Published online Feb 7, 2016. doi: 10.3748/wjg.v22.i5.1911
Peer-review started: April 1, 2015
First decision: June 19, 2015
Revised: October 6, 2015
Accepted: November 13, 2015
Article in press: November 13, 2015
Published online: February 7, 2016
Processing time: 295 Days and 17.4 Hours
AIM: To evaluate the relationship between glutathione S-transferase M1 (GSTM1) polymorphism and susceptibility to esophageal cancer (EC).
METHODS: A comprehensive search of the United States National Library of Medicine PubMed database and the Elsevier, Springer, and China National Knowledge Infrastructure databases for all relevant studies was conducted using combinations of the following terms: “glutathione S-transferase M1”, “GSTM1”, “polymorphism”, and “EC” (until November 1, 2014). The statistical analysis was performed using the SAS software (v.9.1.3; SAS Institute, Cary, NC, United States) and the Review Manager software (v.5.0; Oxford, England); crude odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the association between the GSTM1 null genotype and the risk of EC.
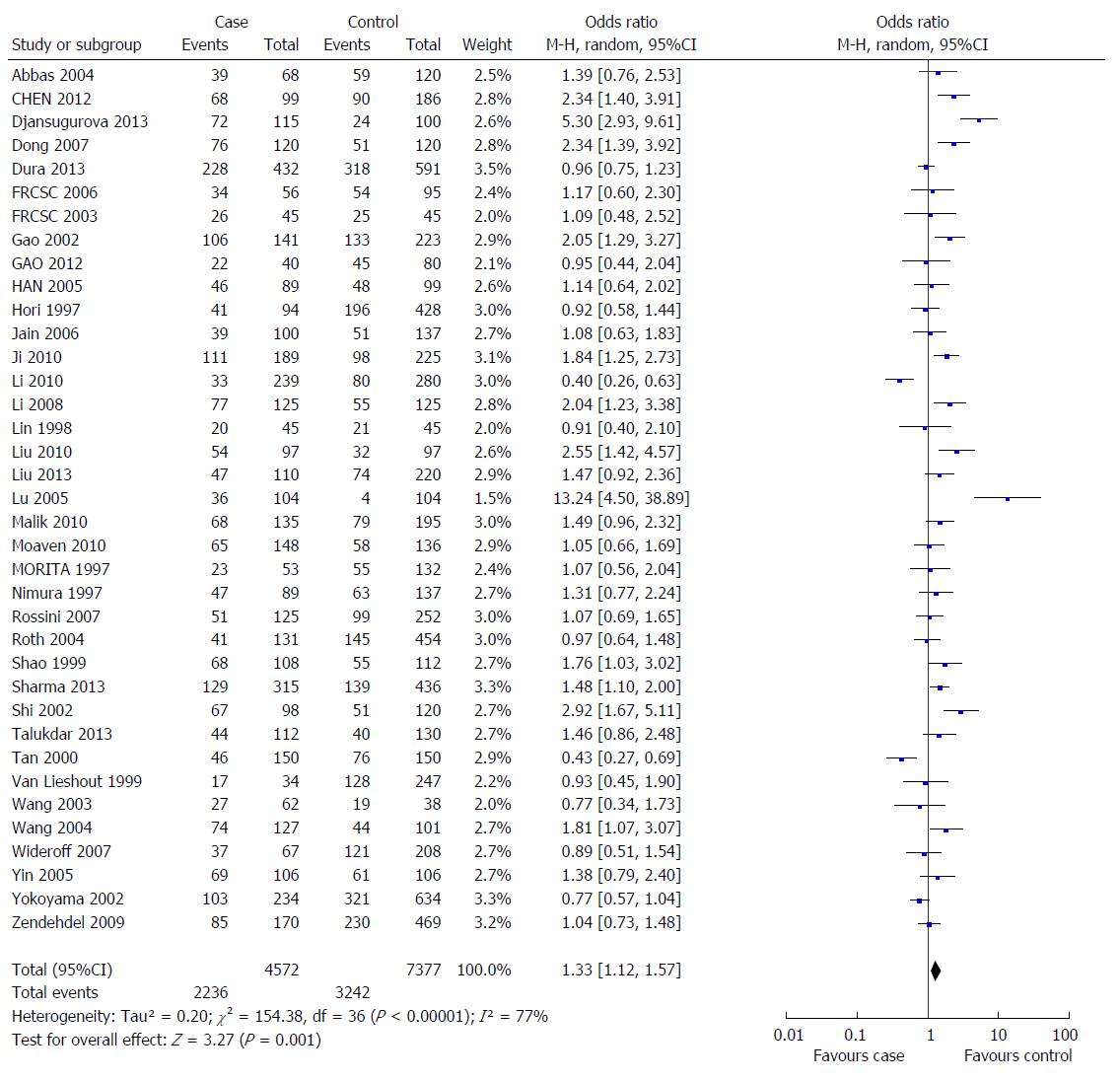
RESULTS: A total of 37 studies involving 2236 EC cases and 3243 controls were included in this meta-analysis. We observed that the GSTM1 null genotype was a significant risk factor for EC in most populations (OR = 1.33, 95%CI: 1.12-1.57, Pheterogeneity < 0.000001, and I2 = 77.0%), particularly in the Asian population (OR = 1.53, 95%CI: 1.26-1.86, Pheterogeneity < 0.000001, and I2 = 77.0%), but not in the Caucasian population (OR = 1.02, 95%CI: 0.87-1.19, Pheterogeneity = 0.97, and I2 = 0%).
CONCLUSION: The GSTM1 null polymorphism may be associated with an increased risk for EC in Asian but not Caucasian populations.
Core tip: Many previous studies have investigated the association between the glutathione S-transferase M1 (GSTM1) null genotype and the risk of esophageal cancer (EC), but these studies have provided controversial findings. The present study represents the largest meta-analysis to estimate the association between the GSTM1 polymorphism and EC risk. We investigated these two genotypes (GSTM1 null or GSTM1 present) in terms of EC morbidity.
- Citation: Lu QJ, Bo YC, Zhao Y, Zhao EJ, Sapa WB, Yao MJ, Duan DD, Zhu YW, Lu WQ, Yuan L. Glutathione S-transferase M1 polymorphism and esophageal cancer risk: An updated meta-analysis based on 37 studies. World J Gastroenterol 2016; 22(5): 1911-1918
- URL: https://www.wjgnet.com/1007-9327/full/v22/i5/1911.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i5.1911
Esophageal cancer (EC), which is the sixth leading cause of cancer-associated death worldwide, has two major histological types: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EADC)[1]. The five-year survival rate for EC is less than 20%[2]. A growing body of epidemiological evidence suggests that environmental factors together with genetic factors play important roles in the risk of developing EC[3,4]. The major risk factors for EC include alcohol consumption, smoking tobacco, and micronutrient deficiency[5]. Various factors and multiple processes lead to EC development. In addition to the above mentioned factors, genetic factors also account for EC cases.
Previous studies have suggested that glutathione S-transferases (GSTs) are phase II metabolizing enzymes that detoxify free radicals and other carcinogens[6]. Therefore, individual variation in phases II enzyme activity may contribute to varying susceptibility to EC progression. The GST family plays an important role in the detoxification of a variety of electrophilic carcinogens through conjugation with glutathione, and there is a widely variable organ distribution of the four classes of GSTs, although all of these display esophageal expression: GSTA (a), GSTM (m), GSTP (p), and GSTT (h)[7,8]. Homozygous deletions of GSTM1 have been associated with the loss of enzymatic activity for the detoxification of carcinogens, which consequently confers a risk for some cancers, such as colorectal, pancreatic, esophageal, and head and neck cancers[9-12]. Therefore, the null genotype of GSTM1 might be associated with an increased risk of EC[13]. Many previous studies have investigated the association between the GSTM1 null genotype and the risk of EC, but these studies have provided controversial findings[8,14-18]. It remains uncertain whether the GSTM1 polymorphism is a risk factor for EC. Considering these controversial results, we conducted a meta-analysis summarizing reported case-control or prospective studies to assess the risk of EC.
We conducted a comprehensive search of the US National Library of Medicine PubMed database and the Elsevier, Springer, and China National Knowledge Infrastructure databases for all relevant studies using combinations of the following terms: “glutathione S-transferase M1”, “GSTM1”, “polymorphism”, and “EC” (until November 1, 2014). Additional eligible studies were identified through references that were cited in the relevant articles. The full text of each potentially relevant paper was scrutinized to ensure that the following inclusion criteria were met: (1) the articles clearly described studies concerning the association of EC with GSTM1 polymorphism; (2) The study design should be observational (case-control or prospective); (3) Sufficient data for estimating the odds ratios (ORs) and 95% confidence intervals (CIs) were present; and (4) If more than one publication reported on the same population, we selected the study with the largest sample size.
Two researchers independently extracted the following data from each study that met the inclusion criteria: first author’s surname, year of publication, country, ethnicity of the subjects (stratified into Asian, Caucasian, and African populations), sources of the controls (categorized as population-based studies and hospital-based studies), histological type (adenocarcinoma and squamous cell carcinoma), number of different genotypes in cases and controls, smoking status, and the frequency of different genotypes in the cases and controls. Individuals with “present” genotype were defined as carriers with at least one of the functional alleles in accordance with the definition used in most studies, whereas individuals carrying none of the alleles were classified as the “null” genotype.
Crude ORs with 95%CIs were used to estimate the strength of the relationship between the GSTM1 polymorphism and EC risk. The pooled ORs were evaluated for null vs present genotypes. The heterogeneity was assessed using a χ2 analysis based on the Q-test[19]. The heterogeneity was considered significant for P < 0.05. In the presence of significant heterogeneity, a random-effects model (the DerSimonian and Laird method)[20] was used to calculate pooled estimates; otherwise, a fixed-effects model (the Mantel-Haenszel method) was used[21]. These two models provided similar results in the absence of heterogeneity. The potential publication bias was assessed using a funnel plot and linear regression asymmetry test[22].The statistical analyses were performed using the SAS (v.9.1.3; SAS Institute, Cary, NC, United States) and Review Manager software (v.5.0; Oxford, England) with two-sided P values and a 0.05 significance level.
A total of 37 studies involving 2236 EC cases and 3243 controls were finally included in this meta-analysis[8,12-18,23-51]. The main characteristics of these studies are presented in Table 1. Among these studies, one case-control study was nested within a cohort study[51], and 25 studies provided data of the histological type of the EC cases. The smoking statuses of the cases and controls were recorded in six studies.
| Ref. | Year | Ethnicity | Country | Source of controls | Genotype distribution | |||
| Case | Control | |||||||
| Null | Present | Null | Present | |||||
| Morita et al[49] | 1997 | Asian | Japan | PB | 23 | 30 | 55 | 77 |
| Nimura et al[43] | 1997 | Asian | China | HB | 47 | 42 | 63 | 74 |
| Hori et al[31] | 1997 | Asian | Japan | PB | 41 | 53 | 196 | 232 |
| Lin et al[27] | 1998 | Asian | China | PB | 20 | 25 | 21 | 24 |
| Shao et al[36] | 1999 | Asian | China | HB | 68 | 40 | 55 | 57 |
| van Lieshout et al[30] | 1999 | Caucasian | The Netherland | PB | 17 | 17 | 128 | 119 |
| Tan et al[14] | 2000 | Asian | China | PB | 46 | 104 | 76 | 74 |
| Shi et al[40] | 2002 | Asian | China | HB | 67 | 31 | 51 | 69 |
| Yokoyama et al[15] | 2002 | Asian | Japan | HB | 103 | 131 | 321 | 313 |
| Gao et al[42] | 2002 | Asian | China | PB | 106 | 35 | 133 | 90 |
| Casson et al[48] | 2003 | Caucasian | Canada | PB | 26 | 19 | 25 | 20 |
| Wang et al[50] | 2003 | Asian | China | PB | 27 | 35 | 19 | 19 |
| Wang et al[44] | 2004 | Asian | China | HB | 74 | 53 | 44 | 57 |
| Abbas et al[33] | 2004 | Caucasian | French | PB | 39 | 29 | 59 | 61 |
| Roth et al[51] | 2004 | Asian | China | Nest | 41 | 90 | 145 | 309 |
| Han et al[39] | 2005 | Asian | China | HB | 46 | 43 | 48 | 51 |
| Lu et al[28] | 2005 | Asian | China | PB | 36 | 68 | 4 | 100 |
| Yin et al[35] | 2005 | Asian | China | HB | 69 | 37 | 61 | 45 |
| Casson et al[45] | 2006 | Caucasian | Canada | HB | 34 | 22 | 54 | 41 |
| Jain et al[32] | 2006 | Asian | India | HB | 39 | 61 | 51 | 86 |
| Dong et al[47] | 2007 | Asian | China | HB | 76 | 44 | 51 | 69 |
| Wideroff et al[41] | 2007 | Caucasian | United States | PB | 37 | 30 | 121 | 87 |
| Rossini et al[29] | 2007 | Caucasian | Brazil | HB | 51 | 74 | 99 | 153 |
| Li et al[34] | 2008 | Asian | China | PB | 77 | 48 | 55 | 70 |
| Zendehdel et al[23] | 2009 | Caucasian | Sweden | PB | 85 | 85 | 230 | 239 |
| Ji et al[38] | 2010 | Asian | China | PB | 111 | 78 | 98 | 127 |
| Malik et al[26] | 2010 | Asian | India | HB | 68 | 67 | 79 | 116 |
| Liu et al[24] | 2010 | Asian | China | PB | 54 | 43 | 32 | 65 |
| Moaven et al[12] | 2010 | Asian | Iran | HB | 65 | 83 | 58 | 78 |
| Li et al[16] | 2010 | Black | Africa | HB | 33 | 206 | 80 | 200 |
| Gao et al[37] | 2012 | Asian | China | HB | 22 | 18 | 45 | 35 |
| Chen et al[17] | 2012 | Asian | China | HB | 68 | 31 | 90 | 96 |
| Liu et al[46] | 2013 | Asian | China | HB | 47 | 63 | 74 | 146 |
| Talukdar et al[25] | 2013 | Asian | India | PB | 44 | 68 | 40 | 90 |
| Sharma et al[13] | 2013 | Asian | India | PB | 129 | 186 | 139 | 297 |
| Dura et al[8] | 2013 | Caucasian | The Netherland | PB | 228 | 204 | 318 | 273 |
| Djansugurova et al[18] | 2013 | Asian | Kazakhstan | PB | 72 | 43 | 24 | 76 |
Considering the obvious heterogeneity among the 37 included studies (P < 0.001, I2 = 77%), the random-effects model (DerSimonian-Laird method) was used to calculate the pooled ORs for the GSTM1 null vs GSTM1 present genotypes. Individuals with GSTM1 null genotypes were significantly associated with an increased risk for EC compared those carrying the GSTM1 present genotype (OR = 1.33, 95%CI: 1.12-1.57, Figure 1). In the sensitivity analysis, individual studies were sequentially removed. The results indicated that no individual study significantly affected the pooled OR, suggesting that these results were statistically robust.
In the subgroup analysis based on ethnicity, a positive correlation was observed between the GSTM1 null genotype and the EC risk in the Asian population (OR = 1.53, 95%CI: 1.26-1.86) but not in the Caucasian population (OR = 1.02, 95%CI: 0.87-11.19). However, the results of the stratified analysis based on histological type showed that the GSTM1 null genotype increased the risk of EC in patients whose histological type was unknown, but no statistically significant association was observed for either the ESCC patients or the EADC patients. Moreover, the heterogeneity was significantly reduced among Caucasian populations and studies based on the histological type of adenocarcinoma. Because only one study (14EADC, 137 ESCC) reported an association between the GSTM1 polymorphism and EADC in Asian populations, we only analyzed the data according to ESCC and EADC in Caucasian populations, and the results showed no statistically significant association between the GSTM1 polymorphism and ESCC or EADC. The main results of this meta-analysis and the heterogeneity test are shown in Table 2.
| Null vs present | ||||
| No. of studies | OR | 95%CI | P value | |
| Total | 37 | 1.33 | 1.12-1.57 | 0.00001 |
| Ethnicity | ||||
| Asian | 27 | 1.53 | 1.26-1.86 | 0.00001 |
| Caucasian | 8 | 1.02 | 0.87-1.19 | 0.97 |
| Histological type | ||||
| ESCC | 22 | 1.15 | 0.91-1.45 | 0.00001 |
| EADC | 8 | 0.98 | 0.81-1.18 | 0.93 |
| NR | 12 | 1.82 | 1.58-2.09 | 0.007 |
| Smoking status | ||||
| Smokers | 6 | 0.97 | 0.53-1.77 | 0.00001 |
| Nonsmokers | 6 | 0.97 | 0.57-1.64 | 0.001 |
| Histological type of Caucasian | ||||
| ESCC | 5 | 1.15 | 0.91-1.45 | 0.33 |
| EADC | 8 | 0.98 | 0.81-1.18 | 0.93 |
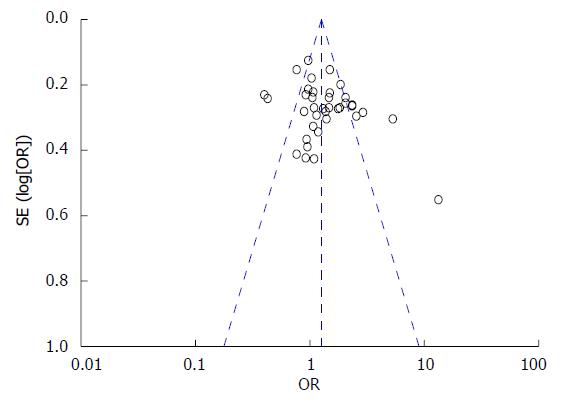
A funnel plot was used to graphically estimate the publication bias of the literature. As shown in Figure 2, the shape of the funnel plot was symmetrical in the overall population, suggesting the absence of publication bias. The results of Egger’s test showed statistical evidence for funnel plot symmetry (t = 1.76, P = 0.0873).
GSTM1 is a member of the family of cytosolic GSTs, which are phase II xenobiotic-metabolizing enzymes. These enzymes play a crucial role in the detoxification and elimination of electrophilic carcinogens through conjugation with glutathione[12]. Many studies have investigated the association between the GSTM1 null genotype and various types of cancer, such as colorectal carcinoma, lung cancer, liver cancer, and EC, but the findings are controversial, particularly those obtained for EC[52,53]. The results of this meta-analysis showed that the GSTM1 null genotype is significantly associated with an increased risk of EC in the overall population. Furthermore, in the subgroup analysis by ethnicity, we detected a significant association between the GSTM1 polymorphism and EC risk in Asians but not in Caucasians, suggesting that the GSTM1 null polymorphism might contribute to increased susceptibility to EC in Asians. Similar results have been obtained in several previous meta-analyses[54,55]. However, other studies have shown conflicting results. A pooled analysis of 20 studies from the Archives of Medical Research revealed that there was no evidence of increased risk of EC associated with the GSTM1 null genotype[56]. The result might reflect a relatively small sample size and, to a lesser extent, different ethnicities, different histological types and the source of the controls.
In the present meta-analysis, most of the included studies concerned Asian populations. This phenomenon might be attributed to the occurrence of EC, which displays a remarkable geographical difference. Specifically, the ‘‘EC belt’’, which stretches from North Central China westward through Central Asia and northern Iran, exhibits a particularly high EC incidence in Asian populations[57], which explains why many of the studies were conducted in Asian countries.
In the subgroup analysis based on histological type, no significant association was detected between the GSTM1 polymorphism and ESCC or EADC risk, indicating that histological type might affect the statistical correlation between the GSTM1 polymorphism and EC. Similar results have been reported in previous studies[23,55,58], indicating that further clarification of the histological type might avoid the interference of some confounding factors.
Several potential limitations of the present meta-analysis should also be acknowledged. Only one of the included studies was conducted in Africa, and it did not provide sufficient data for the subgroup analysis based on ethnicity. Therefore, we could not include the African population in the subgroup analysis based on ethnicity. Moreover, only published studies were included in the present meta-analysis, which might have biased the results.
In conclusion, this meta-analysis demonstrated that the GSTM1 null polymorphism might be associated with an increased risk for EC in Asian populations but not in Caucasian populations. Larger well-designed epidemiological studies are warranted to verify these findings.
Esophageal cancer (EC), which is the sixth leading cause of cancer-associated death worldwide, has two major histological types: esophageal squamous cell carcinoma and esophageal adenocarcinoma. The five-year survival rate for EC is less than 20%. Previous studies have suggested that glutathione S-transferase (GSTs) are phase II metabolizing enzymes that detoxify free radicals and other carcinogens. Therefore, individuals with low phase II activity might have a higher risk of developing cancer. The GST family plays an important role in the detoxification of a variety of electrophilic carcinogens through conjugation with glutathione, and there is a widely variable organ distribution of the four classes of GSTs, namely, GSTA (a), GSTM (m), GSTP (p), and GSTT (h), although all show esophageal expression.
A growing body of epidemiological evidence suggests that environmental factors together with genetic factors play important roles in the risk of developing esophageal carcinoma: alcohol consumption, smoking tobacco, and micronutrient deficiency are considered the major risk factors for EC. The GSTM1 null genotype has been associated with an increased risk of EC. Many previous studies have investigated the association between the GSTM1 null genotype and the risk of esophageal carcinoma, but these studies provide controversial findings.
The results of the present study indicated that the GSTM1 null polymorphism might be associated with an increased risk of EC in Asian populations but not in Caucasian populations, which would be helpful for the identification of individuals at an increased risk of developing EC.
The present study enhances the current understanding of the effects of GSTM1 on EC. Larger well-designed epidemiological studies are warranted to confirm the precise mechanism underlying the involvement of the GSTM1 gene in EC progression.
GSTM1 is a primary member of the GST family, which comprises enzymes that play important roles in the detoxification of a variety of electrophilic carcinogens through conjugation with glutathione. Homozygous deletions of GSTM1 might disrupt enzymatic detoxification of carcinogens and consequently confer risk for some cancers, such as colorectal, pancreatic, esophageal, and head and neck cancers.
The present study analyzed the effect of the GST1 polymorphism on EC risk. The meta-analysis of 37 studies showed that the GSTM1 null polymorphism is associated with a significantly increased risk of EC.
P- Reviewer: Corrales FJ, Ghiorzo P, Nagahara H S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Cai Y, Wang J. Significant association of glutathione S-transferase T1 null genotype with esophageal cancer risk: a meta-analysis. Mol Biol Rep. 2013;40:2397-2403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25543] [Article Influence: 1824.5] [Reference Citation Analysis (7)] |
| 3. | Denlinger CE, Thompson RK. Molecular basis of esophageal cancer development and progression. Surg Clin North Am. 2012;92:1089-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Zheng S, Vuitton L, Sheyhidin I, Vuitton DA, Zhang Y, Lu X. Northwestern China: a place to learn more on oesophageal cancer. Part two: gene alterations and polymorphisms. Eur J Gastroenterol Hepatol. 2011;23:1087-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 2009;24:729-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 271] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 6. | Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2409] [Cited by in RCA: 2419] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 7. | Di Pietro G, Magno LA, Rios-Santos F. Glutathione S-transferases: an overview in cancer research. Expert Opin Drug Metab Toxicol. 2010;6:153-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 157] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 8. | Dura P, Salomon J, Te Morsche RH, Roelofs HM, Kristinsson JO, Wobbes T, Witteman BJ, Tan AC, Drenth JP, Peters WH. No role for glutathione S-transferase genotypes in Caucasian esophageal squamous cell or adenocarcinoma etiology: an European case-control study. BMC Gastroenterol. 2013;13:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Duell EJ, Holly EA, Bracci PM, Wiencke JK, Kelsey KT. A population-based study of the Arg399Gln polymorphism in X-ray repair cross- complementing group 1 (XRCC1) and risk of pancreatic adenocarcinoma. Cancer Res. 2002;62:4630-4636. [PubMed] |
| 10. | Economopoulos KP, Sergentanis TN. GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: a comprehensive meta-analysis. Eur J Cancer. 2010;46:1617-1631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 104] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 11. | Lourenço GJ, Silva EF, Rinck-Junior JA, Chone CT, Lima CS. CYP1A1, GSTM1 and GSTT1 polymorphisms, tobacco and alcohol status and risk of head and neck squamous cell carcinoma. Tumour Biol. 2011;32:1209-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Moaven O, Raziee HR, Sima HR, Ganji A, Malekzadeh R, A’rabi A, Abdollahi A, Memar B, Sotoudeh M, Naseh H. Interactions between Glutathione-S-Transferase M1, T1 and P1 polymorphisms and smoking, and increased susceptibility to esophageal squamous cell carcinoma. Cancer Epidemiol. 2010;34:285-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Sharma A, Das BC, Sehgal A, Mehrotra R, Kar P, Sardana S, Phukan R, Mahanta J, Purkayastha J, Saxena S. GSTM1 and GSTT1 polymorphism and susceptibility to esophageal cancer in high- and low-risk regions of India. Tumour Biol. 2013;34:3249-3257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Tan W, Song N, Wang GQ, Liu Q, Tang HJ, Kadlubar FF, Lin DX. Impact of genetic polymorphisms in cytochrome P450 2E1 and glutathione S-transferases M1, T1, and P1 on susceptibility to esophageal cancer among high-risk individuals in China. Cancer Epidemiol Biomarkers Prev. 2000;9:551-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Yokoyama A, Kato H, Yokoyama T, Tsujinaka T, Muto M, Omori T, Haneda T, Kumagai Y, Igaki H, Yokoyama M. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis. 2002;23:1851-1859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 169] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 16. | Li D, Dandara C, Parker MI. The 341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genet. 2010;11:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 17. | Chen Y, Zhang H, Yin D, Wang H, Wang Y, Deng Y, Ma Y. Relationship between GSTM1 gene polymorphism and interaction of gene environment and susceptibility of esophageal cancer. Xiandai Yufang Yixue. 2012;39:110-113. |
| 18. | Djansugurova LB, Perfilyeva AV, Zhunusova GS, Djantaeva KB, Iksan OA, Khussainova EM. The determination of genetic markers of age-related cancer pathologies in populations from Kazakhstan. Front Genet. 2013;4:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1858] [Cited by in RCA: 1996] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 20. | DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26739] [Cited by in RCA: 30428] [Article Influence: 780.2] [Reference Citation Analysis (0)] |
| 21. | Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719-748. [PubMed] |
| 22. | Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34245] [Cited by in RCA: 40566] [Article Influence: 1448.8] [Reference Citation Analysis (2)] |
| 23. | Zendehdel K, Bahmanyar S, McCarthy S, Nyren O, Andersson B, Ye W. Genetic polymorphisms of glutathione S-transferase genes GSTP1, GSTM1, and GSTT1 and risk of esophageal and gastric cardia cancers. Cancer Causes Control. 2009;20:2031-2038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Liu R, Yin L, Pu Y, Li Y, Liang G, Zhang J, Li X. Functional alterations in the glutathione S-transferase family associated with enhanced occurrence of esophageal carcinoma in China. J Toxicol Environ Health A. 2010;73:471-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Talukdar FR, Ghosh SK, Laskar RS, Mondal R. Epigenetic, genetic and environmental interactions in esophageal squamous cell carcinoma from northeast India. PLoS One. 2013;8:e60996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 26. | Malik MA, Upadhyay R, Mittal RD, Zargar SA, Mittal B. Association of xenobiotic metabolizing enzymes genetic polymorphisms with esophageal cancer in Kashmir Valley and influence of environmental factors. Nutr Cancer. 2010;62:734-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | Lin DX, Tang YM, Peng Q, Lu SX, Ambrosone CB, Kadlubar FF. Susceptibility to esophageal cancer and genetic polymorphisms in glutathione S-transferases T1, P1, and M1 and cytochrome P450 2E1. Cancer Epidemiol Biomarkers Prev. 1998;7:1013-1018. [PubMed] |
| 28. | Lu XM, Zhang YM, Lin RY, Arzi G, Wang X, Zhang YL, Zhang Y, Wang Y, Wen H. Relationship between genetic polymorphisms of metabolizing enzymes CYP2E1, GSTM1 and Kazakh’s esophageal squamous cell cancer in Xinjiang, China. World J Gastroenterol. 2005;11:3651-3654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 29. | Rossini A, Rapozo DC, Soares Lima SC, Guimarães DP, Ferreira MA, Teixeira R, Kruel CD, Barros SG, Andreollo NA, Acatauassú R. Polymorphisms of GSTP1 and GSTT1, but not of CYP2A6, CYP2E1 or GSTM1, modify the risk for esophageal cancer in a western population. Carcinogenesis. 2007;28:2537-2542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | van Lieshout EM, Roelofs HM, Dekker S, Mulder CJ, Wobbes T, Jansen JB, Peters WH. Polymorphic expression of the glutathione S-transferase P1 gene and its susceptibility to Barrett’s esophagus and esophageal carcinoma. Cancer Res. 1999;59:586-589. [PubMed] |
| 31. | Hori H, Kawano T, Endo M, Yuasa Y. Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and human esophageal squamous cell carcinoma susceptibility. J Clin Gastroenterol. 1997;25:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 83] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Jain M, Kumar S, Rastogi N, Lal P, Ghoshal UC, Tiwari A, Pant MC, Baiq MQ, Mittal B. GSTT1, GSTM1 and GSTP1 genetic polymorphisms and interaction with tobacco, alcohol and occupational exposure in esophageal cancer patients from North India. Cancer Lett. 2006;242:60-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Abbas A, Delvinquiere K, Lechevrel M, Lebailly P, Gauduchon P, Launoy G, Sichel F. GSTM1, GSTT1, GSTP1 and CYP1A1 genetic polymorphisms and susceptibility to esophageal cancer in a French population: different pattern of squamous cell carcinoma and adenocarcinoma. World J Gastroenterol. 2004;10:3389-3393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 67] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 34. | Li Y, Zhu W, Lin Z, Wu H, Ye Z. Correlation between smoking and the polymorphism of gene GSTM1 and esophageal carcinoma. Heilongjiang Yixue Zazhi. 2008;32:18-20. |
| 35. | Yin L, Pu Y, Zhu Z, Hu X, Liu Y, Kai H. Polymorphisms of susceptible genes for esophageal cancer risk in Huaian population in Jiangsu Province. Tumor. 2005;25:357-367. |
| 36. | Shao G, Hu Z, Li E, Li J, Wen B. Relationship between the GSTM1 genetic polymorphism and susceptibility to squamous cell carcinoma of esophagus. Shantou Daxue Yixueyuan Xuebao. 1999;12:1-2. |
| 37. | Gao P, Tian Y, Ye X, Ge J, Zhang D, Xu W. Study of CTPIA1, GSTT1, GSTM1 polymorphisms and susceptibility on esophageal carcinoma in Ningxia Hui nationality. Ningxia Yixue Zazhi. 2012;34:196-199. |
| 38. | Ji R, Wu Jg, ZHou Y, ZHANG B, ZHANG Z, Yang Z. Relationship between CYP1A1, GSTM1 and GSTT1 genetic polymorphisms and susceptibility of esophageal cancer in Wuwei, Gansu Province. Lanzhou Daxue Xuebao (Yixueban). 2010;36:29-34. |
| 39. | Han Y, Feng X, Li P, Niu Z. Case-control study of relationship of CYP1A1 and GSTM1 polymorphisms and susceptibility to esophageal squamous carcinoma. Zhongguo Gonggong Weisheng. 2005;21:3-6. |
| 40. | Shi Y, Zhou X, Zhou Y, Ren S. Analysis of CYP2E1, GSTM1 genetic polymorphisms in relation to human lung cancer and esophageal carcinoma. Huazhong Keji Daxue Xuebao. 2002;31:14-17. |
| 41. | Wideroff L, Vaughan TL, Farin FM, Gammon MD, Risch H, Stanford JL, Chow WH. GST, NAT1, CYP1A1 polymorphisms and risk of esophageal and gastric adenocarcinomas. Cancer Detect Prev. 2007;31:233-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 42. | Gao CM, Takezaki T, Wu JZ, Li ZY, Liu YT, Li SP, Ding JH, Su P, Hu X, Xu TL. Glutathione-S-transferases M1 (GSTM1) and GSTT1 genotype, smoking, consumption of alcohol and tea and risk of esophageal and stomach cancers: a case-control study of a high-incidence area in Jiangsu Province, China. Cancer Lett. 2002;188:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 84] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 43. | Nimura Y, Yokoyama S, Fujimori M, Aoki T, Adachi W, Nasu T, He M, Ping YM, Iida F. Genotyping of the CYP1A1 and GSTM1 genes in esophageal carcinoma patients with special reference to smoking. Cancer. 1997;80:852-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 44. | Wang AH, Sun CS, Li LS, Huang JY, Chen QS, Xu DZ. Genetic susceptibility and environmental factors of esophageal cancer in Xi’an. World J Gastroenterol. 2004;10:940-944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 19] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 45. | Casson AG, Zheng Z, Porter GA, Guernsey DL. Genetic polymorphisms of microsomal epoxide hydroxylase and glutathione S-transferases M1, T1 and P1, interactions with smoking, and risk for esophageal (Barrett) adenocarcinoma. Cancer Detect Prev. 2006;30:423-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 46. | Liu L. The relationship between CYPIA1, GSTT1, GSTM1, GSTP1 genetic Polymorphisms and susceptibility of Ningxia Han People esophageal carcinoma. Ningxia Medical University. 2013. Available from: http://www.cnki.net/KCMS/detail/detail.aspx?QueryID=0&CurRec=1&recid=&filename=1013046360.nh&dbname=CMFD201401&dbcode=CMFD&pr=&urlid=&yx=&v=MDYzMzRxVHJXTTFGckNVUkwrZVorVnZGeW5rVkwzUFZGMjZIYk84R05MS3I1RWJQSVI4ZVgxTHV4WVM3RGgxVDM=. |
| 47. | Dong CX, Wu J, Jin Y, Zhang J. Correlation between genetic polymorphism of CYP2E1, GSTM1 and esophageal cancer in Gansu. Weichangbingxue and Ganbingxue Zazhi. 2007;16:115-118. |
| 48. | Casson AG, Zheng Z, Chiasson D, MacDonald K, Riddell DC, Guernsey JR, Guernsey DL, McLaughlin J. Associations between genetic polymorphisms of Phase I and II metabolizing enzymes, p53 and susceptibility to esophageal adenocarcinoma. Cancer Detect Prev. 2003;27:139-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 49. | Morita S, Yano M, Shiozaki H, Tsujinaka T, Ebisui C, Morimoto T, Kishibuti M, Fujita J, Ogawa A, Taniguchi M. CYP1A1, CYP2E1 and GSTM1 polymorphisms are not associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 1997;71:192-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 50. | Wang LD, Zheng S, Liu B, Zhou JX, Li YJ, Li JX. CYP1A1, GSTs and mEH polymorphisms and susceptibility to esophageal carcinoma: study of population from a high- incidence area in north China. World J Gastroenterol. 2003;9:1394-1397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 51] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Roth MJ, Abnet CC, Johnson LL, Mark SD, Dong ZW, Taylor PR, Dawsey SM, Qiao YL. Polymorphic variation of Cyp1A1 is associated with the risk of gastric cardia cancer: a prospective case-cohort study of cytochrome P-450 1A1 and GST enzymes. Cancer Causes Control. 2004;15:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 52. | Darazy M, Balbaa M, Mugharbil A, Saeed H, Sidani H, Abdel-Razzak Z. CYP1A1, CYP2E1, and GSTM1 gene polymorphisms and susceptibility to colorectal and gastric cancer among Lebanese. Genet Test Mol Biomarkers. 2011;15:423-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 53. | Yadav DS, Devi TR, Ihsan R, Mishra AK, Kaushal M, Chauhan PS, Bagadi SA, Sharma J, Zamoawia E, Verma Y. Polymorphisms of glutathione-S-transferase genes and the risk of aerodigestive tract cancers in the Northeast Indian population. Genet Test Mol Biomarkers. 2010;14:715-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 54. | Zhong S, Zhao W, Lu C, Li B, Yuan Y, Guo D, Chang Z, Jiao B, Yang L. Glutathione S-transferase M1 null genotype contributes to increased risk of esophageal carcinoma in Chinese population. Tumour Biol. 2013;34:2403-2407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 55. | Zhang C, Chai Y, Wang P, Yun Y, Dai L, Wang K, Zhang J. Meta-analysis on glutathione S-transferase M1 polymorphisms and the risk of esophageal cancer. Zhongguo Gonggong Weisheng. 2011;27:241-243. |
| 56. | Zhuo WL, Zhang YS, Wang Y, Zhuo XL, Zhu B, Cai L, Chen ZT. Association studies of CYP1A1 and GSTM1 polymorphisms with esophageal cancer risk: evidence-based meta-analyses. Arch Med Res. 2009;40:169-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 57. | Akbari MR, Malekzadeh R, Nasrollahzadeh D, Amanian D, Sun P, Islami F, Sotoudeh M, Semnani S, Boffeta P, Dawsey SM. Familial risks of esophageal cancer among the Turkmen population of the Caspian littoral of Iran. Int J Cancer. 2006;119:1047-1051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Bull LM, White DL, Bray M, Nurgalieva Z, El-Serag HB. Phase I and II enzyme polymorphisms as risk factors for Barrett’s esophagus and esophageal adenocarcinoma: a systematic review and meta-analysis. Dis Esophagus. 2009;22:571-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |