Published online Apr 28, 2017. doi: 10.3748/wjg.v23.i16.2819
Peer-review started: September 19, 2016
First decision: October 28, 2016
Revised: November 16, 2016
Accepted: March 15, 2017
Article in press: March 15, 2017
Published online: April 28, 2017
Processing time: 225 Days and 7.2 Hours
RNA sequencing is the use of high throughput next generation sequencing technology to survey, characterize, and quantify the transcriptome of a genome. RNA sequencing has been used to analyze the pathogenesis of several malignancies such melanoma, lung cancer, and colorectal cancer. RNA sequencing can identify differential expression of genes (DEG’s), mutated genes, fusion genes, and gene isoforms in disease states. RNA sequencing has been used in the investigation of several colorectal diseases such as colorectal cancer, inflammatory bowel disease (ulcerative colitis and Crohn’s disease), and irritable bowel syndrome.
Core tip: RNA sequencing is the use of high throughput next generation sequencing technology to survey, characterize, and quantify the transcriptome of a genome. RNA sequencing has been used in the investigation of several colorectal diseases such as colorectal cancer, inflammatory bowel disease (ulcerative colitis and Crohn’s disease), and irritable bowel syndrome.
- Citation: Gao M, Zhong A, Patel N, Alur C, Vyas D. High throughput RNA sequencing utility for diagnosis and prognosis in colon diseases. World J Gastroenterol 2017; 23(16): 2819-2825
- URL: https://www.wjgnet.com/1007-9327/full/v23/i16/2819.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i16.2819
RNA Sequencing Basics: RNA sequencing is the use of high throughput next generation sequencing technology to survey, characterize, and quantify the transcriptome of a genome[1]. In contrast to previous methods, RNA sequencing utilizes sequencing by synthesis technology to define the nucleotide sequences and quantify RNA molecules in a sample[2]. Next generation sequencing (NGS) can process this data in hours to days with high fidelity, making it the preferred technique for RNA analysis amongst many researchers[3]. The utilization of this technology in research and literature has been exploding in popularity. There are many promising potential clinical applications of RNA sequencing with recent discoveries using RNA sequencing in many disease states[4,5].
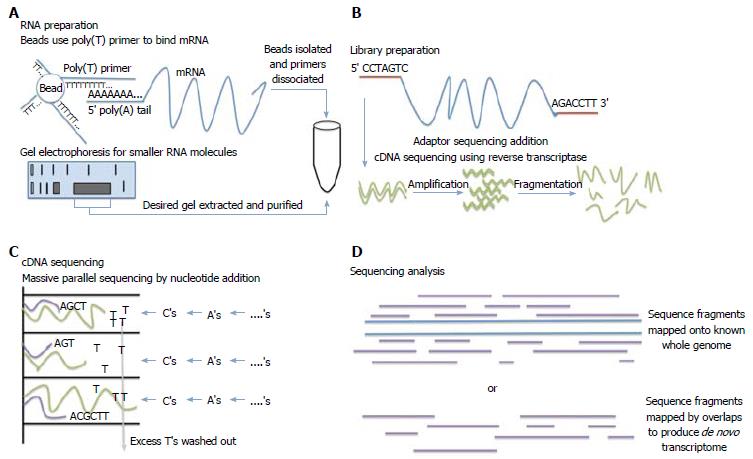
Several commercial RNA sequencing kits are available for any sample. Most follow similar processing steps, but ultimately depend on experimental considerations[6]. Total RNA, mRNA, and small RNA analysis can be done with most kits. For mRNA isolation, poly(T) primers attached to beads or magnets are used to bind mRNA and isolate these strands. For small RNA molecules or non-coding RNA, gel electrophoresis is used to isolate these molecules. Total RNA isolation utilizes a combination of these two techniques[7]. Adaptors are then ligated to the 5’ end, 3’ end, or both. Once RNA is isolated, cDNA is generated, amplified, and then fragmented. Some kits provide direct RNA sequencing without the need to create cDNA. rRNA can be removed since it makes up a significant proportion of the total RNA but is of little research interest. These samples are then sequenced through massive parallel next generation sequencing technologies that utilize sequencing by synthesis of short DNA strands complimentary to the cDNA. Once the reads are produced, software is available to analyze the sequence reads and correspond the reads to portions of the genome. Mapping gene fragments together with sequencing analysis software can also produce de novo transcriptome maps (Figure 1). Using the total of number of reads for each gene product, proportional gene expression can be quantified[8].
Advantages over previous attempts at transcriptome investigation have prompted the recent increased utilization of RNA sequencing. Two prominent techniques were available before NGS RNA sequencing. Hybridization of cDNA probes attached to microarrays allowed for transcriptome analysis but was limited by the requirement for extensive knowledge of the genome, transcription products, alternative splicing, and exons. Resolution was also limited during attempts to quantify gene expression because of background noise produced by cross- hybridization. The other technology was Sanger sequencing, which utilized chain termination methods to determine nucleotide sequences. In contrast to NGS, Sanger methods are more expensive, more time consuming, and only limited portions of transcripts could be analyzed[2,3,8].
Both the discovery of non-coding RNA, such as miRNA (miRNA), and the discovery of post- transcriptional mRNA expression regulation has necessitated the creation of an assay that survey these small non-coding RNAs along with variant mRNAs with high throughput and resolution[9]. RNA sequencing technology allows researchers to perform both those tasks as well as quantifying RNA expression and thus gene expression with a single assay. Because of the high throughput nature of RNA sequencing, transcriptomes can be analyzed and compared between time, different tissue samples, and different environmental factors such as disease states and pharmacologic interventions in an efficient manner. Because of the possibility of de novo transcriptome synthesis, prior genomic and transcriptional knowledge of the sample is not needed, allowing analysis and discovery of novel products. The resolution of RNA sequencing also allows for the identification of single nucleotide variants, novel post-transcriptional modification, novel alternative splicing patterns, and non-coding RNA molecules that have not been previously identified. RNA sequencing provides an accurate quantification of mRNA expression as compared with real-time PCR experiments[10-13].
Using RNA sequencing, we can look at the molecular basis for disease susceptibility, cancer pathogenesis/progression, and response to therapy. RNA Sequencing has been used to analyze the pathogenesis of several malignancies such melanoma, lung cancer, and colorectal cancer. RNA sequencing can identify differential expression of genes (DEG’s), mutated genes, fusion genes, and gene isoforms in disease states. RNA sequencing has the potential for diagnostic and therapeutic applications as well. Current research in colorectal disease using RNA sequencing are unlocking new discoveries that may help clinicians treating patients with colorectal disease in the future.
RNA sequencing has been used in the investigation of several colorectal diseases such as colorectal cancer, inflammatory bowel disease (ulcerative colitis and Crohn’s disease), and irritable bowel syndrome (IBS). RNA sequencing has been used to identify genomic mutations such as fusion transcripts in colon cancer[14], as well as the pathogenesis of colorectal cancer[15,16]. Attempts to discover a unique transcript marker for colorectal cancer[17,18] and inflammatory bowel disease have also been attempted for quicker diagnosis than current screening methods[19,20]. RNA sequencing has also been used to investigate treatment response for rectal cancer[21]. Alterations in transcriptional patterns have also been observed in patients with irritable bowel syndrome through RNA sequencing techniques[22].
Colorectal cancer (CRC) is the third most common cancer among men and women, as well as the third leading cause of death from cancer. It is estimated that more than 50000 people died from colorectal cancer in 2014. While screening methods have dramatically dropped the mortality from CRC, prevention of disease can be improved by diagnosing patients at an earlier progression of disease[23]. The genomic mutation progression of CRC is well documented[24], but clinicians are still left without a clear molecular disease marker. CRC still poses a significant disease burden on public health[25].
Inflammatory bowel disease poses significant morbidity, and even possible mortal complications, to patients that are inflicted. Surgical intervention is oftentimes needed to control disease or prevent carcinoma from developing. An estimated 1.5 million people in North America are inflicted with IBD. While the incidence has recently been stabilizing in North America and Europe, incidence has been increasing in the Middle East and Asia. New molecular insights are needed to find more effective diagnosis, prognosis[26], and treatments[27].
While irritable bowel syndrome poses less of a risk to public health than colorectal cancer or inflammatory bowel disease, it is one of the most common colorectal diseases. It is estimated that as many as 20% of the adult population may be inflicted. Despite IBS’s high prevalence, diagnosis and treatment of this disease still elude researchers[28].
Research shows that certain RNA sequences are upregulated or downregulated in colorectal diseases, opening the possibility of using RNA sequencing to screen for, diagnose, and assess the prognosis of colorectal cancers[29]. Given the increase in treatment resistance to standard chemotherapy regimens[30,31], RNA sequencing also allows for the detection of those that are treatment resistant. Table 1 provides a summary of the most recently studied markers in colorectal cancer and ulcerative colitis.
| Name | Level | Disease | Biomarker | Ref. |
| miR-143 | ↓ | CRC | Diagnosis | [32] |
| miR-20a | ↑ | CRC | Diagnosis, prognosis | [33,37] |
| miR-21 | ↑ | CRC | Diagnosis | [35] |
| miR-132 | ↓ | CRC | Prognosis | [36] |
| DANCR | ↑ | CRC | Prognosis | [38] |
| miR-4299 | ↑ | CRC | Chemoresistance | [39] |
| miR-196b | ↓ | CRC | Chemoresistance | [39] |
| miR-214 | ↑ | Active UC and CAC | Diagnosis, prognosis | [40] |
Wang et al[32] provides a review of the various diagnostic biomarkers that are altered including stool miRNA, serum miRNA, piwi-interacting RNA, and long non-coding RNA (lncRNA) while Hollis et al[29] provides a more thorough review of the miRNA biomarkers used for early detection, prognosis, and chemosensitivity of CRC. More recently, Yang et al[33] used 16 cancer tissues to find that miR-143 acts as a tumor suppressor and is downregulated in CRC tissues and can be used to diagnose CRC. Using 40 CRC tumor tissues and 595 fecal samples (198 CRC, 199 adenoma, 198 healthy subjects), Yau et al[34] found that miR-20a is upregulated in tumors and fecal samples and can also be used to diagnosis CRC. Sun et al[35] validated that miR-21 is upregulated and miR-143 is downregulated in CRC and are the most important miRNAs in CRC.
Qin et al[36] found through 6 human CRC cell lines that miR-132 acts as a tumor suppressor and that hypermethylation of this causes its downregulation and is a thus marker for poor prognosis. Xu et al[37] used 30 samples of CRC tumors to show that increased miR-20a is also associated with tumor invasion and lymph node metastasis. Using 104 CRC specimens, Liu et al[38] found that lncRNA DANCR is upregulated in CRC tissues. It is correlated with TNM stage, histologic grade, lymph node metastasis, shorter overall survival and disease-free survival.
Hu et al[39], through a retrospective analysis of 126 patients with colon adenocarcinoma, found that miR-4299 and miR-196b are potential novel biomarkers for XELOX chemoresistance. Downregulation of miR-4299 and upregulation of miR-196b is correlated with better survival.
In regards to ulcerative colitis (UC) and colitis-associated cancer (CAC), Polytarchou et al[40] used 401 colon specimens of patients with UC, Crohn’s, IBS, sporadic CRC, and CAC to show that miR-214 is upregulated in active UC and CAC. Its expression is correlated with UC activity and disease duration and could serve as a biomarker for identifying patients at risk for malignant transformation[40].
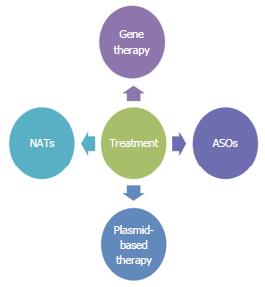
Through specific targeting, RNA sequencing allows for the development of new therapeutic approaches to colorectal diseases. The various RNA seq-based approaches to therapy of various diseases include gene therapy[41], natural antisense transcripts (NATs), antisense oligonucleotides (ASOs), and plasmid based therapy (Figure 2).
In terms of gene therapy techniques, Wang et al[42] found that tumor suppressor long intergenic non-coding RNA (lincR-p21), downregulated in CRC, administered exogenously can suppress the stem-like traits of colorectal cancer stem cells. An adenoviral vector with the miRNA responsive element of miR-451 delivers the lincR-p21 into cells that have low miR-451 levels. This inhibits β-catenin signaling and attenuates the viability, self-renewal, and glycolysis of CRC in vitro. It also suppresses the self-renewal potential and tumorigenicty of CRC in nude mice[42].
Davis et al[43] in 2010 entered phase 1 clinical trial testing to systemically administer small interfering RNA (siRNA) to patients with solid cancers using a targeted, nanoparticle delivery system. The siRNA is designed to reduce the expression of the M2 subunit of ribonucleotide reductase RRM2.
Another treatment being studied is Resveratrol. It has been found that Resveratrol, extracted from Chinese herbal medicine Polygonum cuspidatum, downregulates lncRNA MALAT1 which decreases nuclear localization of β-catenin thus diminishing the wnt/β-catenin signaling, ultimately inhibiting CRC invasion and metastasis[44].
Phase 1 clinical trials have shown that CEQ508 is a possible medical treatment for familial adenomatosis polyposis. Through transkingdown RNA interference, nonpathogenic E. coli produce and deliver short hairpin RNA (shRNA) against β-catenin to target cells, again inhibiting intestinal cell growth and polyp growth. It has been found to be safe and well-tolerated in nonhumans[45].
Therapies using ASO, NAT, and plasmid based therapy have not yet been studied with colorectal diseases[46-49]. However, these techniques are available and should be studied with colorectal diseases in the future.
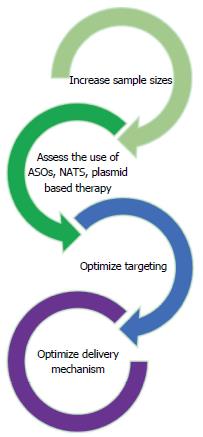
Despite the advances that are being made in using RNA sequencing for diagnosis and treatment of colorectal diseases, there are still limitations including targeting and delivery of treatment. Figure 3 summarizes future directions to address these limitations.
RNA Sequencing itself has many limitations and problems. NGS requires sequences shorter than most mRNA sequences to process in a parallel manner, requiring fragmentation of either the RNA strand or cDNA strand. This requirement can introduce bias to the strands as RNA fragment leads to decreased amplification of 5’ and 3’ ends of the strand and cDNA fragmentation leads to preferential amplification to the 3’ end of a strand. cDNA synthesis of small RNA molecules can also be biased based on the adaptors used, specific G/C-content, and complex tertiary and quaternary structures of these molecules[50,51]. NGS also produces large amounts of data that presents a problem for storage and retrieval of said data[1]. Alternative splicing, trans-splicing, and fragments that correspond to multiple genomic locations also present a problem for the analysis of the transcriptome[1].
Other challenges include library construction, bioinformatics challenges, and coverage vs cost. In constructing a cDNA library, there are many manipulation stages to go through which can get complicated in profiling all the types of transcripts. Bioinformatic challenges include storing, retrieving, and processing the large amounts of data that is garnered through RNA sequencing. In terms of coverage vs cost, greater coverage requires more sequencing depth and thus greater cost to detect rare transcripts or variants[2].
As seen in the previous studies described above, small sample sizes were used for most research conducted. While some studies used 400 samples, many had sample sizes that ranged from 6 to 30. Larger samples sizes can help to yield more significant results[52]. Another study with promising results that are not significant due to sample size is Cohen et al[53]’s study on the predictive value of Target Now. Target Now uses immunostaining and RNA expression on tumor samples to identify potentially beneficial or ineffective drugs. The results of this study were not statistically significant due to its small sample size of 19 patients. Despite the promising results of the Target Now study, the small sample size exemplifies the limitations of much of the RNA Sequencing literature and its application in colorectal diseases.
Complications and side effects have been seen when siRNA has been used as a therapeutic agent, further limiting the usage of RNA sequencing in colorectal disease. A phase 1 drug candidate that targeted apoB was withdrawn because of the immune response elicited by its cationic lipid-based formulation that delivers siRNA into endosomes where immune receptors are most dense. This caused one patient to have severe flu-like symptoms typical of an immune response[47].
In response, dual targeting siRNA is being studied to reduce the potential for off-target gene silencing. Theoretically, fewer strands compete for RISC entry which helps avoid the innate immune response. However, more research needs to be conducted in this area[54].
The biodistribution of siRNA in vivo has also been a limitation of siRNA application. van de Water et al[55] found that intravenous siRNA accumulates in the kidney of rats rather than being absorbed in the GI tract. There, it acts to suppress the gene function in proximal tubules, limiting the application of siRNA in colorectal disease. Further research is needed to manipulate the localization of siRNA for therapeutic colorectal applications.
To conclude, despite the large amount of research dedicated to using RNA sequencing to diagnose and screen for colorectal diseases, further studies need to be conducted on using these techniques for treatment of these colorectal diseases. With more research, RNA sequencing could be the next novel treatment for colorectal diseases.
The authors are grateful and indebted to Dr. Vyas and the surgical faculty and staff at Texas Tech University Health Science Center at Permian Basin for all of their support and career guidance.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Crea F S- Editor: Gong ZM L- Editor: A E- Editor: Wang CH
| 1. | Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 260] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 2. | Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10137] [Cited by in RCA: 8284] [Article Influence: 517.8] [Reference Citation Analysis (0)] |
| 3. | Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 906] [Article Influence: 90.6] [Reference Citation Analysis (0)] |
| 4. | Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, Luo L, Zhang XH, Xiao J, Alekseyev YO. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res (Phila). 2011;4:803-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 5. | Nalpas NC, Magee DA, Conlon KM, Browne JA, Healy C, McLoughlin KE, Rue-Albrecht K, McGettigan PA, Killick KE, Gormley E. RNA sequencing provides exquisite insight into the manipulation of the alveolar macrophage by tubercle bacilli. Sci Rep. 2015;5:13629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Chu Y, Corey DR. RNA sequencing: platform selection, experimental design, and data interpretation. Nucleic Acid Ther. 2012;22:271-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 7. | Tuch BB, Laborde RR, Xu X, Gu J, Chung CB, Monighetti CK, Stanley SJ, Olsen KD, Kasperbauer JL, Moore EJ. Tumor transcriptome sequencing reveals allelic expression imbalances associated with copy number alterations. PLoS One. 2010;5:e9317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 8. | Han Y, Gao S, Muegge K, Zhang W, Zhou B. Advanced Applications of RNA Sequencing and Challenges. Bioinform Biol Insights. 2015;9:29-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 134] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 9. | Burroughs AM, Ando Y, Aravind L. New perspectives on the diversification of the RNA interference system: insights from comparative genomics and small RNA sequencing. Wiley Interdiscip Rev RNA. 2013;5:141-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 10. | de Klerk E, ‘t Hoen PA. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015;31:128-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 251] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 11. | Scarpato M, Federico A, Ciccodicola A, Costa V. Novel transcription factor variants through RNA-sequencing: the importance of being “alternative”. Int J Mol Sci. 2015;16:1755-1771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | de Klerk E, den Dunnen JT, ‘t Hoen PA. RNA sequencing: from tag-based profiling to resolving complete transcript structure. Cell Mol Life Sci. 2014;71:3537-3551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Derks KW, Misovic B, van den Hout MC, Kockx CE, Gomez CP, Brouwer RW, Vrieling H, Hoeijmakers JH, van IJcken WF, Pothof J. Deciphering the RNA landscape by RNAome sequencing. RNA Biol. 2015;12:30-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Nome T, Thomassen GO, Bruun J, Ahlquist T, Bakken AC, Hoff AM, Rognum T, Nesbakken A, Lorenz S, Sun J. Common fusion transcripts identified in colorectal cancer cell lines by high-throughput RNA sequencing. Transl Oncol. 2013;6:546-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Peltekova VD, Lemire M, Qazi AM, Zaidi SH, Trinh QM, Bielecki R, Rogers M, Hodgson L, Wang M, D’Souza DJ. Identification of genes expressed by immune cells of the colon that are regulated by colorectal cancer-associated variants. Int J Cancer. 2014;134:2330-2341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Liu F, Ji F, Ji Y, Jiang Y, Sun X, Lu Y, Zhang L, Han Y, Liu X. Dissecting the mechanism of colorectal tumorigenesis based on RNA-sequencing data. Exp Mol Pathol. 2015;98:246-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Løvf M, Nome T, Bruun J, Eknaes M, Bakken AC, Mpindi JP, Kilpinen S, Rognum TO, Nesbakken A, Kallioniemi O. A novel transcript, VNN1-AB, as a biomarker for colorectal cancer. Int J Cancer. 2014;135:2077-2084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Ji H, Chen M, Greening DW, He W, Rai A, Zhang W, Simpson RJ. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS One. 2014;9:e110314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 166] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 19. | Lin J, Welker NC, Zhao Z, Li Y, Zhang J, Reuss SA, Zhang X, Lee H, Liu Y, Bronner MP. Novel specific microRNA biomarkers in idiopathic inflammatory bowel disease unrelated to disease activity. Mod Pathol. 2014;27:602-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Holgersen K, Kutlu B, Fox B, Serikawa K, Lord J, Hansen AK, Holm TL. High-resolution gene expression profiling using RNA sequencing in patients with inflammatory bowel disease and in mouse models of colitis. J Crohns Colitis. 2015;9:492-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 21. | Lopes-Ramos C, Koyama FC, Habr-Gama A, Salim AC, Bettoni F, Asprino PF, França GS, Gama-Rodrigues J, Parmigiani RB, Perez RO. Comprehensive evaluation of the effectiveness of gene expression signatures to predict complete response to neoadjuvant chemoradiotherapy and guide surgical intervention in rectal cancer. Cancer Genet. 2015;208:319-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 22. | Camilleri M, Carlson P, Acosta A, Busciglio I, Nair AA, Gibbons SJ, Farrugia G, Klee EW. RNA sequencing shows transcriptomic changes in rectosigmoid mucosa in patients with irritable bowel syndrome-diarrhea: a pilot case-control study. Am J Physiol Gastrointest Liver Physiol. 2014;306:G1089-G1098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Vyas D, Garthe CC, Vyas A. Limitations of current timing and frequency of screening colonoscopy and possible future direction. J Laparoendosc Adv Surg Tech A. 2013;23:271-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Gambhir S, Vyas D, Hollis M, Aekka A, Vyas A. Nuclear factor kappa B role in inflammation associated gastrointestinal malignancies. World J Gastroenterol. 2015;21:3174-3183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 92] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (2)] |
| 25. | Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1848] [Cited by in RCA: 2073] [Article Influence: 188.5] [Reference Citation Analysis (0)] |
| 26. | Joshi SS, Vyas AK, Vyas D, Kalla R. Reclassifying inflammatory bowel disease with capsule endoscopy in children. J Pediatr (Rio J). 2013;89:514-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 27. | Malik TA. Inflammatory Bowel Disease: Historical Perspective, Epidemiology, and Risk Factors. Surg Clin North Am. 2015;95:1105-1122, v. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 28. | Sayuk GS, Gyawali CP. Irritable bowel syndrome: modern concepts and management options. Am J Med. 2015;128:817-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284-8292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 94] [Cited by in RCA: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 30. | Balakrishnan A, Vyas A, Deshpande K, Vyas D. Pharmacological cyclin dependent kinase inhibitors: Implications for colorectal cancer. World J Gastroenterol. 2016;22:2159-2164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Vyas D, Laput G, Vyas AK. Chemotherapy-enhanced inflammation may lead to the failure of therapy and metastasis. Onco Targets Ther. 2014;7:1015-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 234] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 32. | Wang J, Song YX, Ma B, Wang JJ, Sun JX, Chen XW, Zhao JH, Yang YC, Wang ZN. Regulatory Roles of Non-Coding RNAs in Colorectal Cancer. Int J Mol Sci. 2015;16:19886-19919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 33. | Yang F, Xie YQ, Tang SQ, Wu XB, Zhu HY. miR-143 regulates proliferation and apoptosis of colorectal cancer cells and exhibits altered expression in colorectal cancer tissue. Int J Clin Exp Med. 2015;8:15308-15312. [PubMed] |
| 34. | Yau TO, Wu CW, Tang CM, Chen Y, Fang J, Dong Y, Liang Q, Ng SS, Chan FK, Sung JJ. MicroRNA-20a in human faeces as a non-invasive biomarker for colorectal cancer. Oncotarget. 2016;7:1559-1568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 35. | Sun G, Cheng YW, Lai L, Huang TC, Wang J, Wu X, Wang Y, Huang Y, Wang J, Zhang K. Signature miRNAs in colorectal cancers were revealed using a bias reduction small RNA deep sequencing protocol. Oncotarget. 2016;7:3857-3872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Qin J, Ke J, Xu J, Wang F, Zhou Y, Jiang Y, Wang Z. Downregulation of microRNA-132 by DNA hypermethylation is associated with cell invasion in colorectal cancer. Onco Targets Ther. 2015;8:3639-3648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Xu T, Jing C, Shi Y, Miao R, Peng L, Kong S, Ma Y, Li L. microRNA-20a enhances the epithelial-to-mesenchymal transition of colorectal cancer cells by modulating matrix metalloproteinases. Exp Ther Med. 2015;10:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 38. | Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8:11480-11484. [PubMed] |
| 39. | Hu J, Xu Y, Cai S. Specific microRNAs as novel biomarkers for combination chemotherapy resistance detection of colon adenocarcinoma. Eur J Med Res. 2015;20:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 40. | Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, van der Meulen-de Jong AE, Oikonomopoulos A, van Deen WK, Vorvis C. MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology. 2015;149:981-92.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 41. | Deshpande K, Vyas A, Balakrishnan A, Vyas D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Genetic Engineering: Robotic Genetic Surgery. Am J Robot Surg. 2015;2:49-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 42. | Wang J, Lei ZJ, Guo Y, Wang T, Qin ZY, Xiao HL, Fan LL, Chen DF, Bian XW, Liu J. miRNA-regulated delivery of lincRNA-p21 suppresses β-catenin signaling and tumorigenicity of colorectal cancer stem cells. Oncotarget. 2015;6:37852-37870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 43. | Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067-1070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2129] [Cited by in RCA: 1884] [Article Influence: 125.6] [Reference Citation Analysis (0)] |
| 44. | Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L, Sun J, Cai J, Qin J, Ren J. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/β-catenin signal pathway. PLoS One. 2013;8:e78700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 305] [Cited by in RCA: 303] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 45. | Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6:1130-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 333] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 46. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1291] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 47. | Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J Pathol. 2012;226:365-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 341] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 48. | Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453-459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 466] [Cited by in RCA: 524] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 49. | Mizrahi A, Czerniak A, Levy T, Amiur S, Gallula J, Matouk I, Abu-lail R, Sorin V, Birman T, de Groot N. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 50. | Liu D, Graber JH. Quantitative comparison of EST libraries requires compensation for systematic biases in cDNA generation. BMC Bioinformatics. 2006;7:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 51. | Raabe CA, Tang TH, Brosius J, Rozhdestvensky TS. Biases in small RNA deep sequencing data. Nucleic Acids Res. 2014;42:1414-1426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 163] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 52. | Vyas D, Balakrishnan A, Vyas A. The Value of the P Value. Am J Robot Surg. 2015;2:53-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 53. | Cohen JE, Cohen Y, Peretz T, Hubert A. Retrospective Study of the Predictive Value of Target Now in Systemic Therapy for Metastatic Colorectal and Gastric Carcinomas. Isr Med Assoc J. 2015;17:612-615. [PubMed] |
| 54. | Tiemann K, Höhn B, Ehsani A, Forman SJ, Rossi JJ, Saetrom P. Dual-targeting siRNAs. RNA. 2010;16:1275-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 55. | van de Water FM, Boerman OC, Wouterse AC, Peters JG, Russel FG, Masereeuw R. Intravenously administered short interfering RNA accumulates in the kidney and selectively suppresses gene function in renal proximal tubules. Drug Metab Dispos. 2006;34:1393-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 168] [Article Influence: 8.8] [Reference Citation Analysis (0)] |