Published online Apr 28, 2017. doi: 10.3748/wjg.v23.i16.2891
Peer-review started: December 28, 2016
First decision: January 19, 2017
Revised: February 14, 2017
Accepted: March 30, 2017
Article in press: March 30, 2017
Published online: April 28, 2017
Processing time: 125 Days and 22.5 Hours
To investigate the use of droplet digital polymerase chain reaction (ddPCR) for detecting host mRNA markers in stools as a non-invasive test for colorectal cancer screening.
ddPCR and quantitative PCR were compared side by side for their performance in the detection of ITGA6 and ITGA6A transcripts in stool samples obtained from patients with various types of colorectal lesions (advanced adenomas and stage II-IV colorectal cancers) and control (patients displaying no pathological findings) using duplex TaqMan reactions for both methods. ITGA6 and ITGA6A were chosen for this proof-of-concept study based on their relative medium and low abundance in stool samples, respectively, as established in a previous study.
We found that the ddPCR and qPCR methods performed equally well in this TaqMan duplex assay for the detection of ITGA6 and ITGA6A transcripts in stools of patients with colorectal lesions. For ITGA6, receiver operating characteristic (ROC) curve analysis showed comparable areas under the curve of 0.91 (P < 0.0001) and 0.89-0.90 (P < 0.0001) for the prediction of advanced adenomas and colorectal cancers, respectively. ITGA6A, which was detected at very low levels in control patients, was found to be significantly elevated (over 40 times) in stage II and III colorectal cancers (P < 0.0002). Comparison of the two sets of data revealed a strong correlation of the copy numbers obtained by ddPCR and qPCR for both ITGA6 and ITGA6A.
We found that ITGA6 and ITGA6A detection in stools of patients with colorectal cancers with ddPCR is comparable to that of qPCR using TaqMan assays.
Core tip: We investigated the use of droplet digital polymerase chain reaction (ddPCR) for detecting host mRNA markers in stools as a non-invasive test for colorectal cancer screening using ITGA6 and ITGA6A as established medium and low abundance stool biomarkers. Our side by side comparison of ddPCR and qPCR on stool samples obtained from patients with advanced adenomas and colorectal cancers and controls using duplex TaqMan reactions revealed that both methods performed equally well.
- Citation: Herring E, Kanaoka S, Tremblay É, Beaulieu JF. Droplet digital PCR for quantification of ITGA6 in a stool mRNA assay for the detection of colorectal cancers. World J Gastroenterol 2017; 23(16): 2891-2898
- URL: https://www.wjgnet.com/1007-9327/full/v23/i16/2891.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i16.2891
Colorectal cancer (CRC) is one of the most common causes of cancer-related mortality worldwide, now accounting for more than 600000 deaths annually just in the United States[1]. Early detection is a key factor in reducing mortality from CRC[2-4]. The extent of the spread of the disease in terms of local invasion as well as to lymph nodes and distal organs at the time of diagnosis is an important prognostic factor, with five year survival rates of more than 90% for localized CRC but only about 10% for CRC having metastasized to distant organs[5]. Several screening regiments for CRC are recommended such as fecal occult blood testing and colonoscopy. While colonoscopy remains the gold standard for the detection of colorectal lesions (up to 95% sensitivity for cancer and 76% for adenomas), compliance is low owing to discomfort and unpleasant preparation procedures[6]. The risk of complications, cost and access are other limitations of this procedure. On the other hand, the improved immunological version of fecal occult blood testing, which detects human hemoglobin, has been used for some time but poor precursor lesion detection rates (66%-80% sensitivity for cancer but only 10%-28% for adenomas) albeit an excellent specificity (95%) limit its utility[3,7-9]. It is therefore imperative to explore alternate or complementary strategies with the potential to improve CRC screening performance, especially for the detection of cancers at their early stages.
Non-invasive stool testing represents an advantageous as well as a biologically rational approach based on the high rate of tumor cell exfoliation into the lumen for neoplasm detection through DNA- and RNA-based approaches[10-15]. Detection of host mRNA in stools has been reported to be particularly challenging considering that human mRNA is expected to represent less than 1% of total stool RNA[16] and that stool mRNA may be particularly sensitive to degradation[3,17]. Nevertheless, the potential usefulness of detecting specific transcripts in feces for CRC screening was demonstrated more than two decades ago, using first generation PCR[18,19]. More recently, using TaqMan quantitative PCR (qPCR), a number of transcripts such as PTGS2 (COX-2), B2M, CEACAM5, CDH1, ITGA6, MYC and GADD45B are now listed among the stool mRNA known to be up-regulated in patients with CRC[10,20]. Interestingly, a few other genes such as PTPRC (CD45) and ITGA6A have been detected at low levels, but only in patients with CRC[10,20], making them of particular interest for their specificity. One may now wonder if a more sensitive assay for mRNA detection could improve the reliability of these specific biomarkers in a non-invasive test for CRC detection.
In the present study, we addressed this question in the context of the development of droplet digital PCR (ddPCR), a now commercially available technology identified as third generation PCR, claimed to be of an unprecedented sensitivity[21,22]. Besides the obvious interest in the detection of rare mutations of various kinds, ddPCR technology displays a number of additional advantages for general nucleic acid quantification[23,24] such as the capability to obtain absolute quantification without the need of standard dilution curves and reaction efficiency determination[22,23]. Another potential advantage of ddPCR technology, being an end-point approach, is the reaction tolerance to PCR inhibitors[22], an important advantage when using clinical samples[25].
The integrin α6 subunit has been extensively characterized in CRC cell lines and primary tumors as being up-regulated[26,27] together with its β4 partner[28] to form a pro-proliferative α6β4 laminin receptor[27,29]. In this proof-of-concept comparative study, we used ITGA6 (integrin α6 subunit) and ITGA6A (its α6A variant) as representative medium and low detectable stool target genes, respectively, as demonstrated previously[20]. Our results show that when analyzed side by side using duplex TaqMan reactions, qPCR and ddPCR methods performed with comparable sensitivity for the detection of these two target genes in stools of patients with colorectal lesions.
Detailed information pertaining to the samples used in this work has been provided in a previous study[20]. Briefly, the study cohort used herein included 24 patients with advanced adenomas (Ad; defined as being larger than 1 cm at the greatest dimension) as well as 65 patients with stage II-IV CRC (32 stage II; 22 stage III and 11 stage IV) that were diagnosed by colonoscopy and histology. A total of 30 patients with no pathological findings were used as controls. For control patients and patients with adenomas, stools samples were collected before colonoscopy while patients with CRC provided samples 2-4 wk after colonoscopy and biopsy (but before surgery)[18].
Isolation of RNA from fecal specimens was performed according to a procedure described previously[10,18]. cDNA was synthesized using M-MLV Reverse Transcriptase, RNase H Minus (Takara Bio Inc., Otsu, Japan) with 0.375 μg total RNA from stools and 750 ng random hexamers in a final reaction volume of 60 μL. For preamplification, the TaqMan PreAmp Master Mix Kit (Life Technologies) was used to provide unbiased, multiplex preamplification of specific amplicons for analysis with TaqMan gene expression assays. Commercially available TaqMan primer and probe mixtures were used for the amplification of ITGA6 (Hs01041011_m1 labelled with the reporter dye VIC) and ITGA6A (Hs01041013_m1 labelled with the reporter dye FAM). Briefly, 20 × TaqMan gene expression assays were pooled at a final concentration of 0.2 × in 1 × TE, combined with 5 μL fecal cDNA and 2 × TaqMan PreAmp Master Mix in a total volume of 20 μL, then preamplified for 14 PCR cycles of 15 s at 95 °C and 4 min at 60 °C. Linear amplification of the cDNA samples using the preamplification kit under these conditions has been previously verified[20]. Preamplification products were diluted 1:20 with 1 × TE buffer, aliquoted and stored at -80 °C.
Duplex qPCR reactions were prepared by combining 1 μL of the 20 × diluted preamplification products with 10 μL TaqMan Gene Expression Master Mix and 1 μL of each 20 × TaqMan Gene Expression Assay in a total volume of 20 μL. qPCR was performed for 60 cycles of 30 s 95 °C, 1 min 60 °C in an Mx3000P Real Time PCR machine (Stratagene, Mississauga, Ontario) and read in both the FAM and HEX channels.
Twenty microliters of each reaction mix containing 10 μL ddPCR Supermix for Probes (BioRad), 1 μL of each 20 × TaqMan Gene Expression Assay for ITGA6A and ITGA6 and 1 μL of each 20 × diluted preamplification product was converted to droplets using the QX200 droplet generator (BioRad). Droplet-partitioned samples were then transferred to a 96-well plate, sealed and cycled in a MyCycler Thermocycler (BioRad). DNA polymerase activation was at 94 °C for 10 min, followed by 45 cycles of 94 °C for 30 s and 55 °C for 1 min and a post-cycling step of 98 °C for 10 min for enzyme inactivation. The cycled plate was then transferred and read in the FAM and HEX channels using the QX200 reader (BioRad).
Stool mRNA detection results were presented as copy number per microlitre for both PCR assays. For each gene, a standard reference curve was generated for ITGA6 and ITGA6A using serial 2 fold dilutions of a cDNA stock solution of the target sequence quantified using a NanoDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE, United States).
For statistics, data were analyzed as previously described[20] using Prism 6 software (GraphPad). Correlations of stool mRNA detection by qPCR and ddPCR were evaluated using the nonparametric Spearman correlation test. Comparisons of mRNA expression in stool samples from controls and patients with lesions were expressed as median with interquartile range and analyzed by the Kruskal-Wallis test followed by Dunn’s multiple comparison tests. Areas under the receiver operating characteristic (ROC) curves were calculated using Prism 6. Sensitivities and specificities were expressed in % with a 95% confidence interval (CI). Optimal cutoff values were calculated with Cutoff Finder[30]. Statistical significance was defined as P < 0.05.
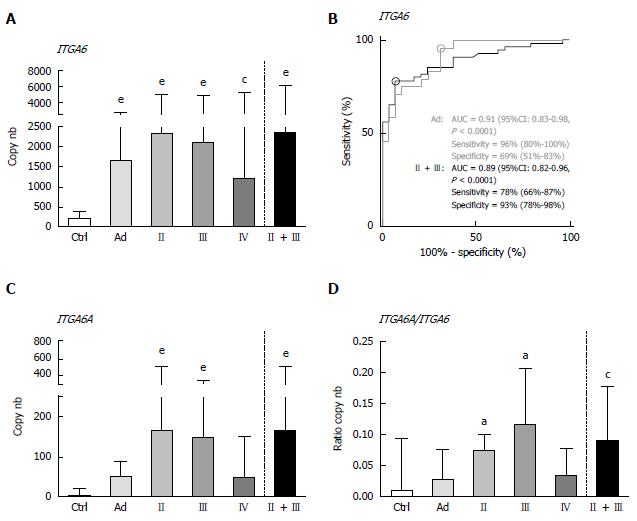
The mRNA levels of ITGA6 and ITGA6A were evaluated in the same sets of stool samples obtained from patients diagnosed with Ad or CRC and controls in duplex for both PCR methods. For each group, results were expressed as median transcript copy number.
By ddPCR, the median levels of ITGA6 transcript detection in stools of patients with colorectal lesions were found to be statistically significantly increased as compared to those of the controls (Figure 1A). As compared to controls, the ITGA6 mRNA levels were approximately 8 times higher for Ad (P < 0.0001) and 6-11 times for CRC, depending on the stage (P < 0.002-0.0001; 11 times when stages II + III are considered, P < 0.0001). ROC analysis of ITGA6 showed an area under the curve (AUC) of 0.89 (P < 0.0001) for the prediction of stages II and III CRC with 78% sensitivity and 93% specificity and of 0.91 (P < 0.0001) for the prediction of Ad with 96% sensitivity and 69% specificity (Figure 1B). The detection of ITGA6A, a pro-proliferative ITGA6 splicing variant up-regulated in CRC cells[26,27], was also investigated in the stools of patients with colorectal lesions. ITGA6A levels detected in the stools of control patients were very low while found to be significantly more elevated (over 40 times) in the stools of patients with stages II and III CRC (P < 0.0002). Analyses of the ITGA6A/ITGA6 ratios revealed that ITGA6A represented less than 1% and 3% of the total ITGA6 copy number in the stools of control or Ad patients, respectively, but approximately 10% in the stools of patients with stage II or III CRC (Figure 1D).
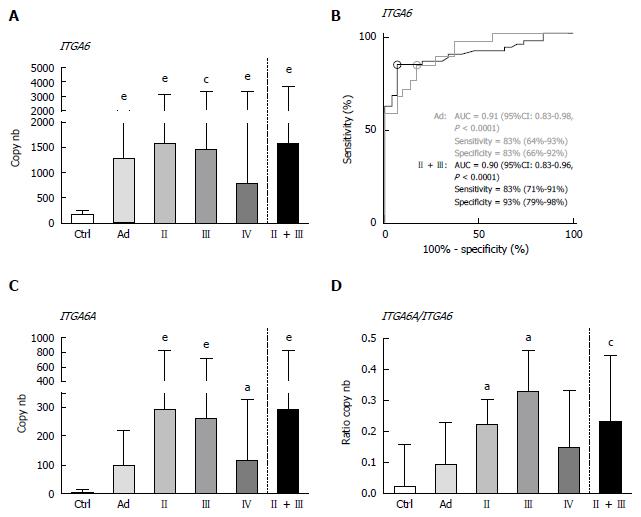
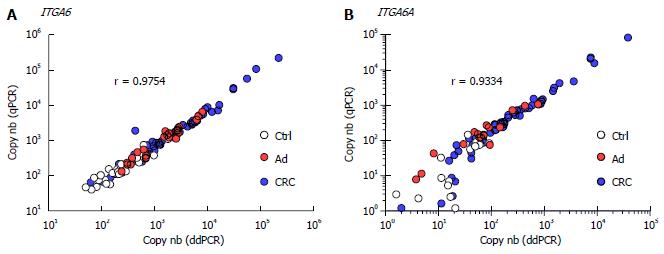
Overall, the results using duplex ddPCR for ITGA6 and ITGA6A were found to be comparable to those reported previously with conventional qPCR[20] albeit with an apparent underestimation of the general ITGA6A/ITGA6 ratio. To ensure a valid comparison of the PCR methods, the same samples as above were run in parallel in duplex qPCR. As shown in Figure 2, the results were very similar to those obtained with ddPCR. For instance, ROC analysis of ITGA6 showed comparable AUC of 0.90 (P < 0.0001) and 0.91 (P < 0.0001) for the prediction of stage II + III and Ad, respectively (Figure 2B). In fact the only notable distinction in the data obtained using qPCR was the more elevated ITGA6A/ITGA6 ratios resulting in an estimation of the proportion of ITGA6A at 2% and 9% of the total ITGA6 for control and Ad patients and up to 25% in the stools of patients with stage II and III CRC. This appears consistent with the apparent slight under-estimation of ITGA6 and over-estimation of ITGA6A. To verify this phenomenon, the linearity between the two sets of samples generated with ddPCR and qPCR was evaluated using nonparametric Spearman analysis. As shown in Figure 3, a significant correlation was obtained for both ITGA6 (r = 0.9754) and ITGA6A (r = 0.9334) confirming that the inter-variations between the ddPCR and qPCR analyses were uniform and not the result of a subset of divergent samples.
In the present study, we have compared side by side ddPCR and qPCR for the detection of ITGA6 and ITGA6A transcripts in stool samples obtained from patients with various types of colorectal lesions and controls and have shown that both methods performed with comparable sensitivity for the two target genes. While expected for ITGA6, which is a relatively abundant transcript ubiquitously expressed in normal and cancer intestinal epithelial cells[26,31], the comparable sensitivity in the detection of its ITGA6A variant was more surprising considering that it is less abundant, its expression being restricted to the lower crypt cells which do not extrude into the lumen[31] and to colorectal cancer cells[26]. Indeed, ddPCR has been shown to be more sensitive to detect low expression targets and to be more tolerant to PCR inhibitors[21-24], two advantages that could have increased stool ITGA6A detection. For instance, ddPCR was recently successfully used for the measurement of inflammatory transcripts in stools of children confirming that this method can be used in clinical investigation[16].
Recent studies compared quantitative to digital PCR for the detection of pathogens in clinical stool samples. The ddPCR assay was reported to perform better than qPCR for the detection of human cytomegalovirus, mainly because it was less susceptible to PCR inhibitors[25]. In another study designed for the enumeration of Cryptosporidium oocysts, ddPCR was also reported to be less affected by the presence of inhibitors but overall, once qPCR data were corrected for pipetting and DNA losses, the sensitivity of both methods was considered to be comparable[32]. In a third study, digital PCR outperformed SYBR green-based qPCR for the detection of fecal enterotoxigenic Bacteroides fragilis but provided similar results compared to TaqMan qPCR[33]. It is noteworthy that we also used TaqMan Gene Expression Assays for ITGA6 and ITGA6A in order to allow direct comparison between duplex ddPCR and qPCR.
Use of TaqMan assays has been favored for the detection of host mRNA in stools for better sensitivity[10,20] and also because they are not susceptible to the generation of primer-dimers, a phenomenon frequently observed for low expression targets in complex biological samples by the SYBR green method[34]. TaqMan qPCR may also be less affected by PCR inhibitors than SYBR green-based qPCR[33]. In this context, it is noteworthy that the specificities for ITGA6A detection in patients with CRC vs control patients were over 96% for both PCR methods (ddPCR: 96.6 %; qPCR: 96.7%). With such low levels of false positives combined with a sensitivity of 65% for both methods, ITGA6A appears to be an excellent candidate biomarker for inclusion in a multiplex stool RNA assay for CRC screening. It is interesting to note that the failure of ITGA6A to detect patients with Ad is consistent with our previous observation that ITGA6A is only significantly overexpressed in CRC lesions[26,27]. On the other hand, the areas under the ROC curves for ITGA6 determined by ddPCR and qPCR were 0.91 for Ad and 0.89 and 0.90, respectively, for CRC, confirming our previous findings on the usefulness of ITGA6 in the detection of colorectal lesions in a non-invasive assay.
Taken together, these data indicate that both TaqMan qPCR and ddPCR perform well in this duplex assay comprised of the moderately abundant target transcript ITGA6 and its scarcer variant ITGA6A. One of the main advantages of ddPCR is that it provides absolute quantification of the target DNA in copy number to a point where it has been proposed for the construction of calibration curves[22,23,35]. Direct comparison of the results obtained with the two techniques revealed strong correlations between ddPCR and qPCR measurements. Interestingly, the yields observed for ddPCR were slightly higher for ITGA6 but lower for ITGA6A than for qPCR resulting in an apparent 2-3-fold overestimation of ITGA6A relative to the total ITGA6 by qPCR in the stools of patients with CRC. These differences in target quantitation between ddPCR and qPCR are comparable to those reported by other authors[22,36].
In conclusion, we found that ITGA6 and ITGA6A detection with ddPCR was comparable to that of qPCR using TaqMan assays. Considering the cost of ddPCR is estimated to be five times that of TaqMan qPCR[33] while qPCR allows the processing of more samples per run, we agree with the suggestion that ddPCR is optimal for calibrating targets for subsequent use on other platforms such as qPCR[22,23,35].
The authors thank Bio-Rad Laboratories Canada and Dr. Katia Nadeau for the prolonged trial period with the Q200 droplet generator and reader and Dr. Sean C. Taylor for scientific advice on ddPCR.
Because colorectal cancer can be successfully treated before the occurrence of metastasis, early and efficient diagnosis of the colorectal lesions is important. Currently approved non-invasive methods based on the detection of biomarkers in the stool include fecal occult blood and more recently, stool host DNA. Unfortunately, the sensitivity of these methods for detecting precancerous lesions remains low. Detection of host mRNA in stools has been also investigated over the last 2 decades and lead to the identification of a number of transcripts up-regulated in patients with colorectal cancers.
Detection of host mRNA in stools appears challenging considering its low proportion in regard to total stool RNA and its particular sensitivity to degradation. Efficient mRNA detection protocols are thus needed to insure the reliability of this approach.
In this proof of concept study, we compared side by side droplet digital polymerase chain reaction (PCR) to quantitative PCR using duplex TaqMan reactions for the detection of the two previously characterized stool colorectal cancer markers ITGA6 and ITGA6A. These results show that the two methods performed well, showing comparable sensitivity for the detection of the two targets genes in the stools of patient with colorectal lesions.
These findings suggest that although droplet digital PCR has some advantages, its sensitivity for detection seems comparable to the less expensive quantitative PCR when using duplex TaqMan reactions.
Droplet digital PCR is a now commercially available technology for DNA amplification considered as third generation PCR, relative to its previous generation, the quantitative PCR. TaqMan assays consist of the use of a pair of unlabeled primers with a specific labeled probe, which increase specificity of PCR.
This is an interesting and well written study on droplet digital PCR for quantification of ITGA6 in a stool mRNA assay for the early detection of colorectal neoplasms.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Canada
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Crea F, Uppara M S- Editor: Gong ZM L- Editor: A E- Editor: Wang CH
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12135] [Cited by in RCA: 12983] [Article Influence: 1442.6] [Reference Citation Analysis (2)] |
| 2. | Maratt JK, Saini SD. Colorectal cancer screening in the 21st century: where do we go from here? Am J Manag Care. 2015;21:e447-e449. [PubMed] |
| 3. | Robertson DJ, Imperiale TF. Stool Testing for Colorectal Cancer Screening. Gastroenterology. 2015;149:1286-1293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 4. | Willyard C. Screening: Early alert. Nature. 2015;521:S4-S5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Manfredi S, Bouvier AM, Lepage C, Hatem C, Dancourt V, Faivre J. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg. 2006;93:1115-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 235] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 6. | Schroy PC, Lal S, Glick JT, Robinson PA, Zamor P, Heeren TC. Patient preferences for colorectal cancer screening: how does stool DNA testing fare? Am J Manag Care. 2007;13:393-400. [PubMed] |
| 7. | Allison JE, Fraser CG, Halloran SP, Young GP. Population screening for colorectal cancer means getting FIT: the past, present, and future of colorectal cancer screening using the fecal immunochemical test for hemoglobin (FIT). Gut Liver. 2014;8:117-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 8. | Hundt S, Haug U, Brenner H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann Intern Med. 2009;150:162-169. [PubMed] |
| 9. | Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med. 2014;160:171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 485] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 10. | Hamaya Y, Yoshida K, Takai T, Ikuma M, Hishida A, Kanaoka S. Factors that contribute to faecal cyclooxygenase-2 mRNA expression in subjects with colorectal cancer. Br J Cancer. 2010;102:916-921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Ahlquist DA, Harrington JJ, Burgart LJ, Roche PC. Morphometric analysis of the “mucocellular layer” overlying colorectal cancer and normal mucosa: relevance to exfoliation and stool screening. Hum Pathol. 2000;31:51-57. [PubMed] |
| 12. | Koga Y, Yasunaga M, Katayose S, Moriya Y, Akasu T, Fujita S, Yamamoto S, Baba H, Matsumura Y. Improved recovery of exfoliated colonocytes from feces using newly developed immunomagnetic beads. Gastroenterol Res Pract. 2008;2008:605273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Rengucci C, De Maio G, Menghi M, Scarpi E, Guglielmo S, Fusaroli P, Caletti G, Saragoni L, Casadei Gardini A, Zoli W. Improved stool DNA integrity method for early colorectal cancer diagnosis. Cancer Epidemiol Biomarkers Prev. 2014;23:2553-2560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Yu YJ, Majumdar AP, Nechvatal JM, Ram JL, Basson MD, Heilbrun LK, Kato I. Exfoliated cells in stool: a source for reverse transcription-PCR-based analysis of biomarkers of gastrointestinal cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:455-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;371:187-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 16. | Stauber J, Shaikh N, Ordiz MI, Tarr PI, Manary MJ. Droplet digital PCR quantifies host inflammatory transcripts in feces reliably and reproducibly. Cell Immunol. 2016;303:43-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Reck M, Tomasch J, Deng Z, Jarek M, Husemann P, Wagner-Döbler I. Stool metatranscriptomics: A technical guideline for mRNA stabilisation and isolation. BMC Genomics. 2015;16:494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 18. | Kanaoka S, Yoshida K, Miura N, Sugimura H, Kajimura M. Potential usefulness of detecting cyclooxygenase 2 messenger RNA in feces for colorectal cancer screening. Gastroenterology. 2004;127:422-427. [PubMed] |
| 19. | Takai T, Kanaoka S, Yoshida K, Hamaya Y, Ikuma M, Miura N, Sugimura H, Kajimura M, Hishida A. Fecal cyclooxygenase 2 plus matrix metalloproteinase 7 mRNA assays as a marker for colorectal cancer screening. Cancer Epidemiol Biomarkers Prev. 2009;18:1888-1893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Beaulieu JF, Herring E, Kanaoka S, Tremblay É. Use of integrin alpha 6 transcripts in a stool mRNA assay for the detection of colorectal cancers at curable stages. Oncotarget. 2016;7:14684-14692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Huggett JF, Whale A. Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin Chem. 2013;59:1691-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 22. | Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003-1005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 927] [Cited by in RCA: 1090] [Article Influence: 90.8] [Reference Citation Analysis (0)] |
| 23. | Cao L, Cui X, Hu J, Li Z, Choi JR, Yang Q, Lin M, Ying Hui L, Xu F. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron. 2017;90:459-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 186] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 24. | Zec H, Shin DJ, Wang TH. Novel droplet platforms for the detection of disease biomarkers. Expert Rev Mol Diagn. 2014;14:787-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Sedlak RH, Kuypers J, Jerome KR. A multiplexed droplet digital PCR assay performs better than qPCR on inhibition prone samples. Diagn Microbiol Infect Dis. 2014;80:285-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 26. | Dydensborg AB, Teller IC, Groulx JF, Basora N, Paré F, Herring E, Gauthier R, Jean D, Beaulieu JF. Integrin alpha6Bbeta4 inhibits colon cancer cell proliferation and c-Myc activity. BMC Cancer. 2009;9:223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Groulx JF, Giroux V, Beauséjour M, Boudjadi S, Basora N, Carrier JC, Beaulieu JF. Integrin α6A splice variant regulates proliferation and the Wnt/β-catenin pathway in human colorectal cancer cells. Carcinogenesis. 2014;35:1217-1227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Ni H, Dydensborg AB, Herring FE, Basora N, Gagné D, Vachon PH, Beaulieu JF. Upregulation of a functional form of the beta4 integrin subunit in colorectal cancers correlates with c-Myc expression. Oncogene. 2005;24:6820-6829. [PubMed] |
| 29. | Beaulieu JF. Integrin α6β4 in colorectal cancer. World J Gastrointest Pathophysiol. 2010;1:3-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 783] [Cited by in RCA: 1011] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 31. | Dydensborg AB, Teller IC, Basora N, Groulx JF, Auclair J, Francoeur C, Escaffit F, Paré F, Herring E, Ménard D. Differential expression of the integrins alpha6Abeta4 and alpha6Bbeta4 along the crypt-villus axis in the human small intestine. Histochem Cell Biol. 2009;131:531-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 32. | Yang R, Paparini A, Monis P, Ryan U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int J Parasitol. 2014;44:1105-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 33. | Purcell RV, Pearson J, Frizelle FA, Keenan JI. Comparison of standard, quantitative and digital PCR in the detection of enterotoxigenic Bacteroides fragilis. Sci Rep. 2016;6:34554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | Tajadini M, Panjehpour M, Javanmard SH. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv Biomed Res. 2014;3:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 119] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 35. | Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015;61:79-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 323] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 36. | Campomenosi P, Gini E, Noonan DM, Poli A, D’Antona P, Rotolo N, Dominioni L, Imperatori A. A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol. 2016;16:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |