Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2314
Peer-review started: June 15, 2015
First decision: October 14, 2015
Revised: October 28, 2015
Accepted: November 30, 2015
Article in press: November 30, 2015
Published online: February 21, 2016
Processing time: 231 Days and 2.4 Hours
AIM: To investigate the driver gene mutations associated with colorectal cancer (CRC) in the Taiwanese population.
METHODS: In this study, 103 patients with CRC were evaluated. The samples consisted of 66 men and 37 women with a median age of 59 years and an age range of 26-86 years. We used high-resolution melting analysis (HRM) and direct DNA sequencing to characterize the mutations in 13 driver genes of CRC-related pathways. The HRM assays were conducted using the LightCycler® 480 Instrument provided with the software LightCycler® 480 Gene Scanning Software Version 1.5. We also compared the clinicopathological data of CRC patients with the driver gene mutation status.
RESULTS: Of the 103 patients evaluated, 73.79% had mutations in one of the 13 driver genes. We discovered 18 novel mutations in APC, MLH1, MSH2, PMS2, SMAD4 and TP53 that have not been previously reported. Additionally, we found 16 de novo mutations in APC, BMPR1A, MLH1, MSH2, MSH6, MUTYH and PMS2 in cancerous tissues previously reported in the dbSNP database; however, these mutations could not be detected in peripheral blood cells. The APC mutation correlates with lymph node metastasis (34.69% vs 12.96%, P = 0.009) and cancer stage (34.78% vs 14.04%, P = 0.013). No association was observed between other driver gene mutations and clinicopathological features. Furthermore, having two or more driver gene mutations correlates with the degree of lymph node metastasis (42.86% vs 24.07%, P = 0.043).
CONCLUSION: Our findings confirm the importance of 13 CRC-related pathway driver genes in the development of CRC in Taiwanese patients.
Core tip: In Taiwan, colorectal cancer (CRC) has had the highest incidences among cancers recently. In a study of 103 patients with CRC, we identified 18 novel mutations in APC, MLH1, MSH2, PMS2, SMAD4 and TP53. We assessed the frequency of non-pathological somatic mutations during oncogenesis, which has not been explored before. Our results indicated 16 de novo mutations that have been previously described in a public database and were detected in cancerous tissues only, but not in the patient’s blood cells. We suggest these mutation sites may belong to a frequent mutational hotspot in both germline and cancerous tissues.
- Citation: Chang YC, Chang JG, Liu TC, Lin CY, Yang SF, Ho CM, Chen WTL, Chang YS. Mutation analysis of 13 driver genes of colorectal cancer-related pathways in Taiwanese patients. World J Gastroenterol 2016; 22(7): 2314-2325
- URL: https://www.wjgnet.com/1007-9327/full/v22/i7/2314.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i7.2314
Colorectal cancer (CRC) is one of the major causes of mortality and morbidity in Western and in Asian countries. In 2014, an estimated 136830 new cases were diagnosed, and 50310 deaths were due to CRC, making it the third most common cancer among men and women in the United States[1]. The lifetime risk of CRC is 6%, and the average age at diagnosis is 66 years old in the United States[2]. CRC has also become the third leading cause of cancer-related death in the Taiwanese population[3].
Inherited CRCs can be attributed to hereditary nonpolyposis CRC (HNPCC), familial adenomatous polyposis (FAP), and closely related variant syndromes[4]. Approximately 15%-30% of the patients may fall into this category, and their first- or second-degree relatives may have CRC[5]. New or de novo germ-line mutations of adenomatous polyposis coli (APC) occur in approximately 25% of FAP cases. The lifetime incidence of CRC in untreated FAP patients is approaching 100%[6]. The most common germ-line APC mutations are located at codons 1061 and 1309[7]. HNPCC is the result of germline mutations in DNA mismatch repair (MMR) genes, including mutL homolog 1 (MLH1), mutS homolog 2 (MSH2), mutS homolog 6 (MSH6) and PMS1 homolog 2 (PMS2), with mutations in MLH1 and MSH2 being more common than those in other MMR genes[8]. Juvenile polyposis syndrome is caused by mutations in the bone morphogenetic protein receptor, type 1A (BMPR1A) or SMAD family member 4 (SMAD4) tumor suppressor genes[9]. Cowden syndrome is associated with mutations in phosphatase and tensin homolog (PTEN)[10]. Homozygous mutations in the base excision repair (BER) pathway gene mutY DNA glycosylase (MUTYH) cause MUTYH-associated polyposis syndrome, and heterozygous MUTYH mutations are found in some cases of familial CRC[11].
Sporadic CRCs account for approximately 70%-85% of all cases, and these patients have no distinguishable genetic risk factors. The development of sporadic CRC is probably as a result of diet, lifestyle, and environmental factors as well as somatic mutations[12]. Sporadic CRCs have more biological variables compared with hereditary CRCs[13]. Chromosomal instability (CIN), microsatellite instability and CpG island methylator phenotype pathways are the major genetic mechanisms responsible for sporadic CRCs[14]. The CIN pathway implies the progression from adenoma to carcinoma. This pathway suggests a stepwise pattern of mutational inactivation of tumor suppressor genes, such as the APC and tumor protein p53 (TP53), and the activation of oncogenes, such as Kirsten rat sarcoma viral oncogene homolog (KRAS)[15]. Most sporadic CRCs (70%-80%) have APC somatic mutations, and the mutations appear to be enriched in the mutation cluster region (MCR, codons 1309 to 1450)[7]. Approximately 40% of CRCs have KRAS mutations, and almost all of these mutations are located at codons 12, 13 or 61[16,17]. The MSI pathway is characterized by the inactivation of the MMR genes such as MLH1. Inactivation of MMR genes occurs either through MLH1 promoter hypermethylation or point mutations in one of the MMR genes. De novo germ-line mutations or somatic mutations in MMR genes account for a small number of sporadic CRCs[8]. The Serrated Pathway is characterized by the presence of a mutation in the oncogene v-raf murine sarcoma viral oncogene homolog B (BRAF) and the hypermethylation of other genes[18]; 3%-13% of CRC patients have a mutation in the BRAF gene[19].
Multiple previous reports have revealed that several critical genes and pathways are important in the initiation and progression of CRC, these include WNT, RAS-MAPK, PI3K, TGF-β, P53, and DNA MMR pathways[20]. The Cancer Genome Atlas Network project has identified numerous recurrently mutated genes[21].
The aim of our study was to assess the genes known to be implicated in CRC and to compare the clinicopathological data with the molecular genetic profiles of the tumors. We used a high resolution melting (HRM) technique and direct DNA sequencing to analyze the APC exons 1-14 and part of exon 15, BRAF exon 15, KRAS exon 2, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) exon 9 and 20, the complete coding region of BMPR1A, MLH1, MSH2, MSH6, MUTYH, PMS2, PTEN, SMAD4 and TP53 in CRCs from 103 Taiwanese patients.
One hundred and three colorectal adenocarcinomas were collected and analyzed. All samples were tested for sporadic and familial genetic changes in known CRC related genes (APC, BMPR1A, BRAF, KRAS, MLH1, MSH2, MSH6, MUTYH, PIK3CA, PMS2, PTEN, SMAD4 and TP53). DNA was extracted using a commercially available kit (GE Healthcare, Little Chalfont, UK), following the manufacturer’s recommendations. After extraction, DNA was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington DE, United States). This study was approved by the Institutional Review Board of the China Medical University Hospital.
To assess APC, BMPR1A, BRAF, KRAS, MLH1, MSH2, MSH6, MUTYH, PIK3CA, PMS2, PTEN, SMAD4 and TP53 mutations, we performed HRM of small amplicons using a LightCycler® 480 Instrument (Roche Diagnostics, Roche Instrument Center AG, Rotkreuz, Switzerland) in tumor samples from CRC patients. The primers used for HRM analysis are shown in Supplementary material. The amplifications were performed in 10 μL volumes containing 10 ng of genomic DNA, 0.25 μmol/L primers, 2.5 mmol/L MgCl2 and 5 μL 2X LightCycler® 480 High Resolution Melting Master (reference 04909631001, Roche Diagnostics) buffer. Polymerase chain reaction cycling included an initial denaturation at 95 °C for 10 min followed by 45 cycles of 15 s at 95 °C, 15 s at 60 °C, and 15 s at 72 °C. The melting program included three steps: denaturation at 95 °C for 1 min, renaturation at 40 °C for 1 min, and a subsequent melting cycle that consists of a continuous fluorescent reading from 60 to 90 °C at a rate of 25 acquisitions per °C.
Gene scanning analysis of the data using Gene Scanning Software consisted of three steps: (1) normalization of melting curves, which involved setting the initial fluorescence equal to 100% and the remaining fluorescence signal after DNA dissociation to 0%; (2) shifting of the temperature axis of the normalized melting curves to the point where the entire double-stranded DNA was completely denatured; and (3) generation of difference plots, allowing capture of the melting profile difference between the reference sample curves and the test samples.
After HRM analysis, the samples were purified using the PCR-MTM clean up system (VIOGEN, Sunnyvale CA, United States). The sequence reaction was performed using 1 μL of the purified PCR product, 2.5 μmol/L of one of the PCR primers and 1 μL ABI PRISM terminator cycle sequencing kit v3.1 (Applied Biosystems, Foster City, CA) in a final reaction volume of 10 μL. The samples were sequencing using a 25-cycle PCR program (denaturation at 96 °C for 10 s, annealing at 50 °C for 5 s and elongation at 60 °C for 4 min). The sequencing detection was performed using an ABI Prism 3130 Genetic Analyzer (Applied Biosystems).
Due to its size (2545 nucleotides) it was costly and time consuming to screen MSH6 exon 4 using HRM; thus, direct DNA sequencing was performed.
The results were analyzed using SPSS version 17.0 program. P values of less than 0.05 were considered statistically significant.
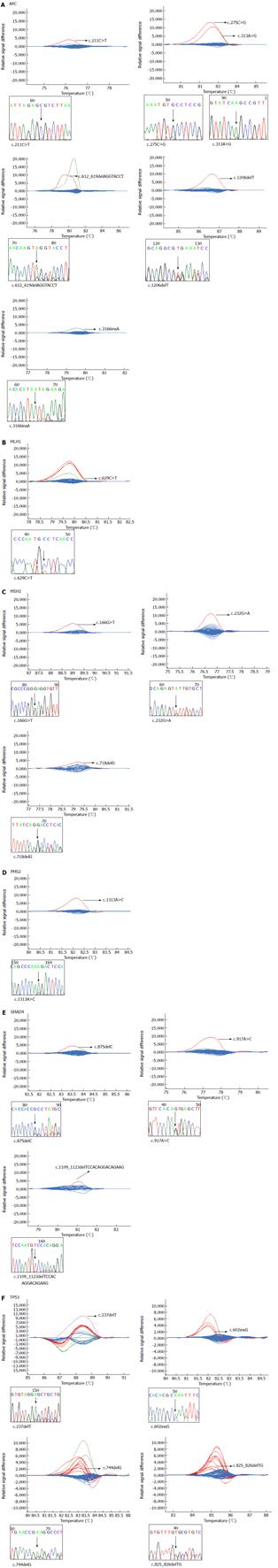
In total, we validated 18 mutations in six genes that have not been previously described in a public database (Figure 1). Six of these mutations occurred in the APC gene (c.211C>T, c.275C>G, c.313A>G, c.612_619delAGGTACCT, c.1206delT and c.3166insA), one in the MLH1 gene (c.629C>T), three in the MSH2 gene (c.166G>T, c.232G>A and c.718delG), one in the PMS2 gene (c.1313A>C), three in the SMAD4 gene (c.875delC, c.917A>G and c.1109_1123delTCCACAGGACAGAAG) and four in the TP53 gene (c.237delT, c.602insG, c.744delG and c.825_826delTG). To our knowledge, these mutations have not been detected in human cancers prior to this study.
In total, we found 16 de novo mutations in seven genes that have been previously described in a public database with the exception of APC c.465A>G, these mutations were detected in cancerous tissues only, and not in the patient’s blood cells (Table 1). Three of these mutations occurred in the APC gene (c.465A>G, c.573T>C and c.1005A>G), one in the BMPR1A gene (c.1578A>G), five in the MLH1 gene (c.462C>T, c.655A>G, c.1151T>A, c.1742C>T and c.2101C>A), two in the MSH2 gene (c.23C>T and c.1886A>G), two in the MSH6 gene (c.3488A>T and c.4065_4066insTTGA), two in the MUTYH gene (c.1422G>C and c.1440C>T) and one in the PMS2 gene (c.1532C>T).
| Gene | Mutation | Protein change | rs number | Number of mutations, n (%) | Minor allele frequency in cancer | Minor allele frequency in Asian | GMAF |
| (global minor allele frequency%) | |||||||
| APC | c.465A>G | p.Lys155= | 1 (0.97) | 0.49% | |||
| c.573T>C | p.Tyr191= | rs185154886 | 1 (0.97) | 0.49% | NA | C = 0.06 | |
| c.1005A>G | p.Leu335= | rs3797704 | 1 (0.97) | 0.49% | G = 0% | G = 0.06 | |
| BMPR1A | c.1578A>G | p.Glu526= | rs202030576 | 1 (0.97) | 0.49% | NA | G = 0.02 |
| MLH1 | c.462C>T | p.Asp154= | rs192938577 | 1 (0.97) | 0.49% | NA | T = 0.02 |
| c.655A>G | p.Ile219Val | rs1799977 | 7 (6.8) | 3.40% | G = 0%-37.5% | G = 12.96 | |
| c.1151T>A | p.Val384Asp | rs63750447 | 4 (3.88) | 1.94% | NA | A = 0.52 | |
| c.1742C>T | p.Pro581Leu | rs63751684 | 1 (0.97) | 0.49% | NA | T = 0.12 | |
| c.2101C>A | p.Gln701Lys | rs63750114 | 1 (0.97) | 0.49% | NA | A = 0.12 | |
| MSH2 | c.23C>T | p.Thr8Met | rs17217716 | 3 (2.91) | 1.46% | T = 0%-5% | T = 0.52 |
| c.1886A>G | p.Gln629Arg | rs61756468 | 3 (2.91) | 1.46% | NA | G = 0.22 | |
| MSH6 | c.3488A>T | p.Glu1163Val | rs63750252 | 2 (1.94) | 0.97% | NA | T = 0.28 |
| c.4065_4066insTTGA | p.Lys1328Aspfs | rs55740729 | 1 (0.97) | 0.49% | NA | TTGA = 0.8 | |
| MUTYH | c.1422G>C | p.Thr474= | rs74318065 | 1 (0.97) | 0.49% | NA | G = 1.04 |
| c.1440C>T | p.Thr480= | rs150269172 | 3 (2.91) | 1.46% | NA | A = 0.4 | |
| PMS2 | c.1532C>T | p.Thr511Met | rs74902811 | 4 (3.88) | 1.94% | NA | A = 3.69 |
In total, we validated 18 polymorphisms in six genes previously described in a public database (Table 2). One of them occurred in the BMPR1A gene (c.4C>A), three in the MSH2 gene (c.471C>A, c.1168C>T and c.1690A>G), two in the MSH6 gene (c.116G>A and c.3306T>A), one in the MUTYH gene (c.1014G>C), ten in the PMS2 gene (c.59G>A, c.288C>T, c.780C>G, c.1408C>T, c.1454C>A, c.1621G>A, c.2253T>C, c.2324A>G, c.2340C>T and c.2570G>C) and one in the TP53 gene (c.215C>G). Some of the frequencies are similar to those in a public database, but some are not, which maybe due to ethnic differences.
| Gene | Mutation | Protein change | rs number | Number of mutations, n (%) | Minor allele frequency in cancer | Minor allele frequency in Asian | GMAF |
| (global minor allele frequency%) | |||||||
| BMPR1A | c.4C>A | p.Pro2Thr | rs11528010 | 41(39.81) | 19.9% | NA | A = 49.98 |
| MSH2 | c.471C>A | p.Gly157= | rs61756463 | 5 (4.85) | 2.43% | NA | A = 0.24 |
| c.1168C>T | p.Leu390Phe | rs17224367 | 5 (4.85) | 2.43% | T = 0%-4.7% | T = 0.28 | |
| c.1690A>G | p.Thr564Ala | rs55778204 | 3 (2.91) | 1.46% | NA | G = 0.06 | |
| MSH6 | c.116G>A | p.Gly39Glu | rs1042821 | 1 (0.97) | 0.49% | NA | A = 20.29 |
| c.3306T>A | p.Thr1102= | rs2020910 | 41 (39.81) | 37.38% | A = 0%-22.9% | A = 4.93 | |
| MUTYH | c.1014G>C | p.Gln338His | rs3219489 | 51 (49.51) | 27.67% | C = 45.2%-46.7% | C = 31.35 |
| PMS2 | c.59G>A | p.Arg20Gln | rs10254120 | 10 (9.71) | 4.85% | NA | T = 7.57 |
| c.288C>T | p.Ala96= | rs12532895 | 58 (56.31) | 33.5% | A = 28.2%-36% | A = 11.36 | |
| c.780C>G | p.Ser260= | rs1805319 | 17 (16.5) | 8.25% | G = 4.8%-8% | G = 16.87 | |
| c.1408C>T | p.Pro470Ser | rs1805321 | 44 (42.72) | 21.36% | T = 0% | T = 35.82 | |
| c.1454C>A | p.Thr485Lys | rs1805323 | 44 (42.72) | 21.36% | NA | T = 11.20 | |
| c.1621G>A | p.Lys541Glu | rs2228006 | 15 (14.56) | 7.28% | A = 4.4%-23% | A = 11.68 | |
| c.2253T>C | p.Phe751= | rs1805325 | 2 (1.94) | 0.97% | NA | NA | |
| c.2324A>G | p.Asn775Ser | rs17420802 | 8 (7.77) | 3.88% | NA | NA | |
| c.2340C>T | p.Pro780= | rs142230276 | 8 (7.77) | 3.88% | NA | A = 0.12 | |
| c.2570G>C | p.Gly857ala | rs1802683 | 1 (0.97) | 0.49% | NA | NA | |
| TP53 | c.215C>G | p.Pro72Arg | rs1042522 | 78 (75.73) | 50% | G = 48.9%-61.4% | G = 45.71 |
In total, we validated 58 mutations in 10 genes that have been previously described in a public database (Table 3). Fifteen of them occurred in the APC gene (c.95A>G, c.646C>T, c.694C>T, c.799G>T, c.832C>T, c.904C>T, c.3907C>T, c.3914C>A, c.3914delC, c.3934G>T, c.3935delG, c.3944C>A, c.3982C>T, c.4012C>T and c.4031C>A), two in the BRAF gene (c.1780G>A and c.1799T>C), six in the KRAS gene (c.34G>C, c.34G>T, c.35G>A, c.35G>C, c.35G>T and c.38G>C), one in the MSH2 gene (c.1480T>C), on in the MUTYH gene (c.74G>A), four in the PIK3CA gene (c.1624G>A, c.1633G>A, c.1636C>G and c.3104A>G), one in the PMS2 gene (c.2437C>T), one in the PTEN gene (c.19G>T), three in the SMAD4 gene (c.1067C>G, c.1069T>C and c.1081C>T) and 24 in the TP53 gene (c.318C>G, c.423C>G, c.440T>G, c.511G>T, c.514G>T, c.524G>A, c.536A>G, c.586C>T, c.638G>T, c.646G>A, c.700T>G, c.734G>A, c.742C>T, c.761T>G, c.772G>A, c.772G>T, c.817C>T, c.818G>A, c.841G>C, c.844C>T, c.853G>A, c.856G>A, c.857A>G and c.1015G>T).
| Gene | Mutation | Protein change | rs number in dbSNP/mutation id in COSMIC | Number of mutations, n (%) | Minor allele frequency in cancer | Minor allele frequency in Asian | GMAF |
| (global minor allele frequency%) | |||||||
| APC | c.95A>G | p.Asn32Ser | rs539108537 | 1 (0.97) | 0.49% | NA | G = 0.02 |
| c.646C>T | p.Arg216Stop | rs62619935 | 1 (0.97) | 0.49% | NA | NA | |
| c.694C>T | p.Arg232Stop | rs397515734 | 1 (0.97) | 0.49% | NA | NA | |
| c.799G>T | p.Gly267Stop | The UMD-APC mutations database | 1 (0.97) | 0.49% | NA | NA | |
| c.832C>T | p.Gln278Stop | The UMD-APC mutations database | 1 (0.97) | 0.49% | NA | NA | |
| c.904C>T | p.Arg302Stop | rs137854568 | 1 (0.97) | 0.49% | NA | NA | |
| c.3907C>T | p.Gln1303Stop | COSM13728 | 1 (0.97) | 0.49% | NA | NA | |
| c.3914C>A | p.Ala1305Glu | COSM1432302 | 1 (0.97) | 0.49% | NA | NA | |
| c.3914delC | p.Ala1305Glufs | COSM19687 | 1 (0.97) | 0.49% | NA | NA | |
| c.3934G>T | p.Gly1312Stop | COSM18817 | 1 (0.97) | 0.49% | NA | NA | |
| c.3935delG | p.Gly1312Glufs | COSM18796 | 1 (0.97) | 0.49% | NA | NA | |
| c.3944C>A | p.Ser1315Stop | COSM18777 | 1 (0.97) | 0.49% | NA | NA | |
| c.3982C>T | p.Gln1328Stop | rs398123121 | 3 (2.91) | 1.46% | NA | NA | |
| c.4012C>T | p.Gln1338Stop | rs121913327 | 3 (2.91) | 1.46% | NA | NA | |
| c.4031C>A | p.Ser1344Stop | COSM19135 | 1 (0.97) | 0.49% | NA | NA | |
| BRAF | c.1780G>A | p.Asp594Asn | rs397516896 | 1 (0.97) | 0.49% | NA | NA |
| c.1799T>C | p.Val600Glu | rs113488022 | 3 (2.91) | 1.46% | NA | NA | |
| KRAS | c.34G>C | p.Gly12Cys | rs121913530 | 2 (1.94) | 0.97% | NA | NA |
| c.34G>T | p.Gly12Ser | rs121913530 | 2 (1.94) | 0.97% | NA | NA | |
| c.35G>A | p.Gly12Ala | rs121913529 | 2 (1.94) | 0.97% | NA | NA | |
| c.35G>C | p.Gly12Asp | rs121913529 | 11 (10.98) | 5.34% | NA | NA | |
| c.35G>T | p.Gly12Val | rs121913529 | 12 (11.65) | 5.83% | NA | NA | |
| c.38G>C | p.Gly13Asp | rs112445441 | 5 (4.85) | 2.43% | NA | NA | |
| MSH2 | c.1480T>C | p.Ser494Pro | rs55653533 | 1 (0.97) | 0.49% | NA | C = 0.02 |
| MUTYH | c.74G>A | p.Gly25Asp | rs75321043 | 1 (0.97) | 0.49% | NA | T = 0.18 |
| PIK3CA | c.1624G>A | p.Glu542Lys | rs121913273 | 1 (0.97) | 0.49% | NA | NA |
| c.1633G>A | p.Glu545Lys | rs104886003 | 1 (0.97) | 0.49% | NA | NA | |
| c.1636C>G | p.Gln546Lys | rs121913286 | 1 (0.97) | 0.49% | NA | NA | |
| c.3140A>G | p.His1047Arg | rs121913279 | 2 (1.94) | 0.97% | NA | NA | |
| PMS2 | c.2437C>T | p.Arg813Trp | rs375968016 | 1 (0.97) | 0.49% | NA | A = 0.02 |
| PTEN | c.19G>T | p.Glu7Stop | COSM5298 | 1 (0.97) | 0.49% | NA | NA |
| SMAD4 | c.1067C>G | p.Pro356Arg | COSM339351 | 1 (0.97) | 0.49% | NA | NA |
| c.1069T>C | p.Ser357Pro | COSM189735 | 1 (0.97) | 0.49% | NA | NA | |
| c.1081C>T | p.Arg361Cys | rs80338963 | 1 (0.97) | 0.49% | NA | NA | |
| TP53 | c.318C>G | p.Ser106Arg | COSM45944 | 1 (0.97) | 0.49% | NA | NA |
| c.423C>G | p.Cys141Trp | COSM44204 | 1 (0.97) | 0.49% | NA | NA | |
| c.440T>G | p.Val147Gly | COSM44309 | 1 (0.97) | 0.49% | NA | NA | |
| c.511G>T | p.Glu171Stop | COSM10996 | 1 (0.97) | 0.49% | NA | NA | |
| c.514G>T | p.Val172Phe | COSM44240 | 1 (0.97) | 0.49% | NA | NA | |
| c.524G>A | p.Arg175His | rs28934578 | 5 (4.85) | 2.43% | NA | NA | |
| c.536A>G | p.His179Arg | COSM10889 | 1 (0.97) | 0.49% | NA | NA | |
| c.586C>T | p.Arg196Stop | rs397516435 | 2 (1.94) | 0.97% | NA | NA | |
| c.638G>T | p.Arg213Leu | COSM43650 | 1 (0.97) | 0.49% | NA | NA | |
| c.646G>A | p.Val216Met | COSM10667 | 1 (0.97) | 0.49% | NA | NA | |
| c.700T>G | p.Tyr234Asp | COSM43768 | 1 (0.97) | 0.49% | NA | NA | |
| c.734G>A | p.Gly245Asp | rs121912656 | 2 (1.94) | 0.97% | NA | NA | |
| c.742C>T | p.Arg248Trp | rs121912651 | 3 (2.91) | 1.46% | NA | NA | |
| c.761T>G | p.Ile254Ser | COSM45035 | 1 (0.97) | 0.49% | NA | NA | |
| c.772G>A | p.Glu258Lys | rs121912652 | 1 (0.97) | 0.49% | NA | NA | |
| c.772G>T | p.Glu258Stop | COSM43568 | 1 (0.97) | 0.49% | NA | NA | |
| c.817C>T | p.Arg273Cys | rs121913343 | 1 (0.97) | 0.49% | NA | NA | |
| c.818G>A | p.Arg273His | rs28934576 | 2 (1.94) | 0.97% | NA | T = 0.02 | |
| c.841G>C | p.Asp281His | COSM10943 | 1 (0.97) | 0.49% | NA | NA | |
| c.844C>T | p.Arg282Trp | rs28934574 | 5 (4.85) | 2.43% | NA | NA | |
| c.853G>A | p.Glu285Lys | rs112431538 | 1 (0.97) | 0.49% | NA | NA | |
| c.856G>A | p.Glu286Lys | COSM10726 | 1 (0.97) | 0.49% | NA | NA | |
| c.857A>G | p.Glu286Gly | COSM43565 | 1 (0.97) | 0.49% | NA | NA | |
| c.1015G>T | p.Glu339Stop | COSM11286 | 1 (0.97) | 0.49% | NA | NA |
To explore the patterns of mutations in the candidate pathways, we divided the 13 driver genes into six pathways: WNT, TGF-β, PI3K, RTK-RAS, P53, and DNA repair pathways. The TP53 gene in the P53 pathway has a relatively high rate of mutation compared with genes in the WNT, TGF-β, PI3K, RTK-RAS, and DNA repair pathways.
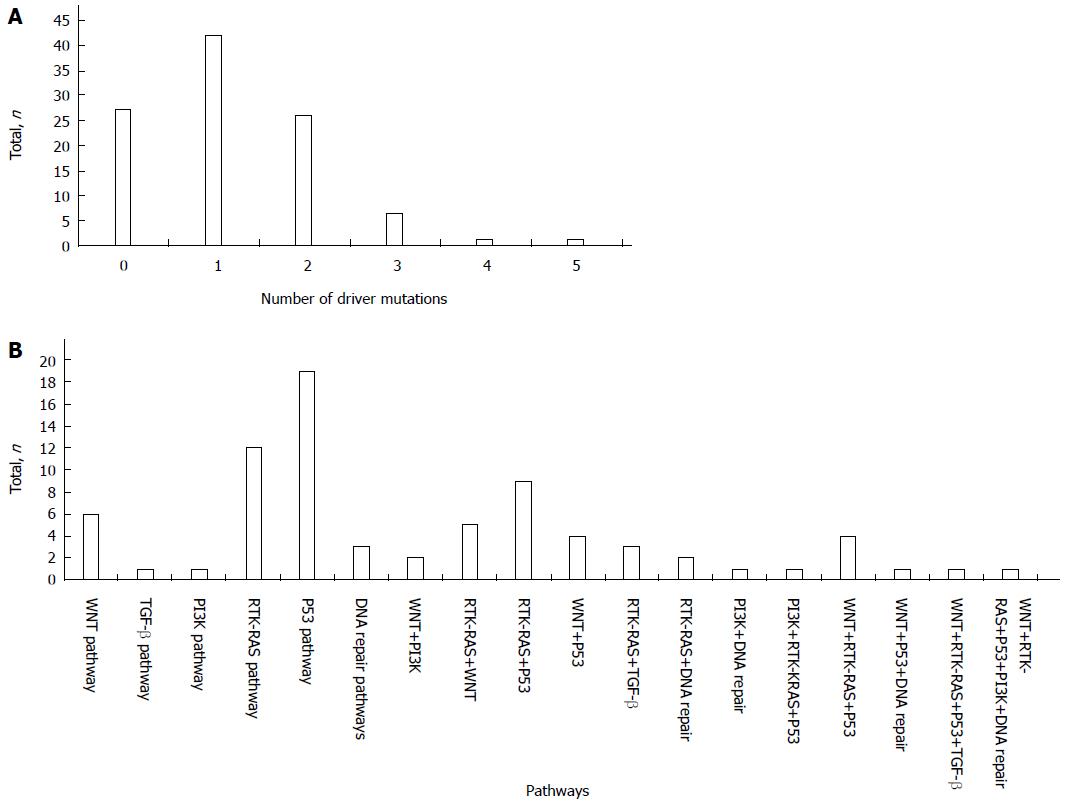
In total, 76 patients (73.79%) had mutations in one of the 13 driver genes; of these, 42 patients (40.78%) had one driver gene mutation, and 34 patients (33.01%) had more than one driver gene mutation, including 26 patients (25.24%) with mutations in two driver genes, 6 patients (5.83%) with mutations in three driver genes, 1 patient (0.97%) with mutations in four driver genes, and 1 patient (0.97%) with mutations in five driver genes (Figure 2A).
The mutation combinations included 9 involved in the RTK-RAS/P53 pathways, 5 in the RTK-RAS/WNT pathways, 4 in the WNT/P53 pathways, 3 in the RTK-RAS/TGF-β pathways, 2 in the WNT/PI3K pathways, 2 in the RTK-RAS/DNA repair pathways, 1 in the PI3K/DNA repair pathway, 4 in the WNT/RTK-RAS/P53 pathways, 1 in the PI3K/RTK-RAS/P53 pathway, 1 in the WNT/P53/DNA repair pathway, 1 in the WNT/RTK-RAS/P53/TGF-β pathway, and 1 in the WNT/RTK-RAS/P53/PI3K/DNA repair pathway (Figure 2B).
APC mutations were significantly correlated with lymph node metastasis (P = 0.009) and cancer stage (P = 0.013) (Table 4). Other mutations did not show any significant correlation with sex, grade, lymph node involvement or stage. In addition, having mutations in two or more driver genes was correlated significantly with the degree of lymph node metastasis (P = 0.043).
| Mutation of APC | Mutations of two or more driver genes | ||||||||
| No | Yes | Total | P value | No | Yes | Total | P value | ||
| Gender | F | 27 | 10 | 37 | 0.503 | 23 | 14 | 37 | 0.435 |
| M | 52 | 14 | 66 | 46 | 20 | 66 | |||
| Grade | Well | 4 | 3 | 7 | 0.443 | 4 | 3 | 7 | 0.579 |
| Moderate | 67 | 19 | 86 | 57 | 29 | 86 | |||
| Poor | 8 | 2 | 10 | 8 | 2 | 10 | |||
| LN | - | 32 | 17 | 49 | 0.009 | 28 | 21 | 49 | 0.043 |
| + | 47 | 7 | 54 | 41 | 13 | 54 | |||
| Stage | I, II | 30 | 16 | 46 | 0.013 | 27 | 19 | 46 | 0.108 |
| III, IV | 49 | 8 | 57 | 42 | 15 | 57 | |||
The advent of next generation sequencing (NGS) has provided a powerful platform to investigate the genetic etiology of diseases. The HRM analysis is not practical for detecting mutations encompassing an entire exon or deletions of entire genes or exons; in contrast, NGS can identify the entire genetic coding sequence. NGS has been comprehensively applied in a variety of ways, including whole genome sequencing, targeted sequencing, chromatin immunoprecipitation sequencing, small RNA sequencing and transcriptome sequencing[22]. Although NGS has become the premier tool in genetic and genomic analyses, this approach can generate a large quantity of genetic data but often with high sequencing errors. Equipment, labor, reagent and supply costs for HRM analysis are significantly lower than those for NGS[23]. The HRM technique does not require complex protocols for experimental steps or data analysis, so it is faster and less expensive than NGS. We used the HRM technique to analyze the mutation profiles in CRCs of Taiwanese patients, and proved the concept.
No studies to date have assessed the frequency of non-pathological somatic mutations during oncogenesis. In our study, a de novo mutation was defined as a genomic alteration that was undetectable in peripheral blood and with nonpathogenic significance; however, the dbSNP database has shown the minor allele frequency for this group. Therefore, we suggest that these nucleotide changes may occur during/after cancer development, and that these mutation sites may belong to a mutational hotspot that occurs frequently in both germline and cancerous tissues.
We identified four point mutations and two deletions in the SMAD4 gene in five CRC cases. SMAD4 plays a unique and pivotal role in the TGF-β pathway by mediating the transcriptional activation of target genes[24]. The mechanism by which mutation alters gene function is still unknown. Ling et al[25] proposed that a mutation in this gene may facilitate CRC progression. In our study, three samples were in the T3 stage and one was in the T4b stage, which may support this idea.
PTEN is a negative regulator of the PI3K pathway that induces cell survival and proliferation. Berg et al[26] found that PTEN mutations were more frequent in young CRC patients (< 50 years). However, in our study, we identified a PTEN nonsense somatic mutation in a male patient aged 66 years.
APC mutations play a critical role in CRC tumorigenesis. Some reports have indicated a potential interdependence of the two hits in APC, both in sporadic and FAP-associated CRCs[27,28]. In our study, one patient had two APC mutations outside the MCR, whereas most of patients had only one APC mutation. From these results, we suggest that one APC mutation is capable of initiating of CRC oncogenesis, similar to a KRAS mutation.
Tomasetti et al[29] showed that only three driver gene mutations are required for the development of advanced cancers in the lung and colon. In addition, they indicated that patients with MMR deficiencies that occurred through the sequential mutation of four or more driver genes significantly increased the incidence of CRC. In this study, we only analyzed 13 driver genes and were unable to confirm their findings, but we determined that 95 patients had fewer than three driver gene mutations. We suggest that further studies using NGS to sequence the exome may solve the discrepancies in these cases.
In conclusion, we identified mutations in genes such as BRAF, KRAS, MUTYH, PIK3CA and PTEN, as well as previously unreported point mutations or frameshift mutations in APC, MLH1, MSH2, PMS2, SMAD4 and TP53 genes in a group of Taiwanese CRC patients.
Previous genetic studies on colorectal cancer (CRC) have revealed multiple critical mutations in candidate pathways; furthermore, statistical analysis has shown that the number of driver gene mutations plays an important role in the development of CRC. However, the genetic mutations associated with CRC in the Taiwanese population are unclear.
Multiple previous reports and The Cancer Genome Atlas database have revealed that several critical genes and pathways are important in the initiation, progression and treatment of CRC, these include WNT, RAS-MAPK, PI3K, TGF-β, P53, and DNA MMR pathways, and some of these mutations may affect the results of treatment.
This is the first study using high-resolution melting analysis (HRM) technique to analyze the mutation profiles in CRCs of Taiwanese patients and evaluating the frequency of non-pathological somatic mutations during oncogenesis.
The studies show that HRM analysis can be used for high-throughput mutation screening for research, as well as for molecular diagnosis and clinical purposes.
HRM analysis is a closed-tube method, indicating that PCR amplification and subsequent analysis are sequentially performed in 1 well.
The manuscript entitled “Mutational analysis of 13 driver genes of colorectal cancer-related pathways in Taiwanese patients” by Chang et al 2015 details the use of HRM and DNA sequencing techniques applied to CRC samples, and details the identification of novel genetic mutations, as well as characterization of the prevalence of other mutations in 13 driver genes of CRC-related pathways. This paper will be of interest to scientists working in the CRC field, recommend publication.
P- Reviewer: Carter WG S- Editor: Yu J L- Editor: A E- Editor: Liu XM
| 1. | Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1848] [Cited by in RCA: 2071] [Article Influence: 188.3] [Reference Citation Analysis (0)] |
| 2. | Hawk ET, Levin B. Colorectal cancer prevention. J Clin Oncol. 2005;23:378-391. [PubMed] |
| 3. | Chang YS, Er TK, Lu HC, Yeh KT, Chang JG. Detection of KRAS codon 12 and 13 mutations by mutant-enriched PCR assay. Clin Chim Acta. 2014;436:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044-2058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 945] [Cited by in RCA: 852] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 5. | Taylor DP, Burt RW, Williams MS, Haug PJ, Cannon-Albright LA. Population-based family history-specific risks for colorectal cancer: a constellation approach. Gastroenterology. 2010;138:877-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 173] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 6. | Galiatsatos P, Foulkes WD. Familial adenomatous polyposis. Am J Gastroenterol. 2006;101:385-398. [PubMed] |
| 7. | Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531-7537. [PubMed] |
| 8. | Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 723] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 9. | Merg A, Howe JR. Genetic conditions associated with intestinal juvenile polyps. Am J Med Genet C Semin Med Genet. 2004;129C:44-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Sampson JR, Jones S, Dolwani S, Cheadle JP. MutYH (MYH) and colorectal cancer. Biochem Soc Trans. 2005;33:679-683. [PubMed] |
| 13. | Vasovcak P, Pavlikova K, Sedlacek Z, Skapa P, Kouda M, Hoch J, Krepelova A. Molecular genetic analysis of 103 sporadic colorectal tumours in Czech patients. PLoS One. 2011;6:e24114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Mundade R, Imperiale TF, Prabhu L, Loehrer PJ, Lu T. Genetic pathways, prevention, and treatment of sporadic colorectal cancer. Oncoscience. 2014;1:400-406. [PubMed] |
| 15. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4464] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 16. | Chang YS, Yeh KT, Chang TJ, Chai C, Lu HC, Hsu NC, Chang JG. Fast simultaneous detection of K-RAS mutations in colorectal cancer. BMC Cancer. 2009;9:179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Chang YS, Yeh KT, Hsu NC, Lin SH, Chang TJ, Chang JG. Detection of N-, H-, and KRAS codons 12, 13, and 61 mutations with universal RAS primer multiplex PCR and N-, H-, and KRAS-specific primer extension. Clin Biochem. 2010;43:296-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 725] [Article Influence: 48.3] [Reference Citation Analysis (1)] |
| 19. | Hsieh LL, Er TK, Chen CC, Hsieh JS, Chang JG, Liu TC. Characteristics and prevalence of KRAS, BRAF, and PIK3CA mutations in colorectal cancer by high-resolution melting analysis in Taiwanese population. Clin Chim Acta. 2012;413:1605-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1163] [Cited by in RCA: 1269] [Article Influence: 90.6] [Reference Citation Analysis (1)] |
| 21. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6647] [Article Influence: 511.3] [Reference Citation Analysis (0)] |
| 22. | Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 677] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 23. | Cousins MM, Ou SS, Wawer MJ, Munshaw S, Swan D, Magaret CA, Mullis CE, Serwadda D, Porcella SF, Gray RH. Comparison of a high-resolution melting assay to next-generation sequencing for analysis of HIV diversity. J Clin Microbiol. 2012;50:3054-3059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Shioda T, Lechleider RJ, Dunwoodie SL, Li H, Yahata T, de Caestecker MP, Fenner MH, Roberts AB, Isselbacher KJ. Transcriptional activating activity of Smad4: roles of SMAD hetero-oligomerization and enhancement by an associating transactivator. Proc Natl Acad Sci USA. 1998;95:9785-9790. [PubMed] |
| 25. | Ling C, Wang L, Wang Z, Xu L, Sun L, Yang H, Li WD, Wang K. A pathway-centric survey of somatic mutations in Chinese patients with colorectal carcinomas. PLoS One. 2015;10:e0116753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Berg M, Danielsen SA, Ahlquist T, Merok MA, Ågesen TH, Vatn MH, Mala T, Sjo OH, Bakka A, Moberg I. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5:e13978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 27. | Lamlum H, Ilyas M, Rowan A, Clark S, Johnson V, Bell J, Frayling I, Efstathiou J, Pack K, Payne S. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: a new facet to Knudson’s ‘two-hit’ hypothesis. Nat Med. 1999;5:1071-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 234] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 28. | Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer WF, Tomlinson IP. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci USA. 2000;97:3352-3357. [PubMed] |
| 29. | Tomasetti C, Marchionni L, Nowak MA, Parmigiani G, Vogelstein B. Only three driver gene mutations are required for the development of lung and colorectal cancers. Proc Natl Acad Sci USA. 2015;112:118-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 298] [Article Influence: 27.1] [Reference Citation Analysis (0)] |