 256 | 1.14 (0.65-2.02) | 0.651 | 1.02 (0.64-1.64) | 0.930 |
| C allele | 169 (29.1) | 211 (28.8) | 1 | 166 (29.4) | 1 | 1 | |||
| G allele | 411 (70.9) | 521 (71.2) | 1.02 (0.80-1.29) | 0.901 | 398 (29.1) | 0.99 (0.76-1.27) | 0.913 | 1.00 (0.80-1.24) | 0.983 |
| Rs5757463 genotype and allele | |||||||||
| Detected number | n = 285 | n = 369 | n = 285 | n = 654 vs n = 285 | |||||
| CC | 241 (84.6) | 308 (83.5) | 1 | 227 (79.6) | 1 | 1 | |||
| CG | 43 (15.1) | 56 (15.2) | 1.02 (0.66-1.57) | 0.944 | 53 (18.6) | 1.34 (0.86-2.10) | 0.202 | 1.16 (0.79-1.71) | 0.456 |
| GG | 1 (0.4) | 5 (1.4) | 1.93 (0.37-10.14) | 0.439 | 5 (1.8) | 2.71 (0.51-14.31) | 0.239 | 2.25 (0.49-10.39) | 0.298 |
| CG + GG | 44 | 61 | 1.06 (0.69-1.62) | 0.799 | 58 | 1.40 (0.91-2.17) | 0.130 | 1.21 (0.83-1.77) | 0.327 |
| C allele | 525 (92.1) | 672 (91.1) | 1 | 507 (88.9) | 1 | 1 | |||
| G allele | 45 (7.9) | 66 (8.9) | 1.15 (0.77-1.70) | 0.500 | 63 (11.1) | 1.45 (0.97-2.17) | 0.069 | 1.28 (0.90-1.82) | 0.194 |
| Rs5757465 genotype and allele | |||||||||
| Detected number | n = 285 | n = 365 | n = 279 | n = 644 vs n = 285 | |||||
| TT | 170 (59.6) | 221 (60.5) | 1 | 169 (60.6) | 1 | 1 | |||
| TC | 101 (35.4) | 129 (35.3) | 1.02 (0.73-1.42) | 0.898 | 92 (33.0) | 0.93 (0.65-1.34) | 0.707 | 0.98 (0.73-1.32) | 0.903 |
| CC | 14 (4.9) | 15 (4.1) | 0.82 (0.38-1.76) | 0.610 | 18 (6.5) | 1.43 (0.68-3.01) | 0.349 | 1.05 (0.55-2.02 | 0.884 |
| TC + CC | 115 | 144 | 0.99 (0.72-1.37) | 0.984 | 110 | 0.99 (0.70-1.39) | 0.953 | 0.99 (0.74-1.32 | 0.945 |
| T allele | 441 (77.4) | 571 (78.2) | 1 | 430 (77.1) | 1 | 1 | |||
| C allele | 129 (22.6) | 159 (21.8) | 0.95 (0.73-1.24) | 0.714 | 128 (22.9) | 1.02 (0.77-1.34) | 0.902 | 0.98 (0.77-1.24) | 0.868 |
| Rs8177832 genotype and allele | |||||||||
| Detected number | n = 291 | n = 369 | n = 287 | n = 656 vs n = 291 | |||||
| AA | 227 (78.0) | 302 (81.8) | 1 | 249 (86.8) | 1 | 1 | |||
| AG | 60 (20.6) | 65 (17.6) | 0.80 (0.54-1.18) | 0.253 | 35 (12.2) | 0.53 (0.33-0.84) | 0.007 | 0.67 (0.47-0.96) | 0.029 |
| GG | 4 (1.4) | 2 (0.5) | 0.40 (0.72-2.26) | 0.302 | 3 (1.0) | 0.75 (0.16-3.43) | 0.711 | 0.55 (0.15-2.07) | 0.374 |
| AG + GG | 64 | 67 | 0.78 (0.53-1.14) | 0.195 | 38 | 0.54 (0.35-0.85) | 0.007 | 0.66 (0.47-0.94) | 0.021 |
| A allele | 514 (88.3) | 669 (90.7) | 1 | 533 (92.9) | 1 | 1 | |||
| G allele | 68 (11.7) | 69 (9.3) | 0.78 (0.55-1.11) | 0.167 | 41 (7.1) | 0.58 (0.39-0.87) | 0.008 | 0.69 (0.50-0.95) | 0.023 |
| Rs2011861 genotype and allele | |||||||||
| Detected number | n = 284 | n = 364 | n = 279 | n = 643 vs n = 284 | |||||
| CC | 53 (18.7) | 55 (15.1) | 1 | 33 (11.8) | 1 | 1 | |||
| CT | 127 (44.7) | 170 (46.7) | 1.25 (0.80-1.95) | 0.328 | 135 (48.4) | 1.69 (1.02-2.80) | 0.042 | 1.42 (0.95-2.13) | 0.085 |
| TT | 104 (36.6) | 139 (38.2) | 1.27 (0.81-2.02) | 0.301 | 111 (39.8) | 1.82 (1.08-3.06) | 0.024 | 1.48 (0.98-2.24) | 0.063 |
| CT + TT | 231 | 309 | 1.26 (0.83-1.91) | 0.277 | 246 | 1.75 (1.08-2.82) | 0.022 | 1.45 (0.99-2.11) | 0.053 |
| C allele | 233 (41.0) | 280 (38.5) | 1 | 201 (36.0) | 1 | 1 | |||
| T allele | 335 (59.0) | 448 (61.5) | 1.11 (0.89-1.39) | 0.350 | 357 (64.0) | 1.24 (0.97-1.57) | 0.085 | 1.16 (0.95-1.42) | 0.140 |
The statistical analyses showed no significant differences among the three groups in terms of sex and age. However, there was a significant difference in smoking between HCC and healthy control patients (P = 0.003) and in alcohol consumption between non-HCC and healthy control patients (P = 0.036). Furthermore, all five SNPs (rs7291971, rs5757463, rs5757465, rs8177832, and rs2011861) among the healthy controls were in equilibrium, as determined by the H-WE test (P = 0.34, P = 0.53, P = 0.84, P = 0.99, and P = 0.20, respectively).
The genotype and allele frequencies of A3G gene polymorphisms among the CHB patients and the healthy controls are shown in Table 3. No significant associations were observed between the genotype and allele frequencies of the A3G gene rs7291971, rs5757463, rs5757465, and rs2011861 polymorphisms and the presence of CHB. We found a significant relationship between the G allele and decreased risk of CHB with an OR of 0.69 (95%CI: 0.50-0.95) for rs8177832. Compared to the AA genotype, the AG genotype was significantly related to a decreased risk of CHB after adjusting for age, sex, tobacco use, and alcohol intake using binary logistic regression analyses (OR = 0.67, 95%CI: 0.47-0.96). The adjusted OR for the AG and GG genotypes combined was 0.66 (95%CI: 0.47-0.94).
The genotype and allele frequencies of A3G gene polymorphisms among the non-HCC patients and the healthy controls are shown in Table 3. No significant differences were found in the frequencies of all alleles and genotypes (rs7291971, rs5757463, rs5757465, rs8177832, rs2011861) between the non-HCC patients and healthy controls.
The genotype and allele frequencies of A3G gene polymorphisms among the HCC and the healthy controls are shown in Table 3. No significant effects were observed between the genotype and allele frequencies of the A3G gene rs7291971, rs5757463, and rs5757465 polymorphisms and the HCC risk after adjusting for sex, age, smoking, and drinking. We found a significant relationship between the G allele of rs8177832 and the risk of HCC with an OR of 0.58 (95%CI: 0.39-0.87). Compared to the AA genotype, the AG genotype and AG plus GG genotype of rs8177832 were significantly related to a decreased risk of HCC (adjusted OR = 0.53, 95%CI: 0.33-0.84; OR = 0.54, 95%CI: 0.35-0.85, respectively). Meanwhile, compared to the CC genotype, the CT and TT genotypes of rs2011861 were significantly related to an increased risk of HCC after adjusting for age, sex, smoking, and drinking (OR = 1.69, 95%CI: 1.02-2.80; OR = 1.82, 95%CI: 1.08-3.06, respectively). The adjusted OR for the CT and TT genotypes combined was 1.75 (95%CI: 1.08-2.82).
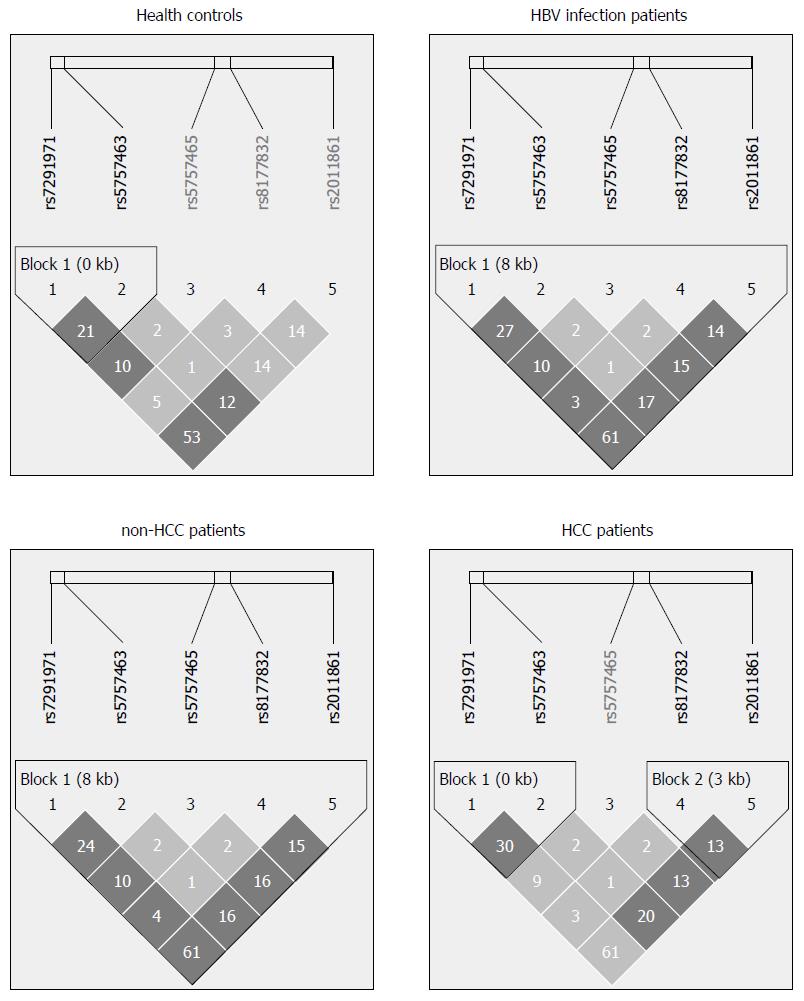
A linkage disequilibrium map among the five SNPs associated with A3G in each group (healthy controls, CHB patients, non-HCC patients, and HCC patients) is shown in Figure 2. We analyzed the differences in haplotype distributions between healthy controls and CHB patients, healthy controls and non-HCC patients, and healthy controls and HCC patients. In the CHB patients and healthy controls, the haplotype C-G allele of rs5757463-rs8177832 was associated with a significantly decreased risk of CHB (OR = 0.71, 95%CI: 0.51-0.98) (Table 4). In healthy controls and HCC patients, the haplotype G-G allele of rs7291971-rs8177832 (OR = 0.61, 95%CI: 0.39-0.95), C-G allele of rs5757463-rs8177832 (OR = 0.61, 95%CI: 0.40-0.91), T-G allele of rs5757465-rs8177832 (OR = 0.58, 95%CI: 0.38-0.88) were significantly associated with a decreased risk of HCC. In contrast, the haplotypes comprised of the C-T, G-C alleles of rs5757463-rs2011861 (OR = 1.41, 95%CI: 1.08-1.83; OR = 1.80, 95%CI: 1.15-2.82, respectively), the C-G-A allele of rs7291971-rs5757463-rs8177832 (OR = 1.64, 95%CI: 1.03-2.61), and the C-G-C allele of rs7291971-rs5757463-rs2011861 (OR = 1.70, 95%CI: 1.05-2.76) were associated with a significantly increased risk of HCC (Table 5).
| Haplotype | Frequency | χ2 | P value | OR (95%CI) | |
| Healthy controls | Chronic HBV infection patients | ||||
| rs5757463-rs8177832 | |||||
| C-A | 455 (80.3) | 1067 (81.7) | 6.07 | 0.048 | 1 |
| C-G | 66 (11.7) | 110 (8.4) | 0.71 (0.51-0.98) | ||
| G-A | 45 (8.0) | 129 (9.9) | 1.22 (0.86-1.75) | ||
| Haplotype | Frequency | χ2 | P value | OR (95%CI) | |
| Healthy controls | HCC | ||||
| rs7291971-rs8177832 | |||||
| C-A | 166 (29.0) | 166 (29.4) | 6.73 | 0.03 | 1 |
| G-A | 339 (59.3) | 357 (63.3) | 1.05 (0.81-1.37) | ||
| G-G | 67 (11.7) | 41 (7.3) | 0.61 (0.39-0.95) | ||
| rs5757463-rs8177832 | |||||
| C-A | 455 (80.4) | 466 (81.8) | 8.96 | 0.01 | 1 |
| C-G | 66 (11.7) | 41 (7.2) | 0.61 (0.40-0.91) | ||
| G-A | 45 (8.0) | 63 (11.1) | 1.37 (0.91-2.05) | ||
| rs5757465-rs8177832 | |||||
| T-A | 371 (66.0) | 390 (69.9) | 6.94 | 0.03 | 1 |
| C-A | 125 (22.2) | 128 (22.9) | 0.97 (0.73-1.30) | ||
| T-G | 66 (11.7) | 40 (7.2) | 0.58 (0.38-0.88) | ||
| rs5757463-rs2011861 | |||||
| C-C | 183 (33.2) | 141 (25.4) | 10.68 | 0.01 | 1 |
| C-T | 326 (59.1) | 354 (63.7) | 1.41 (1.08-1.83) | ||
| G-C | 43 (7.8) | 60 (10.8) | 1.80 (1.15-2.82) | ||
| G-T | 0 (0.0) | 1 (0.2) | 1.00 (0.99-1.02) | ||
| rs7291971-rs5757463-rs8177832 | |||||
| C-C-A | 118 (21.1) | 101 (18.0) | 10.59 | 0.01 | 1 |
| C-G-A | 45 (8.0) | 63 (11.3) | 1.64 (1.03-2.61) | ||
| G-C-A | 332 (59.3) | 355 (63.4) | 1.25 (0.92-1.70) | ||
| G-C-G | 65 (11.6) | 41 (7.3) | 0.74 (0.46-1.18) | ||
| rs7291971-rs5757463-rs2011861 | |||||
| C-C-C | 112 (20.4) | 91 (16.6) | 11.62 | 0.04 | 1 |
| C-C-T | 5 (0.9) | 6 (1.1) | 1.63 (0.42-6.42) | ||
| C-G-C | 43 (7.8) | 59 (10.8) | 1.70 (1.05-2.76) | ||
| C-G-T | 0 (0.0) | 2 (0.4) | 1.02 (0.99-1.05) | ||
| G-C-C | 70 (12.8) | 48(8.8) | 0.85 (0.85-1.35) | ||
| G-C-T | 319 (58.1) | 342 (62.4) | 1.33 (0.97-1.82) | ||
The HBV viral load and rate of HBeAg seroconversion of patients with rs8177832 and rs2011861 polymorphisms in each group are shown in Supplementary Tables 2 and 3. The viral load of rs8177832 GG genotype was lower than AA genotype in the non-HCC group, but the number of cases was small, and the viral loads did not result in statistically significant differences between patients with the mutant gene type and wild-type rs8177832 in the other groups (HCC and CHB). The viral loads of rs2011861 of each genotype also did not result in statistically significant differences among the groups. The AG genotype and AG plus GG genotype of rs8177832 were shown to have high HBeAg seroconversion rates in the non-HCC group, and tended to have high HBeAg seroconversion rates in the CHB group. There was no significant difference between the ratio of HBeAg(+)HBeAb(-)/HBeAg(-)HBeAb(+) in patients with rs2011861 polymorphisms.
Innate immune mechanisms are the first line of defense against invading viruses[21]. The A3 family plays an important role in facilitating innate immunity by restricting many viruses, including HBV[21,22]. With the progression of chronic infection, HBV mutations gradually occur[23]. The mutation rate of HBV DNA caused by A3s in patients with CHB has been shown to be higher than that of acute HBV-infected patients[24]. In CHB patients, the frequency of hypermutated genomes was higher in HBeAg-negative individuals compared to HBeAg-positive cases, and the degree was significantly associated with the extent of fibrosis[24,25]. Also, A3s and their related editing patterns have potential roles in oncogenesis. Recent analyses showed that tumor samples contain hundreds of A3-signature mutations in a wide variety of cancer types. The mutations have implicated A3 cytidine deaminases as significant factors in the mutagenesis of human cancer genomes[26-28]. Overexpression of A3 could lead to induction of DNA breaks and carcinogenic protein mutants through activation of damage responses in a deaminase-dependent manner, as shown in vitro[29,30]. Although the mutations can be highly deleterious in most instances, slightly edited genomes might help the virus evolve, escape from the immune responses and induce drug resistance[12]. Therefore, A3 genetic variation may play an important role in the occurrence of HBV infection and progression of liver disease.
We performed a large case-control study that determined associations between SNPs in the A3G gene and the presence of CHB and HBV-related HCC. The AG genotype and G allele for rs8177832 were significantly associated with a decreased risk of CHB and HCC. Rs8177832 may enhance the effect of APOBEC3G on HBV inhibition and promote seroconversion of HBeAg. A significant correlation was found between the rs2011861 CT, TT genotypes, and increased risk of HCC. Haplotype analyses showed an association between the G-G allele of rs7291971-rs8177832, C-G allele of rs5757463-rs8177832 and T-G allele of rs5757465-rs8177832, and decreased risk of HCC. The three protective haplotypes against HCC development suggested that the primary SNP may be rs8177832 because all three haplotypes share the rs8177832 G allele. However, the haplotypes C-T and G-C allele of rs5757463-rs2011861 compared to the wild-type allele C-C, C-G-A alleles of rs7291971- rs5757463-rs8177832, and the C-G-C allele of rs7291971-rs5757463-rs2011861 were associated with a significantly increased risk of HCC. The four haplotypes associated with HCC development suggested that the rs5757463 G allele and the rs2011861 T allele may have primary effects on an increased risk of HCC. The mechanism by which the differential effects of these genotypes affect the susceptibility to HBV infection and HCC is not clear and requires further study. Rs8177832 caused the substitutions of A to G of exon 4, resulting in an amino acid change from histidine to arginine at codon 186 (H186R). It is speculated that this mutation may change the antiviral function of A3G. Rs2011861 may be associated with other functional genes.
A previous study investigated the relationship between rs8177832 and CHB in 179 HBV chronic carriers and 216 healthy control subjects in a Moroccan population[31]. The results showed that the G genotype tended to be associated with an increased risk of developing CHB (P = 0.254). However, no significant difference was found, perhaps due to the small sample size. There was no evidence of an effect of the rs8177832 mutation on gene expression and anti-HBV ability in vitro. The current report is the first to describe the relationship between rs8177832, rs2011861 polymorphisms, the risk of HBV infection, and the development of HCC.
There are limitations to the current study. The explanation for our findings may have been influenced by many factors involved in the disease. The outcome of HBV infection is closely related to the age at which infection occurred[1]. A maintained long-term response to therapy or a sustained off-treatment response is necessary to prevent liver damage and hepatic decompensation and delay the onset of the long-term complications of CHB, such as HCC[32]. Recruitment of individuals who have cleared an HBV infection, and are also comparable in the duration of the infection, could help clarify whether there is an association with the predisposition to chronicity. Similarly, consistent treatment can help determine the role of genes in disease progression. However, that goal is difficult to achieve in China. Therefore, further comprehensive investigations on large sample populations of different ethnic origin, and with different outcomes of infection, but comparable durations of infection and therapeutic schedule, will be required to confirm and extend our findings. In vitro experiments should be performed to ascertain the effects of SNPs on the changes of gene functions.
In conclusion, the current study provides epidemiological evidence that the A3G locus may mediate host innate resistance to HBV infection and HCC in vivo. The role of inhibiting synthesis and editing of the HBV genome in such defense systems should be further investigated. It is important to continue research on the identification of novel therapeutic targets to stimulate the development of new antiviral agents and immunotherapies. It may be possible to transfect the protective mutation gene into somatic cells, such as liver cells, to express the gene product. Alternatively, the gene product itself could be introduced resulting in therapeutic properties such as anti-viral or anti-HCC effects.
APOBEC3s have one or two catalytic domains that have cytidine deaminase activity, which convert cytosine to uracil in DNA. APOBEC3G (A3G) is a dominant cytidine deaminase that strongly inhibits synthesis and editing of hepatitis B virus (HBV) DNA in vivo.
Several studies identified genetic variants that are associated with risk of human immunodeficiency virus infection and progression to acquired immune deficiency syndrome. Since A3G is an important host factor that may inhibit HBV, we screened the A3G gene for both regulatory and coding region variants that could modify A3G transcription or amino acid sequence.
The current report is the first to describe the relationship between rs8177832 and rs2011861 polymorphisms, the risk of HBV infection, and the development of hepatocellular carcinoma (HCC), suggesting its use as a potential therapeutic target.
Potential use of rs8177832 as therapeutic target in patients with chronic HBV infection (CHB) and HCC.
The A3G rs8177832 polymorphism is associated with a decreased risk of CHB and HCC, while the rs2011861 polymorphism is associated with an increased risk of HCC. Rs8177832 may enhance the effect of APOBEC3G on HBV inhibition and promote seroconversion of hepatitis e antigen.
The manuscript by He et al enrolled chronic HBV-infection patients for studying the association between polymorphisms of the A3G gene and CHB and HBV-related HCC. Authors indicated that the A3G rs8177832 polymorphism is associated with a decreased risk of CHB infection and HCC, while the rs2011861 polymorphism is associated with an increased risk of HCC. The paper is well organized and the results are very straightforward and clear.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Hashimoto N, Tai DI, Wu PF S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Zahng FF
| 1. | Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virol J. 2013;10:239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 221] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 2. | Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20:3884-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 3. | Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 430] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 4. | Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942-1956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 5. | Lee YC, Cohet C, Yang YC, Stayner L, Hashibe M, Straif K. Meta-analysis of epidemiologic studies on cigarette smoking and liver cancer. Int J Epidemiol. 2009;38:1497-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 195] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 6. | McGlynn KA, London WT. Epidemiology and natural history of hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2005;19:3-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 251] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 7. | Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816-1819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 468] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 8. | Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 227] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 9. | Deng Y, Du Y, Zhang Q, Han X, Cao G. Human cytidine deaminases facilitate hepatitis B virus evolution and link inflammation and hepatocellular carcinoma. Cancer Lett. 2014;343:161-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Nguyen DH, Gummuluru S, Hu J. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J Virol. 2007;81:4465-4472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 143] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 11. | Rösler C, Köck J, Kann M, Malim MH, Blum HE, Baumert TF, von Weizsäcker F. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 123] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 12. | Vartanian JP, Henry M, Marchio A, Suspène R, Aynaud MM, Guétard D, Cervantes-Gonzalez M, Battiston C, Mazzaferro V, Pineau P. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010;6:e1000928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 13. | Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 372] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 14. | Köck J, Blum HE. Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J Gen Virol. 2008;89:1184-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Henry M, Guétard D, Suspène R, Rusniok C, Wain-Hobson S, Vartanian JP. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS One. 2009;4:e4277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 74] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:8321-8326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 259] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 17. | Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474-9485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 285] [Cited by in RCA: 275] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 18. | Valcke HS, Bernard NF, Bruneau J, Alary M, Tsoukas CM, Roger M. APOBEC3G genetic variants and their association with risk of HIV infection in highly exposed Caucasians. AIDS. 2006;20:1984-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | An P, Bleiber G, Duggal P, Nelson G, May M, Mangeat B, Alobwede I, Trono D, Vlahov D, Donfield S. APOBEC3G genetic variants and their influence on the progression to AIDS. J Virol. 2004;78:11070-11076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 145] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 20. | Singh KK, Wang Y, Gray KP, Farhad M, Brummel S, Fenton T, Trout R, Spector SA. Genetic variants in the host restriction factor APOBEC3G are associated with HIV-1-related disease progression and central nervous system impairment in children. J Acquir Immune Defic Syndr. 2013;62:197-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int. 2013;2013:683095. [PubMed] |
| 22. | Janahi EM, McGarvey MJ. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J Viral Hepat. 2013;20:821-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Tran A, Kremsdorf D, Capel F, Housset C, Dauguet C, Petit MA, Brechot C. Emergence of and takeover by hepatitis B virus (HBV) with rearrangements in the pre-S/S and pre-C/C genes during chronic HBV infection. J Virol. 1991;65:3566-3574. [PubMed] |
| 24. | Noguchi C, Ishino H, Tsuge M, Fujimoto Y, Imamura M, Takahashi S, Chayama K. G to A hypermutation of hepatitis B virus. Hepatology. 2005;41:626-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 126] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 25. | Beggel B, Münk C, Däumer M, Hauck K, Häussinger D, Lengauer T, Erhardt A. Full genome ultra-deep pyrosequencing associates G-to-A hypermutation of the hepatitis B virus genome with the natural progression of hepatitis B. J Viral Hepat. 2013;20:882-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 812] [Cited by in RCA: 930] [Article Influence: 77.5] [Reference Citation Analysis (0)] |
| 27. | Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 614] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 28. | Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL. Signatures of mutational processes in human cancer. Nature. 2013;500:415-421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7533] [Cited by in RCA: 7314] [Article Influence: 609.5] [Reference Citation Analysis (1)] |
| 29. | Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 30. | Xu R, Zhang X, Zhang W, Fang Y, Zheng S, Yu XF. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology. 2007;46:1810-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 31. | Ezzikouri S, Kitab B, Rebbani K, Marchio A, Wain-Hobson S, Dejean A, Vartanian JP, Pineau P, Benjelloun S. Polymorphic APOBEC3 modulates chronic hepatitis B in Moroccan population. J Viral Hepat. 2013;20:678-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Zoulim F. Hepatitis B virus resistance to antiviral drugs: where are we going? Liver Int. 2011;31 Suppl 1:111-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |