Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2206
Peer-review started: May 4, 2015
First decision: August 26, 2015
Revised: September 28, 2015
Accepted: November 13, 2015
Article in press: November 13, 2015
Published online: February 21, 2016
Processing time: 278 Days and 14.1 Hours
Inflammatory bowel disease (IBD) is characterized by chronic relapsing inflammatory disorders of the gastrointestinal tract, and includes two major phenotypes: ulcerative colitis and Crohn’s disease. The pathogenesis of IBD is not fully understood as of yet. It is believed that IBD results from complicated interactions between environmental factors, genetic predisposition, and immune disorders. miRNAs are a class of small non-coding RNAs that can regulate gene expression by targeting the 3′-untranslated region of specific mRNAs for degradation or translational inhibition. miRNAs are considered to play crucial regulatory roles in many biologic processes, such as immune cellular differentiation, proliferation, and apoptosis, and maintenance of immune homeostasis. Recently, aberrant expression of miRNAs was revealed to play an important role in autoimmune diseases, including IBD. In this review, we discuss the current understanding of how miRNAs regulate autoimmunity and inflammation by affecting the differentiation, maturation, and function of various immune cells. In particular, we focus on describing specific miRNA expression profiles in tissues and peripheral blood that may be associated with the pathogenesis of IBD. In addition, we summarize the opportunities for utilizing miRNAs as new biomarkers and as potential therapeutic targets in IBD.
Core tip: Inflammatory bowel disease (IBD) is characterized by chronic relapsing inflammation in the gastrointestinal tract, but its pathogenesis remains unclear. Further understanding of the molecular mechanisms of IBD is helpful to find new therapeutic strategies. miRNAs play crucial regulatory roles in immune cellular differentiation and maturation, and maintaining immune homeostasis. Aberrant expression of miRNAs is present in IBD. Here, we summarize how miRNAs regulate autoimmunity and inflammation, and describe specific miRNA expression profiles in IBD. We also discuss the opportunities in utilizing miRNAs as new biomarkers and potential therapeutic targets in IBD.
- Citation: Xu XM, Zhang HJ. miRNAs as new molecular insights into inflammatory bowel disease: Crucial regulators in autoimmunity and inflammation. World J Gastroenterol 2016; 22(7): 2206-2218
- URL: https://www.wjgnet.com/1007-9327/full/v22/i7/2206.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i7.2206
Inflammatory bowel disease (IBD) comprises ulcerative colitis (UC) and Crohn’s disease (CD). IBD is characterized by chronic relapsing inflammation in the gastrointestinal tract, and its incidence and prevalence are increasing[1]. The precise pathogenic mechanism of IBD remains unknown. Accumulated evidence suggests that IBD results from complicated interactions between environmental factors, genetic predisposition, and immune dysregulation. Of these, immune dysregulation is believed to play an important role in the pathogenesis of IBD[2]. Consequently, it is important to uncover the molecular mechanisms that regulate the immune responses in IBD.
miRNAs are a new class of small non-coding RNAs that regulate immune responses in physiologic and pathologic conditions[3]. miRNAs are considered to play significant roles in many biologic processes, including cellular proliferation, differentiation, maturation, and apoptosis[4]. In addition, miRNAs have been implicated in the pathogenesis of common human diseases, such as cardiovascular[5], neurologic[6], and hematologic diseases, cancer[7], and inflammatory and autoimmune diseases[8]. These research findings have led to new insights into IBD pathogenesis.
In this review, we summarize recent findings that miRNAs regulate autoimmunity and inflammation by affecting the differentiation, maturation, and function of various immune cells. We particularly focus on providing evidence of specific miRNA expression profiles in IBD pathogenesis. In addition, we also discuss the possibility for miRNAs as new biomarkers and potential therapeutic targets in IBD.
miRNAs are a new class of small (about 22 nucleotides), endogenous, non-coding single-stranded RNA molecules that can negatively regulate target gene expression at the post-transcriptional level[9]. The first miRNA, lin-4, was identified in 1993 in Caenorhabditis elegans[10]. The miRNA sequence database, miRBase, contains 35,828 mature miRNAs in 223 species at time of publication (http://www.mirbase.org/, Release 21, June 2014)[11].
miRNA genes are located either within intronic sequences of protein-coding genes, within intronic or exonic regions of non-coding RNAs, or within intergenic regions[12]. The biogenesis of miRNAs includes two parts: one is transcription in the nucleus, and the other is generation of mature miRNAs in the cytoplasm. First, miRNA is transcribed from the genome by RNA polymerase II or III to generate primary miRNA[13,14]. The primary miRNA is then cleaved by RNase III-type enzyme Drosha to produce a pre-miRNA of approximately 70 nucleotides with a stem-loop structure in the nucleus[15]. Next, the pre-miRNA is exported to the cytoplasm by Exportin 5[16]. Once in the cytoplasm, the pre-miRNA is cleaved by Dicer in cooperation with protein partners, into an approximately 22-nucleotide miRNA duplex[17]. Then, one strand is selected as a functional miRNA, while the passenger strand is degraded. The functional miRNA is loaded into the RNA-induced silencing complex and acts as a guide strand that recognizes the target mRNA by complementary sequences[18]. Full complementarity occurs in plants, resulting in target mRNA degradation. However, incomplete complementary binding occurs in humans, and this leads to mRNA destabilization and translational inhibition[12].
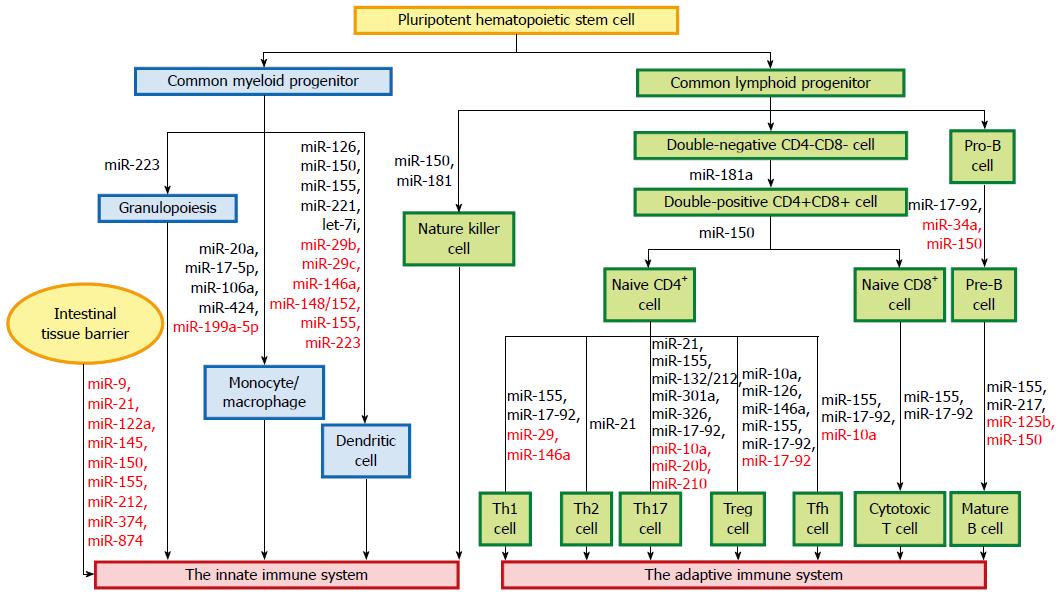
The innate immune system forms the first line of host defense, which is non-specific, and responds to pathogens in a generic way. It is comprised of tissue barriers, immune cells, and immune molecules. The tissue barriers include mechanical (epithelial) barriers, chemical barriers such as antimicrobial peptides, and biologic barriers (commensal flora). The innate immune cells include monocytes/macrophages, dendritic cells (DCs), neutrophils, natural killer (NK) cells, NK T cells, mast cells, eosinophils, and basophils. These cells perform phagocytosis, antigen presentation, and activation of the adaptive immune responses[19]. miRNAs regulate autoimmunity and inflammation by affecting the differentiation, maturation, and function of various immune cells (Figure 1).
The intestinal mucosa forms a barrier that separates luminal contents from the interstitium. Tissue barriers include tight junctions (TJs), adherens junctions, and desmosomes, which regulate the paracellular permeability of epithelial layers across the apical/basolateral axis[20]. The main protein complexes of TJs are composed of transmembrane proteins such as claudins, occludin and junctional adhesion molecules[21]. Disruption of the intestinal barrier has been shown to be an important pathogenic mechanism contributing to the development of intestinal inflammation[22,23]. McKenna et al[24] reported that miRNAs are important for maintaining the function of intestinal barriers.
miR-21, which is overexpressed in chronic UC, induces the degradation of Ras homolog gene family member (Rho)B mRNA and leads to an increase in intestinal epithelial permeability due to the loss of TJ proteins and ultrastructural changes[25]. miR-150 is significantly elevated in colon tissue in dextran-sulfate-sodium-induced murine experimental colitis and active UC patients. Overexpression of miR-150 results in intestinal epithelial disruption through targeting of c-Myb[26]. Both occludin and claudin-1 have been demonstrated to be involved in miR-874-induced intestinal barrier dysfunction by targeting the 3′-untranslated region of aquaporin 3[27]. miR-9 and miR-374 directly target the 3′-untranslated region of claudin-14 mRNA, leading to claudin-14 mRNA translational repression and decay, in a cooperative manner[28]. miR-145 impairs TJ function by repressing junctional adhesion molecule-1 expression[29]. miR-212 impairs the intestinal epithelial barrier by downregulating zonula occludens-1 protein expression, which is another major component of TJs[30].
Tumor necrosis factor (TNF)-α is an essential mediator of inflammation in the gut. Anti-TNF-α therapy induces remission in patients with severe active CD[31], UC[32], and refractory celiac disease[33]. TNF-α-induced upregulation of miR-122a mediates the degradation of occludin mRNA in enterocytes and influences their permeability[34]. TNF-α-induced miR-155 overexpression inhibits synthesis of zonula occludens-1 by downregulating RhoA expression[35].
Monocyte/macrophage differentiation is an essential branch of hematopoiesis, which is under the control of a complex network of regulatory factors[36]. During monocytopoiesis, the transcription factor acute myeloid leukemia (AML)1 is upregulated, while miRNAs-17-5p/20a/106a are downregulated. Monocytopoiesis is regulated by a circuitry comprising sequential miRNAs-17-5p/20a/106a, AML1, and monocyte colony-stimulating factor receptor, whereby miRNAs-17-5p/20a/106a act as a master gene complex that negatively regulates AML1 expression[37]. The transcription factor PU.1 upregulates miR-424 expression, and this induces monocyte differentiation via miR-424-dependent translational inhibition of nuclear factor (NF)I-A. This result indicates an important role of miR-424 and its target NFI-A in controlling monocyte/macrophage differentiation[38]. miR-199a-5p targets the activin A receptor type 1B gene, leading to decreased expression of CCAAT/enhancer binding protein α, and eventually, inhibits monocyte/macrophage differentiation[36].
DCs serve as the most potent antigen-presenting cells, responsible for primary immune responses. Accumulating evidence highlights the importance of specific miRNAs in DC development, antigen-presentation capacity, and cytokine release[39]. miR-146a[40], miR-155[41], and let-7i[42] are involved in the maturation and functional state of DCs, while miR-148/152[43] and miR-223[44] are involved in their antigen-presentation capacity. miR-150 is required for the cross-presentation capacity of Langerhans cells (skin-resident DCs)[45]. In addition, miR-29b, miR-29c[46], miR-126[47], miR-146a[40,48], miR-155, and miR-221[49] have been shown to regulate DC apoptosis and cytokine production.
NK cells are cytotoxic lymphocytes that play a vital role in host defense against infection, and they mediate antitumor responses. Recent advances have demonstrated that miRNAs are crucial in NK cell biology[50]. For instance, miR-150 and miR-181 regulate NK cell development[51,52]. Mice lacking miR-150 are defective in generating mature NK cells. On the contrary, transgenic mice with a gain-of-function miR-150 have enhanced NK cell development[51]. miR-181 promotes NK cell development by targeting Nemo-like kinase, which is a inhibitor of Notch signaling[52].
Invariant NK T cells are a separate subset of T lymphocytes with innate effector functions. A Dicer-dependent miRNA pathway is important in the regulation of invariant NK T cell differentiation, function, and homeostasis[53]. The normal granulocytic differentiation requires the zinc finger protein growth factor independent-1, which is a transcription inhibitor that regulates the expression of miR-21 and miR-196b during myelopoiesis[54]. In addition, miR-223 plays a crucial role in the regulation of granulocyte differentiation and function, and mediates inflammatory responses[55,56].
Pattern recognition receptors are critical for the recognition of microorganisms and the induction of immune and inflammatory responses[57]. The families of these proteins include the membrane-bound Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors, and retinoic acid-inducible gene-I-like receptors[58]. Pattern recognition receptors promote downstream signaling cascades. Emerging evidence indicates that miRNAs regulate these processes.
TLRs: miR-146a expression can be induced through exposure to TLR ligands, such as lipopolysaccharide, peptidoglycan, and flagellin, and this induction is controlled by NF-κB. Mice lacking miR-146a are more likely to develop autoimmune diseases, tumorigenesis, and myeloid cell proliferation. miR-146 targets TNF-receptor-associated factor 6 and interleukin (IL)-1-receptor-associated kinase 1, which are key elements of the myeloid differentiation factor 88 pathway, and form a negative feedback mechanism in TLR signaling[59-61]. miR-155 expression is also induced by TLR signaling[61,62]. Unlike miR-146a, miR-155 promotes the immune response. Mice deficient in miR-155 are highly resistant to experimental autoimmune encephalomyelitis[63].
NOD-like receptors: Of this family of proteins, NOD2 functions as an intracellular sensor that contributes to inflammation and immune defense. It has been identified as the strongest single genetic locus in determining susceptibility in CD[64]. A miRs-NOD interaction has been implicated in IBD. These miRNAs include miR-29[65], miR-122[66], miR-146a[67], and miR-192[68]. For example, polymorphisms in NOD2 impair miR-29 expression in DCs, and this results in exaggerated IL-23-induced inflammation[65]. miR-122 targeting of NOD2 has a crucial role in the damage of intestinal epithelial cells induced by lipopolysaccharide[66].
The adaptive immune system mainly consists of two different lymphocytes (T and B cells), and is highly pathogen specific. The appropriate development and function of these two immune cells are essential when distinguishing foreign from resident antigens. Current studies have indicated that miRNAs play important roles in maintaining the differentiation and function of T and B cells[69].
Increasing evidence shows that some specific miRNAs participate in the regulation of crucial immune functions. These immuno-miRs play significant roles in T-cell development, maturation, activation, differentiation, and aging[70]. For example, miR-150[71] and miR-181a[72] are involved in T-cell development, while miR-21[73] and miR-17-92 cluster[74,75] participate in T-cell activation.
miRNAs have significant effects on T helper (Th) cell differentiation. Naïve T cells can differentiate into Th1, Th2, or Th17 cells after activation[76]. Th1 cells have been associated with the pathogenesis of CD, while Th2 cells have been implicated in UC[19]. miR-155 promotes Th1 differentiation by targeting interferon (IFN)-γ receptor α chain[77]. In contrast, CD4+ T cells deficient in miR-155 display a bias towards Th2 differentiation, which is partly due to increased expression of the Th2-associated transcription factor c-Maf[41,78]. miR-17-92 cluster promotes Th1 differentiation. The function of miR-19b is mediated through phosphatase and tensin homolog, while miR-17 targets transforming growth factor (TGF)-β receptor II and cAMP-responsive element-binding protein 1[79]. miR-29 regulates Th cell differentiation by directly targeting T-box transcription factor T-bet and eomesodermin to suppress IFN-γ production[80]. miR-146a may be a potent inhibitor of Th1 differentiation by targeting protein kinase Cε[81]. miR-21 promotes Th2 differentiation[82].
Th17 cells, a new subset of Th cells capable of producing IL-17, play a vital role in the formation of several autoimmune-mediated inflammatory diseases, including IBD[83,84]. Current studies demonstrate that miR-21[85], miR-155[63], miR-301a[86], miR-326[87], miR-17-92 cluster[88], and miR-132/212 cluster[89] act as positive regulators of Th17 differentiation. For example, miR-155 enhances the development of inflammatory T cells (Th1 and Th17 cells), and facilitates Th17 cell formation through cytokines produced by DCs[63]. miR-10a[90], miR-20b[91], and miR-210[92] act as negative regulators of Th17 differentiation. Deletion of miR-210 promotes Th17 differentiation under hypoxic conditions[92].
Regulatory T (Treg) cells are another subset of CD4+ T cells that can suppress activity of effector T cells and maintain self-tolerance[76,90]. Treg cells can be classified into two populations, naturally-occurring Treg (nTreg) cells that are generated in the thymus, and inducible Treg (iTreg) cells that arise from naïve CD4+ precursors in the periphery[90]. miRNAs play pivotal roles in the regulation of both Treg cell development and function[93,94]. miR-10a is highly expressed in nTreg cells and can be induced by retinoic acid and TGF-β in iTreg cells[90]. Repression of miR-10a in vitro results in reduced forkhead box (Fox)p3 expression levels, while ablation of miR-10a does not affect the phenotype or number of nTreg cells[93]. miR-155-deficient mice display a marked reduction in the number of Treg cells. Additionally, miR-155 maintains homeostasis of Treg cells by targeting suppressor of cytokine signaling 1 via the IL-2 signaling pathway[95]. miR-146a is important for maintaining suppressive function of Treg cells. Treg cells deficient in miR-146a lead to immunologic intolerance by targeting signal transducer and activator of transcription-1[96]. Silencing of miR-126 can influence the expression of Foxp3 on Treg cells and impair their suppressive function via the PI3K/Akt pathway[97]. miR-17-92 cluster is also involved in Treg cell function. Mice with Treg-specific loss of miR-17-92 cluster develop an exacerbated form of experimental autoimmune encephalomyelitis and fail to achieve clinical remission[74]. However, there are conflicting results. For instance, the study from Jiang et al[79] showed that miR-17-92 cluster prevents Treg cell differentiation and promotes Th1 responses.
Follicular helper T (Tfh) cells are a novel subset of CD4+ T cells that can provide help to B cells, and they are important for germinal center formation[98]. Several studies have demonstrated that miRNAs are crucial for Tfh cell differentiation and function[99,100]. Mice with T-cell-specific loss of miR-17-92 cluster exhibit severely compromised Tfh cell differentiation, germinal center formation, and antibody responses. On the contrary, T-cell-specific miR-17-92 cluster transgenic mice spontaneously accumulate Tfh cells[99]. miR-10a attenuates phenotypic conversion of iTreg cells to Tfh cells by simultaneously targeting Bcl-6, a transcription factor critical for Tfh cell differentiation[101], along with the corepressor Ncor2[90]. Moreover, miR-155 has been reported to promote Tfh cell development[102].
CD8+ T cells or cytotoxic T lymphocytes can devastate various intracellular pathogens and malignancies[103]. Dicer is required for CD8+ T-cell survival and accumulation, but not required for the early steps in CD8+ T-cell activation[104,105]. Dicer and miRNAs such as miR-139 and miR-150 also participate in controlling the cytolytic program, as well as other programs of effector cytotoxic T lymphocyte differentiation[106]. miR-155 is demanded for effector CD8+ T-cell responses to viral and intracellular bacterial infection and cancer. miR-155 has the potential to be a target for immunotherapy for infectious diseases and cancer[103,107,108]. miR-17-92 cluster has dynamic regulation of CD8+ T cells differentiating from naïve to effector and memory states[75,109].
Ablation of Dicer in early B-cell progenitors leads to a formative block from the pro-B to pre-B transition[110]. miRNAs are also involved in B-cell development and function[110,111], including miR-34a[112], miR-150[113,114], and miR-17-92 cluster[115]. miR-125b inhibits B-cell differentiation in germinal centers[116]. In addition to regulating B-cell differentiation, miR-150[113], miR-155[78], and miR-217[111] regulate B-cell function, including the establishment of B-cell tolerance, as well as antigen-dependent and -independent antibody repertoire diversification.
Abnormal miRNA expressions exist in some diseases, including IBD (Table 1). This offers a new way to improve our comprehension of the mechanism of this disease. Moreover, some specific miRNAs in IBD may serve as potential biomarkers for diagnosis, evaluation indicators of disease activity, or targets for treatment.
| Sample type | Expression | miRNAs | Ref. |
| Ulcerative colitis vs healthy controls | |||
| Mucosal tissues | Upregulated | miR-7, miR-16, miR-20b, miR-21, miR-23a, miR-24, miR-29a, miR-29b, miR-31, miR-125b-1*, miR-126, miR-126*, miR-127-3p, miR-135b, miR-146a, miR-150, miR-155, miR-195, miR-206, miR-223, miR-324-3p, miR-424, and let-7f | [25,26,117-122] |
| Downregulated | miR-188-5p, miR-192, miR-200b, miR-215, miR-320a, miR-346, miR-375, and miR-422b; miR-124 (pediatric cases) | [117,119,123,124] | |
| Peripheral blood | Upregulated | miR-16, miR-21, miR-28-5p, miR-103-2*, miR-151-5p, miR-155, miR-188-5p, miR-199a-5p, miR-340*, miR-362-3p, miR-378, miR-422a, miR-500, miR-501-5p, miR-532-3p, miR-769-5p, miR-874, and miRplus-E1271 | [125-127] |
| Downregulated | miR-505* | [126] | |
| Crohn’s disease vs healthy controls | |||
| Mucosal tissues | Upregulated | miR-9, miR-21, miR-22, miR-26a, miR-29b, miR-29c, miR-30b, miR-31, miR-34c-5p, miR-106a, miR-106b, miR-126, miR-126*, miR-127-3p, miR-130a, miR-133b, miR-146a, miR-146b-5p, miR-150, miR-155, miR-181c, miR-196a, miR-196, miR-206, miR-324-3p, miR-375, and miR-424 | [119,121,130,131] |
| Downregulated | miR-7 and miR-141 | [128,129] | |
| Peripheral blood | Upregulated | miR-16, miR-23a, miR-29a, miR-106a, miR-107, miR-126, miR-191, miR-199a-5p, miR-200c, miR-362-3p and miR-532-3p; | [125,133] |
| miR-16, miR-20a, miR-21, miR-30e, miR-93, miR-106a, miR-140, miR-192, miR-195, miR-484, and let-7b (pediatric cases) | |||
miRNAs in mucosal tissues: In 2008, the first profiling study of altered expression of miRNAs in IBD patients was published[117]. Wu and colleagues[117] found a specific miRNA expression pattern: three miRNAs (miR-192, miR-375, and miR-422b) were markedly downregulated, whereas eight miRNAs (miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, miR-195, and let-7f) were observably upregulated in active UC tissues. Furthermore, they found that miR-192 participated in the regulation of chemokine production in colonic epithelial cells. Since 2008, some new research in active UC and healthy controls has confirmed the upregulation of miR-21[25,118], miR-29a[119], and miR-126[120], and identified additional upregulated miRNAs, including miR-7, miR-29b, miR-126*, miR-127-3p, miR-135b, miR-223 and miR-324-3p[119], miR-31[119,121], miR-150[26], miR-155[118], miR-146a, miR-206, and miR-424[121], and miR-20b and miR-125b-1*[122]. In contrast, miR-188-5p, miR-215, miR-320a, miR-346[119], and miR-200b[123] were downregulated in colon tissues from active UC patients compared with healthy controls. miR-124 was markedly decreased in pediatric but not in adult UC tissues. Reduced levels of miR-124 in colon tissues appear to increase the expression and activity of signal transducer and activator of transcription-3, and this mediates the pathogenesis of UC in children[124].
miRNAs in peripheral blood: Paraskevi and colleagues[125] found that six miRNAs (miR-16, miR-21, miR-28-5p, miR-151-5p, miR-155, and miR-199a-5p) were remarkably upregulated in blood from UC patients compared with healthy controls. miR-155 had the highest expression level of these six UC-associated miRNAs in peripheral blood. Wu and colleagues[126] found that compared with healthy controls, 12 miRNAs were significantly upregulated, and miRNA-505* was downregulated in blood from active UC patients. Peripheral blood miRNAs may distinguish active UC patients from healthy controls. As compared to active CD patients, ten miRNAs were markedly upregulated, and one miRNA was downregulated in blood from active UC patients[126]. Duttagupta et al[127] completed analyses of miRNA expressions from different hematologic fractions as noninvasive predictors for incidence of UC. They found that seven miRNAs derived from platelets (miR-188-5p, miR-378, miR-422a, miR-500, miR-501-5p, miR-769-5p, and miR-874) were upregulated. This study provides new platelet-derived miRNA biomarkers for clinical application and perception of the potential roles of these miRNAs in the pathogenesis of UC.
miRNAs in mucosal tissues: Most studies of miRNA expression profiles in CD have concentrated on Crohn’s colitis. Fasseu et al[119] found that 23 miRNAs (miR-9, miR-21, miR-22, miR-26a, miR-29b, miR-29c, miR-30b, miR-31, miR-34c-5p, miR-106a, miR-126, miR-126*, miR-127-3p, miR-130a, miR-133b, miR-146a, miR-146b-5p, miR-150, miR-155, miR-181c, miR-196a, miR-324-3p, and miR-375) were remarkably upregulated in colonic tissues from CD patients compared with healthy controls. Five of these miRNAs were specific for patients in an active stage of CD (miR-9, miR-126, miR-130a, miR-181c, and miR-375), whereas the remaining 18 were also upregulated in colonic tissues from inactive CD patients. Huang and colleagues[128] identified that miR-141 was downregulated in inflamed colon tissues from active CD patients. miR-141 inhibited colonic chemokine CXC ligand 12β expression by directly targeting it and blocked colonic immune cell recruitment. Nguyen and colleagues[129] found that only miR-7 was downregulated in eight active colonic CD patients compared to six healthy controls. In addition, miR-206, miR-424[121], miR-106b[130] and miR-196[131] are also upregulated in active colonic CD.
However, is there any tissue-specific miRNA expression profile in the gastrointestinal tract? Wu and colleagues[132] examined miRNA expression patterns in tissues from different intestinal segments in active CD patients. Ten intestine-specific miRNAs (miR-19b, miR-22, miR-23a, miR-26a, miR-31, miR-126, miR-215, miR-320, miR-422b, and let-7d) were identified. Specifically, three of these (miR-22, miR-31, and miR-215) were markedly upregulated in the terminal ileum compared with colon tissue, while miR-19b was downregulated in the terminal ileum. Moreover, miR-23a, miR-26a, miR-126, miR-320, miR-422b, and let-7d showed colon-specific expression. In active colonic CD patients, three miRNAs (miR-23b, miR-106, and miR-191) were upregulated and two (miR-19b and miR-629) were downregulated compared to healthy controls. In active terminal ileal CD patients, four miRNAs (miR-16, miR-21, miR-223, and miR-594) were upregulated in terminal ileal tissues.
miRNAs in peripheral blood: Apart from assessing miRNA expressions in peripheral blood in UC, Paraskevi et al[125] examined miRNA expression patterns in peripheral blood samples from 128 patients with active CD and 162 healthy individuals. Eleven miRNAs (miR-16, miR-23a, miR-29a, miR-106a, miR-107, miR-126, miR-191, miR-199a-5p, miR-200c, miR-362-3p, and miR-532-3p) were markedly upregulated in peripheral blood from CD patients as compared with healthy individuals. There were no significant differences in miRNA expressions in accordance with disease location and phenotype.
Zahm et al[133] examined serum samples from 46 pediatric CD patients and 32 healthy controls by means of a low-density microarray and quantitative reverse transcriptase (qRT) PCR. They found 11 miRNAs (miR-16, miR-20a, miR-21, miR-30e, miR-93, miR-106a, miR-140, miR-192, miR-195, miR-484, and let-7b) that were CD-associated circulating miRNAs. Receiver operating characteristic analyses indicated that these CD-associated miRNAs had promising diagnostic value, with sensitivities of 70%-83% and specificities of 75%-100%. These results demonstrate that circulating miRNAs may be used as novel noninvasive biomarkers in CD.
Iborra and colleagues[134] assessed miRNA expression patterns in serum and tissue samples from nine patients with active UC, nine with inactive UC, nine with active CD, and nine with inactive CD, and serum from 33 healthy subjects. They found that two miRNAs (miR-548a-3p and miR-650) were higher, and three (miR-196b, miR-489, and miR-630) were lower in the mucosa of active UC patients compared with inactive UC patients. There were no differences in serum miRNA expression profiles in patients with active UC compared with inactive UC. However, there were differences in serum miRNA expressions between active and inactive CD patients; two serum miRNAs (miR-188-5p and miR-877) were increased, and four serum miRNAs (miR-18a, miR-128, miR-140-5p, and miR-145) were decreased in patients with active CD. Furthermore, four miRNAs (miR-18a*, miR-140-3p, miR-629*, and let-7b) were higher, and three miRNAs (miR-328, miR-422a, and miR-855-5p) were lower in the mucosa of active CD patients compared with inactive CD patients. These results indicate that there are specific miRNA expression patterns associated with different stages of IBD. Further prospective cohort studies in large samples are necessary to validate these findings.
miRNA-related therapeutic applications may represent a new and fascinating field in IBD treatment. miRNA-related therapy is based on antisense technology and gene therapy; thus, it involves either miRNA antagonists or miRNA mimics.
miRNA antagonists: miRNA antagonists include anti-miRNA oligonucleotides (AMOs), miRNA sponges, and miRNA masks.
AMOs: AMOs are synthetic anti-miRNA oligonucleotides with reverse complementary sequences to their target miRNAs, which suppress miRNA functions. It is believed that AMOs have a promising future in therapeutic applications. Chemical modifications of AMOs can improve their stability and binding affinity. Common modifications include addition of different 2′-ribose modifications to AMOs (2′-O-methyl and 2′-O-methoxyethyl) and 2′,4′-methylene bridge-locked nucleic acid (LNA). LNA-modified AMOs create high-affinity binding to target mRNAs[135,136]. A study by Janssen et al[137] demonstrated that miravirsen, an LNA-anti-miR-122, is designed to target and inhibit miR-122, and this can reduce viral RNA levels in patients with chronic hepatitis C virus infection. This result proves the possibility of miRNA agents in clinical practice.
miRNA sponges: miRNA sponge technology utilizes plasmid or viral vectors to achieve loss-of-function of miRNAs. The strong promoters can be applied in miRNA sponge vectors for generating high-level expression of the competitive inhibitor transcripts for either transient or long-term inhibition of miRNA function. Considering the merit of sharing a common seed sequence by members of a miRNA family, this technology provides a strong approach for coinstantaneous inhibition of multiple miRNAs of interest with a single inhibitor[138].
miRNA mimicry/replacement therapy: In order to restore miRNA activity, miRNA mimics (synthetic oligonucleotides) and miRNA expression gene vectors are used. MRX34 is a double-stranded miRNA mimic of the naturally occurring miR-34a loaded in liposomal nanoparticles to reestablish its tumor suppressor function. MRX34 was the first miRNA mimic introduced into clinical study for primary as well as metastatic liver cancer in 2013[139,140]. Many miRNAs are downregulated in UC and CD. For example, miR-192, miR-375, miR-422b[117], miR-188-5p, miR-215, miR-320a, miR-346[119], and miR-200b[123] are decreased in UC, and miR-19b and miR-629[132] are decreased in Crohn’s colitis tissues. Theoretically, replenishing these decreased miRNAs by miRNA mimics may provide therapeutic restoration of physiologic functions lost in IBD.
In this review, we described the roles of miRNAs as crucial regulators of inflammatory responses and autoimmune disorders, particularly focusing on miRNAs affecting the differentiation, maturation, and function of various immune cells. We also summarized some studies on the current understanding of the connection between miRNAs and IBD. Accumulating evidence suggests that specific miRNA expression profiles exist in IBD, and these miRNAs contribute to the development of inflammation. The definite functions of most miRNAs in IBD have not yet been clarified. Further studies are necessary to validate whether miRNAs could be used to diagnose IBD, distinguish IBD subtypes, and determine the disease activity or location.
P- Reviewer: Lakatos PL S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Liu XM
| 1. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3789] [Cited by in RCA: 3527] [Article Influence: 271.3] [Reference Citation Analysis (5)] |
| 2. | Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1587] [Cited by in RCA: 1537] [Article Influence: 102.5] [Reference Citation Analysis (0)] |
| 3. | O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1160] [Cited by in RCA: 1273] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 4. | Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 650] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 5. | Wang F, Chen C, Wang D. Circulating microRNAs in cardiovascular diseases: from biomarkers to therapeutic targets. Front Med. 2014;8:404-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Wang C, Ji B, Cheng B, Chen J, Bai B. Neuroprotection of microRNA in neurological disorders (Review). Biomed Rep. 2014;2:611-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 7. | Lawrie CH. MicroRNAs in hematological malignancies. Blood Rev. 2013;27:143-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 8. | Iborra M, Bernuzzi F, Invernizzi P, Danese S. MicroRNAs in autoimmunity and inflammatory bowel disease: crucial regulators in immune response. Autoimmun Rev. 2012;11:305-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 9. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 10. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 11. | Van Peer G, Lefever S, Anckaert J, Beckers A, Rihani A, Van Goethem A, Volders PJ, Zeka F, Ongenaert M, Mestdagh P. miRBase Tracker: keeping track of microRNA annotation changes. Database (Oxford). 2014;2014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 12. | Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, Satsangi J. MicroRNAs: new players in IBD. Gut. 2015;64:504-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 13. | Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-4060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2902] [Cited by in RCA: 2997] [Article Influence: 142.7] [Reference Citation Analysis (0)] |
| 14. | Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 938] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 15. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3631] [Article Influence: 165.0] [Reference Citation Analysis (0)] |
| 16. | Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1918] [Cited by in RCA: 1888] [Article Influence: 89.9] [Reference Citation Analysis (0)] |
| 17. | Koscianska E, Starega-Roslan J, Krzyzosiak WJ. The role of Dicer protein partners in the processing of microRNA precursors. PLoS One. 2011;6:e28548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 18. | Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582-3600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 334] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 19. | Wallace KL, Zheng LB, Kanazawa Y, Shih DQ. Immunopathology of inflammatory bowel disease. World J Gastroenterol. 2014;20:6-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 356] [Cited by in RCA: 421] [Article Influence: 38.3] [Reference Citation Analysis (2)] |
| 20. | Cichon C, Sabharwal H, Rüter C, Schmidt MA. MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers. 2014;2:e944446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 21. | Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol. 2009;1:a002584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 652] [Cited by in RCA: 772] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 22. | Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2121] [Cited by in RCA: 2709] [Article Influence: 169.3] [Reference Citation Analysis (0)] |
| 23. | Shen L, Su L, Turner JR. Mechanisms and functional implications of intestinal barrier defects. Dig Dis. 2009;27:443-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 24. | McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654-1664, 1664.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 256] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 25. | Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, Zhang P, Wang F, Yang J, Yang J. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun. 2013;434:746-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 26. | Bian Z, Li L, Cui J, Zhang H, Liu Y, Zhang CY, Zen K. Role of miR-150-targeting c-Myb in colonic epithelial disruption during dextran sulphate sodium-induced murine experimental colitis and human ulcerative colitis. J Pathol. 2011;225:544-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 27. | Zhi X, Tao J, Li Z, Jiang B, Feng J, Yang L, Xu H, Xu Z. MiR-874 promotes intestinal barrier dysfunction through targeting AQP3 following intestinal ischemic injury. FEBS Lett. 2014;588:757-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 28. | Gong Y, Renigunta V, Himmerkus N, Zhang J, Renigunta A, Bleich M, Hou J. Claudin-14 regulates renal Ca++ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J. 2012;31:1999-2012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 207] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 29. | Adammek M, Greve B, Kässens N, Schneider C, Brüggemann K, Schüring AN, Starzinski-Powitz A, Kiesel L, Götte M. MicroRNA miR-145 inhibits proliferation, invasiveness, and stem cell phenotype of an in vitro endometriosis model by targeting multiple cytoskeletal elements and pluripotency factors. Fertil Steril. 2013;99:1346-1355.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 30. | Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 222] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 31. | Akobeng AK. Crohn’s disease: current treatment options. Arch Dis Child. 2008;93:787-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Thukral C, Cheifetz A, Peppercorn MA. Anti-tumour necrosis factor therapy for ulcerative colitis: evidence to date. Drugs. 2006;66:2059-2065. [PubMed] |
| 33. | Gillett HR, Arnott ID, McIntyre M, Campbell S, Dahele A, Priest M, Jackson R, Ghosh S. Successful infliximab treatment for steroid-refractory celiac disease: a case report. Gastroenterology. 2002;122:800-805. [PubMed] |
| 34. | Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323-1333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 250] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 35. | Tian R, Wang RL, Xie H, Jin W, Yu KL. Overexpressed miRNA-155 dysregulates intestinal epithelial apical junctional complex in severe acute pancreatitis. World J Gastroenterol. 2013;19:8282-8291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 41] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Lin HS, Gong JN, Su R, Chen MT, Song L, Shen C, Wang F, Ma YN, Zhao HL, Yu J. miR-199a-5p inhibits monocyte/macrophage differentiation by targeting the activin A type 1B receptor gene and finally reducing C/EBPα expression. J Leukoc Biol. 2014;96:1023-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 37. | Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 350] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 38. | Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA. 2007;104:19849-19854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 235] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 39. | Smyth LA, Boardman DA, Tung SL, Lechler R, Lombardi G. MicroRNAs affect dendritic cell function and phenotype. Immunology. 2015;144:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 40. | Karrich JJ, Jachimowski LC, Libouban M, Iyer A, Brandwijk K, Taanman-Kueter EW, Nagasawa M, de Jong EC, Uittenbogaart CH, Blom B. MicroRNA-146a regulates survival and maturation of human plasmacytoid dendritic cells. Blood. 2013;122:3001-3009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 41. | Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608-611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1554] [Cited by in RCA: 1552] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 42. | Zhang M, Liu F, Jia H, Zhang Q, Yin L, Liu W, Li H, Yu B, Wu J. Inhibition of microRNA let-7i depresses maturation and functional state of dendritic cells in response to lipopolysaccharide stimulation via targeting suppressor of cytokine signaling 1. J Immunol. 2011;187:1674-1683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 43. | Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li N, Cao X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J Immunol. 2010;185:7244-7251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 44. | Mi QS, Xu YP, Wang H, Qi RQ, Dong Z, Zhou L. Deletion of microRNA miR-223 increases Langerhans cell cross-presentation. Int J Biochem Cell Biol. 2013;45:395-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 45. | Mi QS, Xu YP, Qi RQ, Shi YL, Zhou L. Lack of microRNA miR-150 reduces the capacity of epidermal Langerhans cell cross-presentation. Exp Dermatol. 2012;21:876-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 46. | Hong Y, Wu J, Zhao J, Wang H, Liu Y, Chen T, Kan X, Tao Q, Shen X, Yan K. miR-29b and miR-29c are involved in Toll-like receptor control of glucocorticoid-induced apoptosis in human plasmacytoid dendritic cells. PLoS One. 2013;8:e69926. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 47. | Agudo J, Ruzo A, Tung N, Salmon H, Leboeuf M, Hashimoto D, Becker C, Garrett-Sinha LA, Baccarini A, Merad M. The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids. Nat Immunol. 2014;15:54-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 48. | Park H, Huang X, Lu C, Cairo MS, Zhou X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem. 2015;290:2831-2841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 189] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 49. | Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, Blazar BR, Zeng Y, Zhou X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293-4303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 226] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 50. | Leong JW, Sullivan RP, Fehniger TA. microRNA management of NK-cell developmental and functional programs. Eur J Immunol. 2014;44:2862-2868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 51. | Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Exp Med. 2011;208:2717-2731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 193] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 52. | Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171-6175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 153] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 53. | Fedeli M, Napolitano A, Wong MP, Marcais A, de Lalla C, Colucci F, Merkenschlager M, Dellabona P, Casorati G. Dicer-dependent microRNA pathway controls invariant NKT cell development. J Immunol. 2009;183:2506-2512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 54. | Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720-4728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 55. | Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 792] [Cited by in RCA: 793] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 56. | Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 896] [Cited by in RCA: 966] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 57. | Gourbeyre P, Berri M, Lippi Y, Meurens F, Vincent-Naulleau S, Laffitte J, Rogel-Gaillard C, Pinton P, Oswald IP. Pattern recognition receptors in the gut: analysis of their expression along the intestinal tract and the crypt/villus axis. Physiol Rep. 2015;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 58. | Elia PP, Tolentino YF, Bernardazzi C, de Souza HS. The role of innate immunity receptors in the pathogenesis of inflammatory bowel disease. Mediators Inflamm. 2015;2015:936193. [PubMed] |
| 59. | Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481-12486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3121] [Cited by in RCA: 3548] [Article Influence: 186.7] [Reference Citation Analysis (0)] |
| 60. | Saba R, Sorensen DL, Booth SA. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front Immunol. 2014;5:578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 294] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 61. | Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol. 2011;8:388-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 250] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 62. | O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1365] [Cited by in RCA: 1484] [Article Influence: 82.4] [Reference Citation Analysis (0)] |
| 63. | O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607-619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 730] [Cited by in RCA: 736] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 64. | Cuthbert AP, Fisher SA, Mirza MM, King K, Hampe J, Croucher PJ, Mascheretti S, Sanderson J, Forbes A, Mansfield J. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology. 2002;122:867-874. [PubMed] |
| 65. | Brain O, Owens BM, Pichulik T, Allan P, Khatamzas E, Leslie A, Steevels T, Sharma S, Mayer A, Catuneanu AM. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity. 2013;39:521-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 66. | Chen Y, Wang C, Liu Y, Tang L, Zheng M, Xu C, Song J, Meng X. miR-122 targets NOD2 to decrease intestinal epithelial cell injury in Crohn’s disease. Biochem Biophys Res Commun. 2013;438:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 67. | Ghorpade DS, Sinha AY, Holla S, Singh V, Balaji KN. NOD2-nitric oxide-responsive microRNA-146a activates Sonic hedgehog signaling to orchestrate inflammatory responses in murine model of inflammatory bowel disease. J Biol Chem. 2013;288:33037-33048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 68. | Chuang AY, Chuang JC, Zhai Z, Wu F, Kwon JH. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflamm Bowel Dis. 2014;20:126-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 69. | Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2463] [Cited by in RCA: 2548] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 70. | Kroesen BJ, Teteloshvili N, Smigielska-Czepiel K, Brouwer E, Boots AM, van den Berg A, Kluiver J. Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology. 2015;144:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 139] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 71. | Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, Mussolin L, Ruggero K, Bonanno L, Guffanti A. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011;117:7053-7062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 179] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 72. | Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 967] [Cited by in RCA: 938] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 73. | Smigielska-Czepiel K, van den Berg A, Jellema P, Slezak-Prochazka I, Maat H, van den Bos H, van der Lei RJ, Kluiver J, Brouwer E, Boots AM. Dual role of miR-21 in CD4+ T-cells: activation-induced miR-21 supports survival of memory T-cells and regulates CCR7 expression in naive T-cells. PLoS One. 2013;8:e76217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 74. | de Kouchkovsky D, Esensten JH, Rosenthal WL, Morar MM, Bluestone JA, Jeker LT. microRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. J Immunol. 2013;191:1594-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 75. | Wu T, Wieland A, Araki K, Davis CW, Ye L, Hale JS, Ahmed R. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2012;109:9965-9970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 76. | Geginat J, Paroni M, Maglie S, Alfen JS, Kastirr I, Gruarin P, De Simone M, Pagani M, Abrignani S. Plasticity of human CD4 T cell subsets. Front Immunol. 2014;5:630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 217] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 77. | Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol. 2010;40:225-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 215] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 78. | Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1145] [Cited by in RCA: 1199] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 79. | Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li QJ. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487-5497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 243] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 80. | Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 2011;35:169-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 299] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 81. | Möhnle P, Schütz SV, van der Heide V, Hübner M, Luchting B, Sedlbauer J, Limbeck E, Hinske LC, Briegel J, Kreth S. MicroRNA-146a controls Th1-cell differentiation of human CD4+ T lymphocytes by targeting PRKCΕ. Eur J Immunol. 2015;45:260-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 82. | Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol. 2013;54:435-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 83. | Gálvez J. Role of Th17 Cells in the Pathogenesis of Human IBD. ISRN Inflamm. 2014;2014:928461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 84. | Han L, Yang J, Wang X, Li D, Lv L, Li B. Th17 cells in autoimmune diseases. Front Med. 2015;9:10-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 85. | Murugaiyan G, da Cunha AP, Ajay AK, Joller N, Garo LP, Kumaradevan S, Yosef N, Vaidya VS, Weiner HL. MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J Clin Invest. 2015;125:1069-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 86. | Mycko MP, Cichalewska M, Machlanska A, Cwiklinska H, Mariasiewicz M, Selmaj KW. MicroRNA-301a regulation of a T-helper 17 immune response controls autoimmune demyelination. Proc Natl Acad Sci USA. 2012;109:E1248-E1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 164] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 87. | Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 618] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 88. | Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros Family Zinc Finger 4 to promote TH17-mediated inflammation. J Biol Chem. 2014;289:12446-12456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 89. | Nakahama T, Hanieh H, Nguyen NT, Chinen I, Ripley B, Millrine D, Lee S, Nyati KK, Dubey PK, Chowdhury K. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc Natl Acad Sci USA. 2013;110:11964-11969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 90. | Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciumè G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y. TGF-β and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 230] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 91. | Zhu E, Wang X, Zheng B, Wang Q, Hao J, Chen S, Zhao Q, Zhao L, Wu Z, Yin Z. miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORγt and STAT3. J Immunol. 2014;192:5599-5609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 92. | Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol. 2014;15:393-401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 202] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 93. | Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, Lund AH, Bluestone JA. MicroRNA 10a marks regulatory T cells. PLoS One. 2012;7:e36684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 94. | Kelada S, Sethupathy P, Okoye IS, Kistasis E, Czieso S, White SD, Chou D, Martens C, Ricklefs SM, Virtaneva K. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS Pathog. 2013;9:e1003451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 95. | Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 671] [Cited by in RCA: 677] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 96. | Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914-929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 789] [Cited by in RCA: 784] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 97. | Qin A, Wen Z, Zhou Y, Li Y, Li Y, Luo J, Ren T, Xu L. MicroRNA-126 regulates the induction and function of CD4(+) Foxp3(+) regulatory T cells through PI3K/AKT pathway. J Cell Mol Med. 2013;17:252-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 98. | Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol. 2011;29:621-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1946] [Cited by in RCA: 2201] [Article Influence: 157.2] [Reference Citation Analysis (0)] |
| 99. | Kang SG, Liu WH, Lu P, Jin HY, Lim HW, Shepherd J, Fremgen D, Verdin E, Oldstone MB, Qi H. MicroRNAs of the miR-17~92 family are critical regulators of T(FH) differentiation. Nat Immunol. 2013;14:849-857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 100. | Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM. The microRNA cluster miR-17~92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14:840-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 101. | Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006-1010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1361] [Cited by in RCA: 1294] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 102. | Hu R, Kagele DA, Huffaker TB, Runtsch MC, Alexander M, Liu J, Bake E, Su W, Williams MA, Rao DS. miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation. Immunity. 2014;41:605-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 131] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 103. | Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, Mueller YM, Fraietta JA, Wherry EJ, Turner M. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593-602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 223] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 104. | Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. 2010;107:21629-21634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 105. | Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 494] [Cited by in RCA: 508] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 106. | Trifari S, Pipkin ME, Bandukwala HS, Äijö T, Bassein J, Chen R, Martinez GJ, Rao A. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2013;110:18608-18613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 107. | Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, Boudousquie C, Utzschneider DT, Escobar TM, Perret R. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 254] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 108. | Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol. 2013;190:1210-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 109. | Khan AA, Penny LA, Yuzefpolskiy Y, Sarkar S, Kalia V. MicroRNA-17~92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood. 2013;121:4473-4483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 110. | Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 452] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 111. | de Yébenes VG, Bartolomé-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunol Rev. 2013;253:25-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 112. | Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 113. | Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 788] [Cited by in RCA: 811] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 114. | Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080-7085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 483] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 115. | Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1370] [Cited by in RCA: 1311] [Article Influence: 77.1] [Reference Citation Analysis (1)] |
| 116. | Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22:583-592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 117. | Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624-1635.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 404] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 118. | Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, Ando T, Oyamada Y, Isozaki Y, Hongo H. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol. 2010;25 Suppl 1:S129-S133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 119. | Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC, Moreau R. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:e13160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 120. | Feng X, Wang H, Ye S, Guan J, Tan W, Cheng S, Wei G, Wu W, Wu F, Zhou Y. Up-regulation of microRNA-126 may contribute to pathogenesis of ulcerative colitis via regulating NF-kappaB inhibitor IκBα. PLoS One. 2012;7:e52782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 121. | Lin J, Welker NC, Zhao Z, Li Y, Zhang J, Reuss SA, Zhang X, Lee H, Liu Y, Bronner MP. Novel specific microRNA biomarkers in idiopathic inflammatory bowel disease unrelated to disease activity. Mod Pathol. 2014;27:602-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 122. | Coskun M, Bjerrum JT, Seidelin JB, Troelsen JT, Olsen J, Nielsen OH. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J Gastroenterol. 2013;19:4289-4299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 67] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 123. | Chen Y, Xiao Y, Ge W, Zhou K, Wen J, Yan W, Wang Y, Wang B, Qu C, Wu J. miR-200b inhibits TGF-β1-induced epithelial-mesenchymal transition and promotes growth of intestinal epithelial cells. Cell Death Dis. 2013;4:e541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 124. | Koukos G, Polytarchou C, Kaplan JL, Morley-Fletcher A, Gras-Miralles B, Kokkotou E, Baril-Dore M, Pothoulakis C, Winter HS, Iliopoulos D. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842-852.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 191] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 125. | Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis. 2012;6:900-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 199] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 126. | Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, Brant SR, Kwon JH. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17:241-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 191] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 127. | Duttagupta R, DiRienzo S, Jiang R, Bowers J, Gollub J, Kao J, Kearney K, Rudolph D, Dawany NB, Showe MK. Genome-wide maps of circulating miRNA biomarkers for ulcerative colitis. PLoS One. 2012;7:e31241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 128. | Huang Z, Shi T, Zhou Q, Shi S, Zhao R, Shi H, Dong L, Zhang C, Zeng K, Chen J. miR-141 Regulates colonic leukocytic trafficking by targeting CXCL12β during murine colitis and human Crohn’s disease. Gut. 2014;63:1247-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 119] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 129. | Nguyen HT, Dalmasso G, Yan Y, Laroui H, Dahan S, Mayer L, Sitaraman SV, Merlin D. MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem. 2010;285:1479-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 92] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 130. | Lu C, Chen J, Xu HG, Zhou X, He Q, Li YL, Jiang G, Shan Y, Xue B, Zhao RX. MIR106B and MIR93 prevent removal of bacteria from epithelial cells by disrupting ATG16L1-mediated autophagy. Gastroenterology. 2014;146:188-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 131. | Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 458] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 132. | Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16:1729-1738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 239] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 133. | Zahm AM, Thayu M, Hand NJ, Horner A, Leonard MB, Friedman JR. Circulating microRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2011;53:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 134] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 134. | Iborra M, Bernuzzi F, Correale C, Vetrano S, Fiorino G, Beltrán B, Marabita F, Locati M, Spinelli A, Nos P. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin Exp Immunol. 2013;173:250-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 135. | Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 335] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 136. | Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 274] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 137. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1665] [Cited by in RCA: 1702] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 138. | Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 139. | Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1310] [Cited by in RCA: 1551] [Article Influence: 141.0] [Reference Citation Analysis (0)] |
| 140. | Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therap Adv Gastroenterol. 2015;8:4-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |