Published online May 14, 2017. doi: 10.3748/wjg.v23.i18.3252
Peer-review started: January 3, 2017
First decision: February 9, 2017
Revised: February 28, 2017
Accepted: March 30, 2017
Article in press: March 30, 2017
Published online: May 14, 2017
Processing time: 134 Days and 13.7 Hours
To determine the role of hepatitis B virus X protein (HBx), HBx in regulating hepatic progenitor cell (HPC)-like features in hepatocellular carcinoma (HCC) and the underlying molecular mechanisms.
We used a retrovirus vector to introduce wild type HBx or empty vector into HepG2 cells. We then used these cells to analyze cell proliferation, senescence, transformation, and stem-like features. Gene expression profiling was carried out on Affymetrix GeneChip Human U133A2.0 ver.2 arrays according to the manufacturer’s protocol. Unsupervised hierarchical clustering analysis and Class Comparison analysis were performed by BRB-Array Tools software Version 4.2.2. A total of 238 hepatitis B virus (HBV)-related HCC patients’ array data were used for analyzing clinical features.
The histone demethylase KDM5B was significantly highly expressed in HBV-related HCC cases (P < 0.01). In HBV proteins, only HBx up-regulated KDM5B by activating c-myc. Hepatic stem cell (HpSC) markers (EpCAM, AFP, PROM1, and NANOG) were significantly highly expressed in KDM5B-high HCC cases (P < 0.01). KDM5B played an important role in maintaining HpSC-like features and was associated with a poor prognosis. Moreover, inhibition of KDM5B suppressed spheroid formation and cell invasion in vitro.
HBx activates the histone demethylase KDM5B and induces HPC-like features in HCC. Histone demethylases KDM5B may be an important therapeutic target against HBV-related HCC cases.
Core tip: The role of epigenetic regulation in cancer biology has been the subject of several studies. These chromatin structure modifiers have been increasingly shown to facilitate several steps of cancer progression. However, the epigenetic regulation of hepatocellular carcinoma has not been elucidated. We assumed that multifunctional protein hepatitis B virus X (HBx) protein, may affect epigenetic regulation of Hepatocellular carcinoma. We showed that HBx activated the histone demethylase KDM5B and induced HPC-like features in hepatocellular carcinoma (HCC) in this study. Our results suggested that histone demethylases may be an important therapeutic target against HBV-related HCC cases.
- Citation: Wang X, Oishi N, Shimakami T, Yamashita T, Honda M, Murakami S, Kaneko S. Hepatitis B virus X protein induces hepatic stem cell-like features in hepatocellular carcinoma by activating KDM5B. World J Gastroenterol 2017; 23(18): 3252-3261
- URL: https://www.wjgnet.com/1007-9327/full/v23/i18/3252.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i18.3252
Chronic infection by hepatitis B virus (HBV) affects approximately 400 million people worldwide and is a leading cause of hepatocellular carcinoma (HCC), which is considered the sixth most common cancer in the world[1,2].
HBV encodes the nonstructural hepatitis B virus X (HBx) protein, which is conserved among mammalian hepadnaviruses, suggesting an important function. HBx is the most critical protein for viral replication in hepatocytes and for the development of HCC. HBx is a multifunctional protein that has been shown to regulate many transcription factors such as nuclear factor-kappa B, ATF-2, activator protein 1, cAMP response element binding protein[3], Ras-Raf mitogen-activated protein kinase, extracellular signal-regulated kinase, phosphoinositol-3-kinase-protein kinase B/Akt[4], and Wnt/beta-catenin pathway[5]. These pathways are involved in a range of cellular functions including apoptosis, cell proliferation, cell cycle progression, and cytokine production. Therefore, it is proposed that HBx manipulates these cellular signaling pathways, resulting in the transformation of HBV-infected hepatocytes. Although extensive studies have focused on the roles of HBx in malignant transformation[5-10], the molecular mechanisms underlying this process are not well elucidated.
KDM5B, which is also known as JARID1B or PLU1, is an H3K4me3 histone demethylase that is overexpressed in many types of cancer including breast[11], prostate[12], bladder[13], and lung cancer[14]. Histone modifiers, such as KDM5B, and their associated post-translational modifications are thought to be central to the determination of embryonic stem cell fate[15,16]. Embryonic stem cell pluripotency and differentiation depend on the interactions between an array of chromatin modifiers and several downstream transcription factors. This stem cell epigenetic landscape has been implicated in tumor progression and chemoresistance.
The central hypothesis we explored here is that KDM5B maintains cancer stem cells in HCC.
For gene expression analysis, we assessed three already published cohorts. Cohort 1 was 139 caucasian HCC cases. KDM5B expression data were derived from these specimens as described. Survival data linking to this cohort were kindly provided by Dr. Snorri Thorgeirsson at NCI[17]. Cohort 2 was 238 cases who received surgical resection of HBV-related HCC at the Liver Cancer Institute of Fudan University. Their clinicopathologic characteristics and prognostic data were kindly provided by Dr. Xin Wei Wang at NCI[16]. Cohort 3 was 89 Japanese HCC cases who received surgical resection at the Kanazawa University Hospital[18].
Human HCC cell lines (HepG2, Huh7 and Hep3B) were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL, Gaithersburg, MD) containing 10% fetal bovine serum and 1% penicillin-streptomycin. We used two immortalized human hepatocyte cell lines, TTNT16 cells (in which hTERT was introduced)[19,20] and T5B cells [in which the SV40 large T antigen (LT) was introduced][21,22]. These cell lines were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL, Gaithersburg, MD) containing 10% fetal bovine serum and 1% penicillin-streptomycin.
Total RNA was subjected to quantitative real-time reverse-transcription polymerase chain reaction (RT-PCR). mRNAs were analyzed using TaqMan Gene Expression Assays in accordance with the manufacturer’s instructions (Applied Biosystems, Foster City, CA). All RT reactions were run in a GeneAmp PCR 9700 Thermocycler (Applied Biosystems). Probes used for the analyses were as follows: KDM5B, Hs00981910_m1; MYC, Hs00905030_m1 (Applied Biosystems), HBV proteins, shown in Table 1. The experiments were performed in triplicate. The TaqMan gene assay for actin was used to normalize the relative abundance of mRNA.
| PC-C (core) | FW | 5' | CGAGGCAGGTCCCCTAGAA | 3' |
| RV | 5' | CGGCGATTGAGACCTTCGT | 3' | |
| probe | 5'-FAM | AGAACTCCCTCGCCTCG | MGB-3' | |
| PS-S (surface) | FW | 5' | ACCCCAACAAGGATCATTGG | 3' |
| RV | 5' | CGAATGCTCCCGCTCCTA | 3' | |
| probe | 5'-FAM | CAGAGGCAAATCAG | MGB-3' | |
| HBx | FW | 5' | TGTCAACGACCGACCTTGAG | 3' |
| RV | 5' | CCCAACTCCTCCCAGTCCTT | 3' | |
| probe | 5'-FAM | CATACTTCAAAGACTGTTTGTT | MGB-3' | |
| Pol (polymerase) | FW | 5' | TCCGCTGCCGATCCAT | 3' |
| RV | 5' | GCTGCGAGCAAAACAAGCT | 3' | |
| probe | 5'-FAM | CTGCGGAACTCCT | MGB-3' | |
| Pg RNA | FW | 5' | GCTCTGTATCGGGAGGCCTTA | 3' |
| (Pregenomic RNA) | RV | 5' | TGAGTGCTGTATGGTGAGGAGAA | 3' |
| probe | 5'-FAM | AGTCTCCGGAACATT | MGB-3' |
A small interfering RNA (siRNA) specific to KDM5B and a control siRNA were obtained from Nippon Gene. siRNA specific for c-Myc and a control siRNA were obtained from Invitrogen. Transfection was performed with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
Cell migration and invasion were assessed using a Cytoselect 24-Well Cell Migration and Invasion Assay (CELL BIOLABS). For migration and invasion measurements, transfected Huh7 and Hep3B cells were seeded at 5.0 × 105 cells/well. The cells that invaded or migrated to the underside of the chamber were harvested and measured according to the manufacturer’s protocol. Results were determined from triplicate wells in three independent experiments and expressed as a percentage relative to control.
For spheroid formation assays, single cell suspensions of 2.0 × 103 cells were seeded in 6-well Ultra-Low Attachment Microplates (Corning, Corning, NY). The number of spheroids was measured at 10-14 d after seeding. Invasion assays were performed using BD BioCoatTM Matrigel Matrix Cell Culture Inserts and Control Inserts (BD Biosciences, San Jose, CA).
Immunohistochemistry (IHC) was performed using Envision+ kits according to the manufacturer’s instructions. An anti-KDM5B monoclonal antibody was used for detecting KDM5B. The staining area and intensities were evaluated in each sample and graded from 0-3 (0, 0-5%; 1, 5%-25%; 2, 25%-50%; 3, > 50%) and 0-2 (0, negative; 1, weak; 2, strong), respectively. The sum of the area and intensity scores of each marker (IHC score) were calculated. Samples were defined as marker-positive (IHC score ≥ 3) or -negative (IHC score ≤ 2).
Mann-Whitney, χ2, Fisher’s exact, and Kruskal-Wallis tests were used to compare the clinicopathologic characteristics and gene expression data. The correlation of the gene expression data was evaluated by Spearman’s rank correlation coefficient. Kaplan-Meier survival analysis with the log-rank test was performed to compare patients’ survival. All analyses were performed using GraphPad Prism software 5.0.1 (GraphPad Software, San Diego, CA).
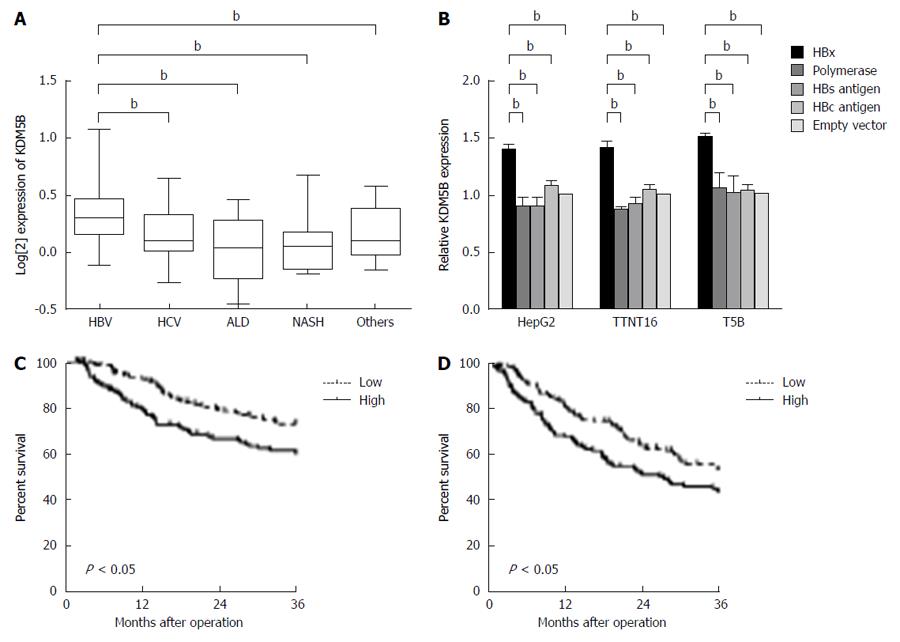
We performed transcriptomic analysis of 139 retrospectively collected HCC (cohort 1) patients to assess whether KDM5B was associated with hepatocarcinogenesis. We evaluated and compared the expression of KDM5B in HCC cases developed from various backgrounds: chronic hepatitis B, chronic hepatitis C, alcoholic liver disease, and nonalcoholic steatohepatitis. As shown in Figure 1A, KDM5B was significantly highly expressed in HBV-related HCC cases compared to other etiological-related HCC cases. Moreover, for HBV proteins, only HBx up-regulated KDM5B in HepG2 cells and immortalized human hepatocytes (TTNT16 and T5b cells) (Figure 1B). These results indicated that HBx was associated with HBV-related hepatocarcinogenesis through the activation of KDM5B.
To understand the effect of KDM5B on the survival of HBV-related HCC cases, we analyzed the microarray data of 238 HCC cases (cohort 2). Kaplan-Meier survival analysis revealed that the KDM5B high expression group had a significantly shorter overall survival (P < 0.05) and recurrence-free survival (P < 0.05) than the KDM5B low expression group (Figure 1C and 1D).
Previously, we identified two HCC subgroups, one resembling the gene expression signatures of hepatic stem cells (HpSCs) (referred to as HpSC-HCC) and the other similar to mature hepatocytes (referred to as MH-HCC). HpSC-HCC had stem cell-like features and a poorer prognosis than MH-HCC cases. We hypothesized that KDM5B induced hepatic progenitor-like features in HBV-related HCC cases.
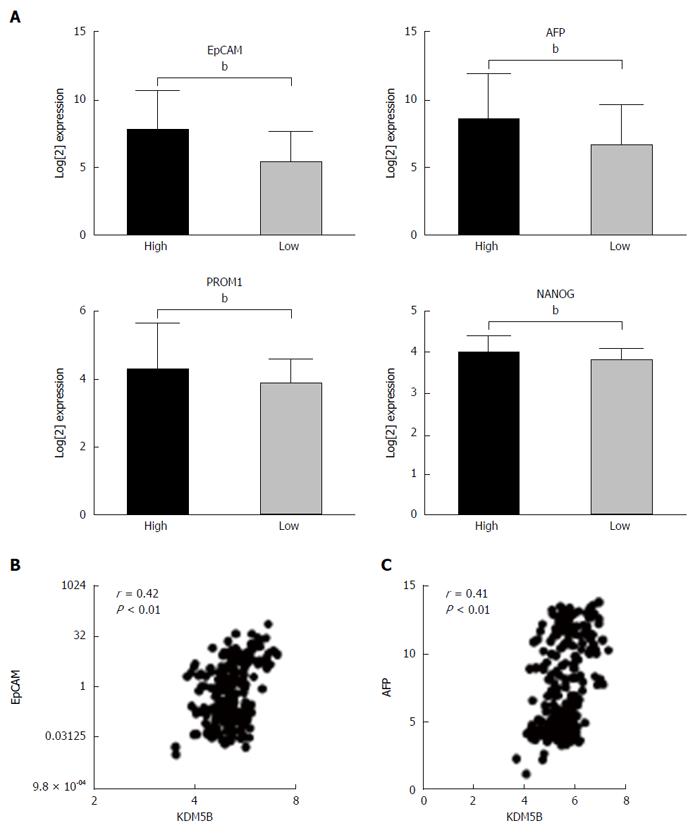
To determine whether KDM5B was functionally linked to the HpSC-like phenotype, we analyzed representative expression levels of HpSC markers in cohort 2 HCC cases. Consistently, HpSC markers, such as EpCAM, AFP, PROM1, and NANOG, were more abundantly expressed in KDM5B high expression HCC cases as compared to KDM5B low expression cases (Figure 2A). Moreover, KDM5B levels were positively correlated with EpCAM or AFP levels in 238 HCC cases (Figure 2B and C).
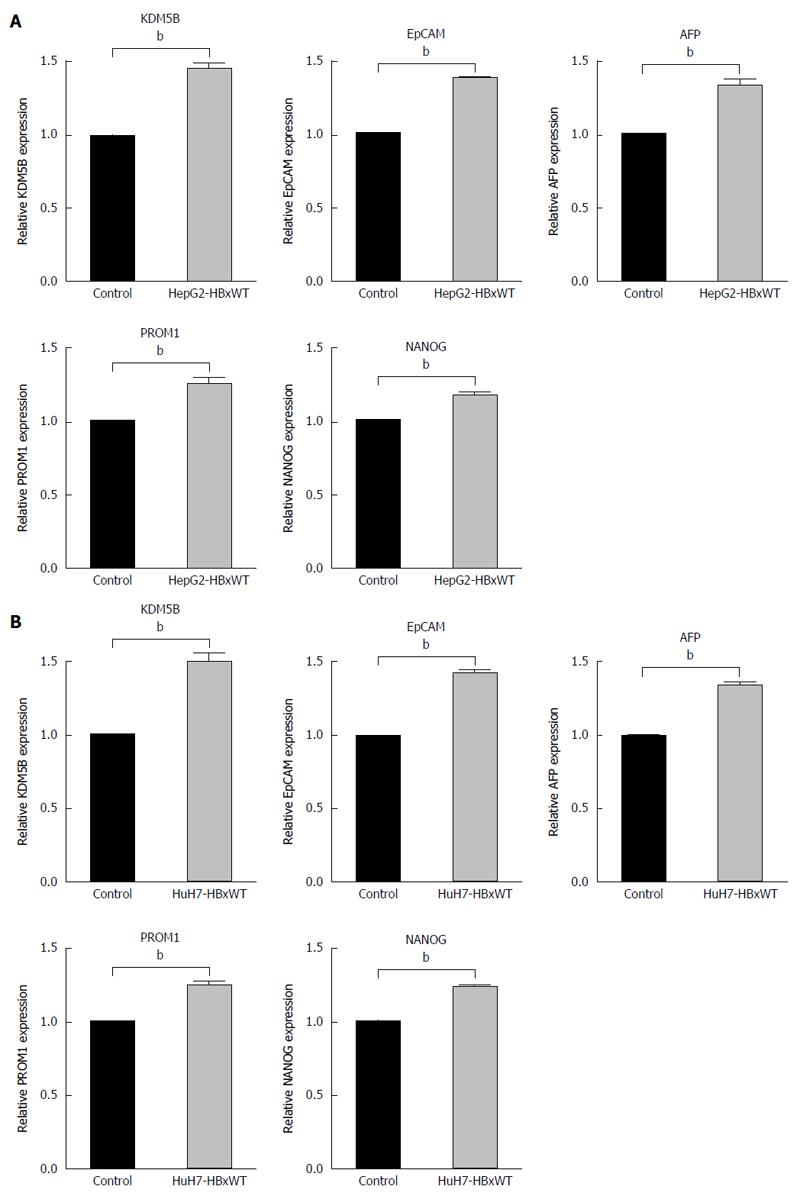
Next, to confirm that HBx upregulated KDM5B and HpSC markers in HCC cells, we introduced HBx-wt in HepG2 and Huh7 cells and evaluate the expression of KDM5B and HpSC markers. Introduction of HBx up-regulated the expression of KDM5B, EpCAM, AFP, PROM1 and NANOG in these cells (Figure 3A and B).
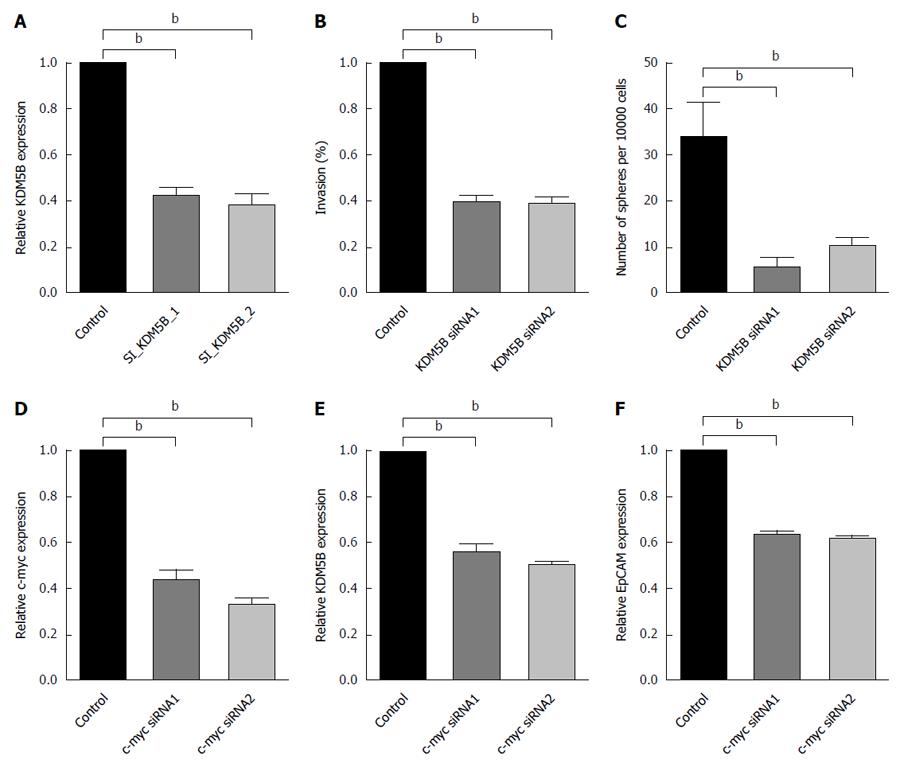
In addition, inhibition of KDM5B suppressed cell invasion (Figure 4A and B) and spheroid formation (Figure 4C) in Hep3B and Huh7 cells. However, KDM5B did not affect cell proliferation or apoptosis in these cells as measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide and transferase-mediated dUTP nick-end labeling assays (data not shown). These data indicated that KDM5B is an important molecule for maintaining HpSC-like features in HBV-related HCC cases.
The available Chip-Seq data (http://genome.ucsc.edu/) revealed that several transcriptional factors, such as c-Myc and TCF, preferentially bound to the immediate 5′ upstream sequence of the predictive transcription initiation site of KDM5B. Moreover, c-Myc is one of the transcription factors known to be activated by HBx. To determine whether c-Myc regulates KDM5B expression, we silenced c-Myc expression with a c-Myc-specific siRNA in Hep3B and Huh7 cells. Consistently, we found that inhibition of c-Myc resulted in the suppression of KDM5B and EpCAM expression (Figure 4D-F).
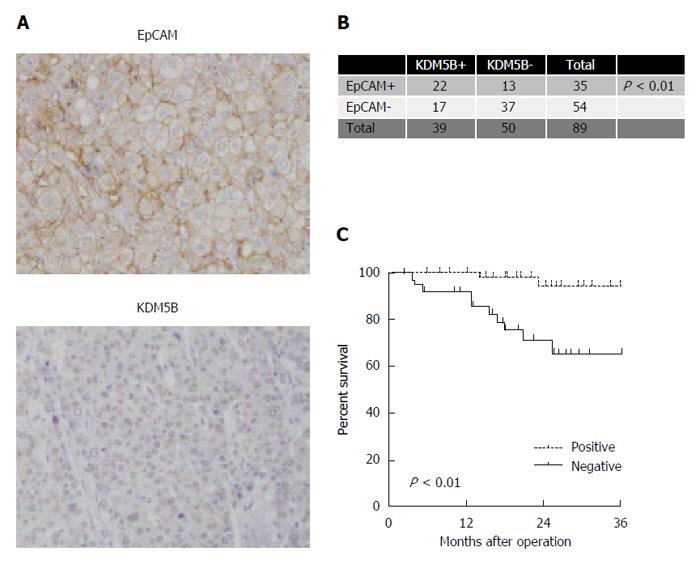
We performed IHC analysis of cohort 3 cases. We confirmed the nuclear accumulation of KDM5B stained by an anti-KDM5B antibody (Figure 5A). After evaluating the clinicopathological characteristics of EpCAM-positive and -negative HCC cases, we found that EpCAM-positive HCCs were associated with a significantly high frequency of KDM5B-positive cases (Figure 5B). We further identified the survival outcome of these cases by Kaplan-Meier survival analysis. KDM5B-positive HCCs were associated with significantly lower overall survival outcomes within three years compared with KDM5B-negative HCCs (P < 0.01) (Figure 5C).
HBV is the smallest human hepatotropic DNA virus, which mainly infects host hepatocytes and causes a spectrum of pathological processes from acute hepatitis and chronic hepatitis, to serious end-stage liver diseases such as hepatic cirrhosis and primary HCC. Studies of its epidemiology and natural history have shown that approximately 25% of chronic hepatitis B patients will develop HCC[23,24]. Although the pathogenesis of HBV-related HCC has not been identified, many studies have suggested that HBx is one of the risk factors and is strongly implicated in hepatocarcinogenesis.
The role of epigenetic regulation in cancer biology, especially that of the histone lysine demethylases (KDMs), has been the subject of several studies[25-27]. These chromatin structure modifiers have been increasingly shown to facilitate several steps of cancer progression[28-30]. Several KDMs have been implicated in tumor growth, angiogenesis, invasion, metastasis, and tumor-related chemoresistance. KDM5B specifically removes methyl residues from methylated lysine 4 of histone 3, consequently repressing gene transcription. The cancer stem-like cell (CSC) hypothesis has drawn much attention[31]. CSCs possess stem cell characteristics, including self-renewal, chemotherapy resistance, and metastasis. On the basis of the role of KDM5B in cancer stem-like features, we hypothesized that KDM5B is actively involved in the poor prognosis of HCC patients.
In this study, we showed that KDM5B were associated with poor prognosis of HBV-related HCC. In various HBV proteins, HBx strongly up-regulated KDM5B expression in HBV-related HCC cases. Moreover, KDM5B expression is associated with the increased presence of CSC features, including enhanced tumor sphere formation, cell migration, and invasion, and is related with poor prognosis. The administration of a KDM5B inhibitor suppressed HpSC-like features in HBV-related cases. Therefore, we consider that KDM5B could be a potential therapeutic target against HBV-related HCC. Many researchers reported that HBx-MYC interaction was associated with HBV-related hepatocarcinogenesis[32-34]. However, its mechanism is not yet clear. Our results indicated that KDM5B was a central molecule of HBx-MYC introduced hepatocarcinogenesis.
Recently, Wang et al[35] reported that KDM5B affected the poor prognosis of HCC cases through regulating p15 and p27. Similarly, in the present study, KDM5B was related to the poor prognosis of HCC patients. Our data show that KDM5B, which is activated by HBx, is a useful marker for poor prognosis in HBV-related HCC cases. Moreover, we demonstrated the possibility that suppression of KDM5B may improve the poor phenotype of HBV-related HCCs.
Our new knowledge is useful for the diagnosis of severe HCC patients. Histone demethylases KDM5B may be an important therapeutic target against HBV-related HCC cases.
In conclusion, HBx induced HpSC-like features in hepatocellular carcinoma by activating KDM5B. Moreover, HBx and KDM5B interaction related poor prognosis of hepatocellular carcinoma cases.
We thank Drs. Xin Wei Wang and Snorri Thorgeirsson at NCI for clinical data of HCC cases. We also thank Dr. Hikari Okada and Takayoshi Shirasaki for help on cell analysis.
The role of epigenetic regulation in cancer biology has been the subject of several studies. These chromatin structure modifiers have been increasingly shown to facilitate several steps of cancer progression. However, the epigenetic regulation of hepatocellular carcinoma has not been elucidated.
Previous study showed that KDM5B affected the poor prognosis of hepatocellular carcinoma (HCC) cases through regulating p15 and p27. However, the relation between HCC background and KDM5B has not yet clear.
This is the first study evaluating that KDM5B, which is activated by HBx, a useful marker for poor prognosis in HBV-related HCC cases. Moreover, the authors demonstrated the possibility that suppression of KDM5B may improve the poor phenotype of HBV-related HCCs.
HBx activated the histone demethylase KDM5B and induced hepatic progenitor cell (HPC)-like features in HCC. Presented results suggested that histone demethylases may be an important therapeutic target against HBV-related HCC cases.
Authors demonstrated that HBx activates the histone demethylase KDM5B and induces HPC-like features in HCC. This is the first study evaluating that KDM5B, which is activated by HBx, a useful marker for poor prognosis in HBV-related HCC cases. Moreover, they demonstrated the possibility that suppression of KDM5B may improve the poor phenotype of HBV-related HCCs.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Ohkoshi S, Qin Y S- Editor: Yu J L- Editor: A E- Editor: Wang CH
| 1. | Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-2128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9500] [Cited by in RCA: 9575] [Article Influence: 736.5] [Reference Citation Analysis (0)] |
| 2. | Hirata A, Hirata T, Takahashi Y, Nakayama T. Surveillance rates for hepatocellular carcinoma among patients with cirrhosis, chronic hepatitis B, and chronic hepatitis C based on Japanese claims database. Hepatol Res. 2017;47:283-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Zhang X, Zhang H, Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. J Lab Clin Med. 2006;147:58-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 156] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 4. | Matsuda Y, Ichida T. Impact of hepatitis B virus X protein on the DNA damage response during hepatocarcinogenesis. Med Mol Morphol. 2009;42:138-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 5. | Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64:S84-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 710] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 6. | Zhang Y, Liu H, Yi R, Yan T, He Y, Zhao Y, Liu J. Hepatitis B virus whole-X and X protein play distinct roles in HBV-related hepatocellular carcinoma progression. J Exp Clin Cancer Res. 2016;35:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang Q, Cai N, Wang H, Liu F, Ye L. Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP. Mol Cancer. 2014;13:128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Oishi N, Shilagardi K, Nakamoto Y, Honda M, Kaneko S, Murakami S. Hepatitis B virus X protein overcomes oncogenic RAS-induced senescence in human immortalized cells. Cancer Sci. 2007;98:1540-1548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 243] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 10. | Hu XM, Yan XH, Hu YW, Huang JL, Cao SW, Ren TY, Tang YT, Lin L, Zheng L, Wang Q. miRNA-548p suppresses hepatitis B virus X protein associated hepatocellular carcinoma by downregulating oncoprotein hepatitis B x-interacting protein. Hepatol Res. 2016;46:804-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274:15633-15645. [PubMed] |
| 12. | Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226-19231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 310] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 13. | Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue H. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer. 2010;9:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 14. | Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473-1484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 15. | Stalker L, Wynder C. Evaluation of histone-modifying enzymes in stem cell populations. Methods Mol Biol. 2012;809:411-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Jia HL, Ye QH, Qin LX, Budhu A, Forgues M, Chen Y, Liu YK, Sun HC, Wang L, Lu HZ. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:1133-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 157] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 17. | Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 748] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 18. | Yamashita T, Honda M, Nakamoto Y, Baba M, Nio K, Hara Y, Zeng SS, Hayashi T, Kondo M, Takatori H. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57:1484-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 227] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 19. | Okitsu T, Kobayashi N, Jun HS, Shin S, Kim SJ, Han J, Kwon H, Sakaguchi M, Totsugawa T, Kohara M. Transplantation of reversibly immortalized insulin-secreting human hepatocytes controls diabetes in pancreatectomized pigs. Diabetes. 2004;53:105-112. [PubMed] |
| 20. | Shirasaki T, Honda M, Shimakami T, Murai K, Shiomoto T, Okada H, Takabatake R, Tokumaru A, Sakai Y, Yamashita T. Impaired interferon signaling in chronic hepatitis C patients with advanced fibrosis via the transforming growth factor beta signaling pathway. Hepatology. 2014;60:1519-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Shinoda M, Tilles AW, Kobayashi N, Wakabayashi G, Takayanagi A, Totsugawa T, Harada H, Obara H, Suganuma K, Berthiaume F. A bioartificial liver device secreting interleukin-1 receptor antagonist for the treatment of hepatic failure in rats. J Surg Res. 2007;137:130-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Pfeifer AM, Cole KE, Smoot DT, Weston A, Groopman JD, Shields PG, Vignaud JM, Juillerat M, Lipsky MM, Trump BF. Simian virus 40 large tumor antigen-immortalized normal human liver epithelial cells express hepatocyte characteristics and metabolize chemical carcinogens. Proc Natl Acad Sci USA. 1993;90:5123-5127. [PubMed] |
| 23. | Buendia MA, Neuveut C. Hepatocellular carcinoma. Cold Spring Harb Perspect Med. 2015;5:a021444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 24. | Saitta C, Tripodi G, Barbera A, Bertuccio A, Smedile A, Ciancio A, Raffa G, Sangiovanni A, Navarra G, Raimondo G. Hepatitis B virus (HBV) DNA integration in patients with occult HBV infection and hepatocellular carcinoma. Liver Int. 2015;35:2311-2317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 25. | Secombe J, Eisenman RN. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle. 2007;6:1324-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 26. | Yamamoto S, Wu Z, Russnes HG, Takagi S, Peluffo G, Vaske C, Zhao X, Moen Vollan HK, Maruyama R, Ekram MB. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 2014;25:762-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 27. | Wang Z, Tang F, Qi G, Yuan S, Zhang G, Tang B, He S. KDM5B is overexpressed in gastric cancer and is required for gastric cancer cell proliferation and metastasis. Am J Cancer Res. 2015;5:87-100. [PubMed] |
| 28. | Li X, Liu L, Yang S, Song N, Zhou X, Gao J, Yu N, Shan L, Wang Q, Liang J. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci USA. 2014;111:7096-7101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 113] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 29. | Shen X, Zhuang Z, Zhang Y, Chen Z, Shen L, Pu W, Chen L, Xu Z. JARID1B modulates lung cancer cell proliferation and invasion by regulating p53 expression. Tumour Biol. 2015;36:7133-7142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Kuo YT, Liu YL, Adebayo BO, Shih PH, Lee WH, Wang LS, Liao YF, Hsu WM, Yeh CT, Lin CM. JARID1B Expression Plays a Critical Role in Chemoresistance and Stem Cell-Like Phenotype of Neuroblastoma Cells. PLoS One. 2015;10:e0125343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 31. | Tan BT, Park CY, Ailles LE, Weissman IL. The cancer stem cell hypothesis: a work in progress. Lab Invest. 2006;86:1203-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 203] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 32. | Shukla SK, Kumar V. Hepatitis B virus X protein and c-Myc cooperate in the upregulation of ribosome biogenesis and in cellular transformation. FEBS J. 2012;279:3859-3871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Lakhtakia R, Kumar V, Reddi H, Mathur M, Dattagupta S, Panda SK. Hepatocellular carcinoma in a hepatitis B ‘x’ transgenic mouse model: A sequential pathological evaluation. J Gastroenterol Hepatol. 2003;18:80-91. [PubMed] |
| 34. | Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, Buendia MA. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 229] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 35. | Wang D, Han S, Peng R, Jiao C, Wang X, Yang X, Yang R, Li X. Depletion of histone demethylase KDM5B inhibits cell proliferation of hepatocellular carcinoma by regulation of cell cycle checkpoints p15 and p27. J Exp Clin Cancer Res. 2016;35:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |