Published online Feb 7, 2016. doi: 10.3748/wjg.v22.i5.1787
Peer-review started: October 8, 2015
First decision: November 5, 2015
Revised: November 12, 2015
Accepted: December 21, 2015
Article in press: December 21, 2015
Published online: February 7, 2016
Processing time: 107 Days and 6.4 Hours
Gallbladder cancer (GBC) is infrequent but most lethal biliary tract malignancy characterized by an advanced stage diagnosis and poor survival rates attributed to absence of specific symptoms and effective treatment options. These necessitate development of early prognostic/predictive markers and novel therapeutic interventions. MicroRNAs (miRNAs) are small, non-coding RNA molecules that play a key role in tumor biology by functioning like tumor suppressor- or onco- genes and their aberrant expression are associated with the pathogenesis of several neoplasms with overwhelming clinical implications. Since miRNA signature is tissue specific, here, we focused on current data concerning the miRNAs abberations in GBC pathogenesis. In GBC, miRNAs with tumor suppressor activity (miR-135-5p, miR-335, miR-34a, miR-26a, miR-146b-5p, Mir-218-5p, miR-1, miR-145, mir-130a) were found downregulated, while those with oncogenic property (miR-20a, miR-182, mir-155) were upregulated. The expression profile of miRNAs was significantly associated with GBC prognosis and prediction, and forced over-expression/ inhibition of these miRNAs was shown to affect tumor growth and development. Further, differential expression of miRNAs in the blood samples of GBC patients suggest miRNAs as promising noninvasive biomarker. Thus, miRNAs represent potential candidate for GBC management, though many hurdles need to be overcome before miRNAs therapy can be clinically applied to GBC prevention and treatment.
Core tip: Emerging evidences have shown a clear link between microRNAs (miRNAs) expression profile and carcinogenesis. In addition, miRNA has been shown a promising biomarker with devastating clinical implications in various cancer. Recently, several studies have investigated miRNA signature or dysregulation in gallbladder cancer (GBC) pathogenesis. In this review, we aimed to amalgamate the available data to predict the clinical significance of miRNA aberration in GBC. Our findings suggested miRNAs as a promising biomarker and therapeutic tool for GBC management, however, there is a long way to go.
- Citation: Chandra V, Kim JJ, Mittal B, Rai R. MicroRNA aberrations: An emerging field for gallbladder cancer management. World J Gastroenterol 2016; 22(5): 1787-1799
- URL: https://www.wjgnet.com/1007-9327/full/v22/i5/1787.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i5.1787
Although it is rare, gallbladder cancer (GBC) is a highly aggressive and fatal disease with wide geographical, ethnic, and gender specific variations in its incidence[1,2]. Since there are no specific signs, symptoms or reliable sensitive markers, GBC is usually diagnosed at advance stages and the outcome is dismal[3,4]. Neither radiation nor conventional chemotherapy have been shown to significantly improve survival or quality of life for GBC patients. The 5-year survival rate is 32% in patients with lesions confined to the gallbladder mucosa, while there is only a 10% one year survival rate for patients with more advanced lesions[5]. Early diagnosis can have a significant curative impact with surgery as the only therapeutic option[6]. However, our knowledge regarding the molecular pathobiology of GBC is rather limited owing to the scarcity of published studies. Thus, exhaustive efforts are needed to identify authentic tumor markers that will facilitate early detection of GBC and add to better therapeutic options.
MicroRNAs (miRNAs) are short (18-25 nucleotides in length) noncoding RNAs that function as master regulators of gene expression regulating nearly 60% of human genes[7,8]. To date, more than 10000 miRNAs have been identified, and shown to regulate various biological processes and numerous developmental and physiological processes[9-11]. Further, mis-expressed miRNAs have also been associated with several pathological conditions[12,13].
MiRNAs are now well established to promote cancer progression by functioning as either tumor suppressors or oncogenes collectively known as “oncomirs”[14]. Oncogenic miRNAs act directly on transcripts with pro-apoptotic or anti-proliferative roles[15]. Conversely, tumor-suppressor miRNAs repress the expression of oncogenes and/or genes that control cell differentiation or apoptosis[16]. Numerous studies have looked for the association of miRNA aberrantions with cancer progression and its prognostic implications[17-19] suggesting circulating and tissue miRNA profiles as superior diagnostic and prognostic biomarkers and therapeutic targets for cancer[20-22]. In addition, tumour-based miRNA signatures were suggested to identify tissue of origin of cancer[23,24]. However, information regarding the role of miRNAs in GBC and systematic evaluations of miRNA panels in GBC is limited. Therefore, in the present review, we aimed to describe the role of miRNA deregulation in tumorigenesis, specifically in GBC to unravel its role in GBC pathophysiology and potential for use in early detection and prognosis.
MiRNAs are endogenous RNA molecules that are ubiquitous in animals, plants, and viruses but, expressed in a tissue-specific manner[25]. These functions mainly as negative regulators of post-transcriptional gene expression by binding to the 3’ untranslated region (UTR) of target mRNAs, thereby either transcriptionally destabilizing/degrading or transnationally inhibiting them (or both) depending on the degree of miRNA-mRNA complementarity[26,27]. Since miRNAs bind to target mRNAs through imperfect pairing, a single miRNA often regulates the expression of multiple genes[28]. Moreover, one gene can be targeted by multiple miRNAs[29].
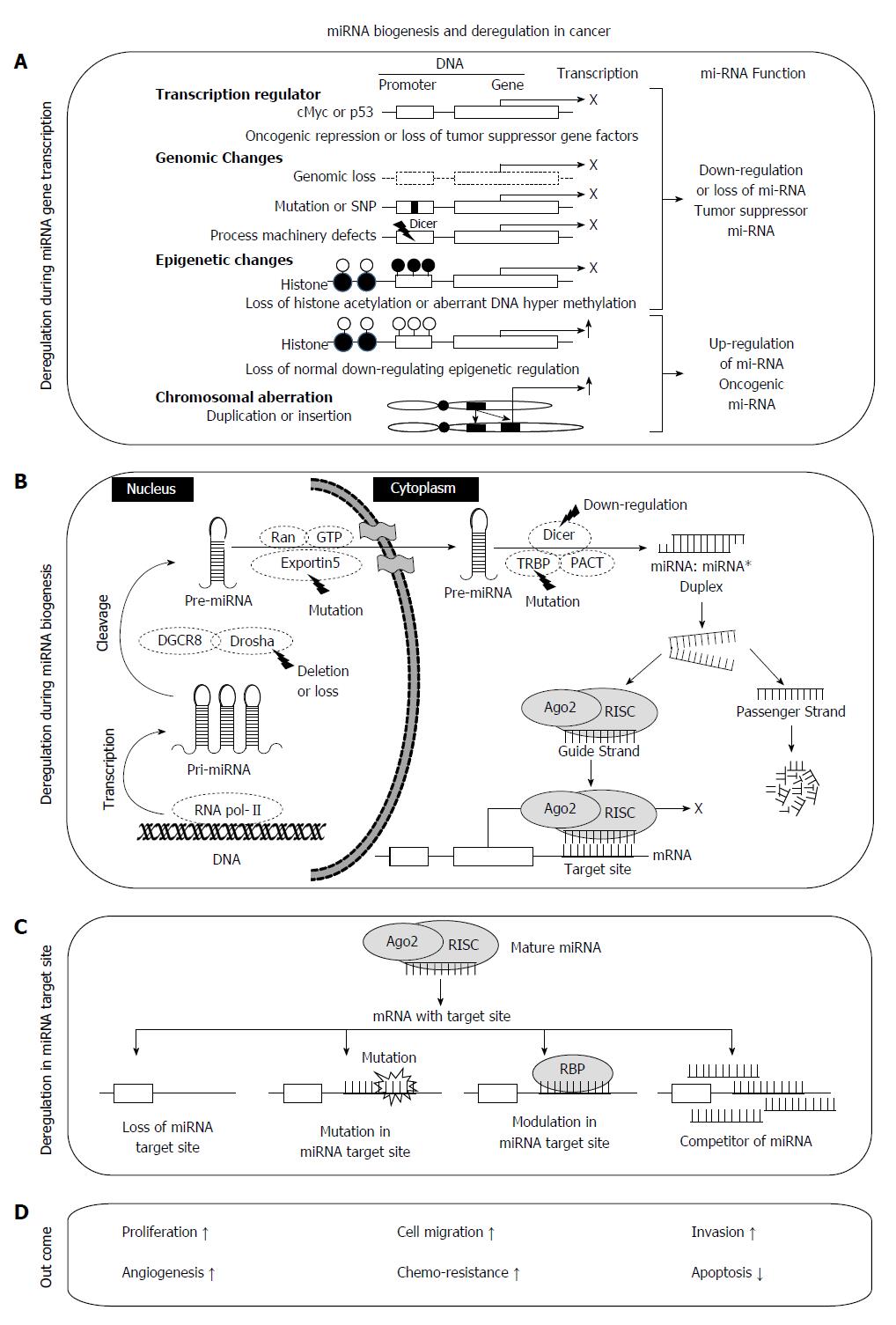
MiRNA biogenesis involves three main steps, transcription, nuclear processing, and nuclear export to the cytoplasm (Figure 1). In the nucleus, RNA polymerase II transcribes a precursor of miRNA (pri-miRNAs) having a stable stem-loop[30] which is cleaved by microprocessor complex (Drosha and Pasha) releasing a hairpin-structured pre-miRNA (60-100nt)[31,32]. Pre-miRNA is exported to the cytoplasm by exportin-5 (XPO5) and subsequently cleaved by Dicer-TRBP (TAR RNA-binding protein)-PACT (or PRKRA) complex producing 20-24 nt miRNA duplexes. These miRNA duplexes, known as miRNA:miRNA*, become associated with argonaute (Ago) proteins, which are central to RNA-induced silencing complex (RISC) function[33,34]. Next, RISC is released and degrades one strand (passenger strand) from miRNA:miRNA* duplex, while another remains associated with RISC in a complex known as mature miRNA (or guide strand miRNA), which interacts and regulate the target genes[35].
The mechanism of miRNA regulation is finely controlled by various nuclear and cytoplasmic factors under three main steps, transcription, biogenesis and binding at target sites. Transcription of miRNA is under the control of various transcription regulators (p53, E2F, or cMyc) with oncogenic or tumor suppressor functions. Epigenetic modifications at the promoter regions of specific miRNA coding genes and genomic changes including chromosomal rearrangements and mutation or SNP may also lead to miRNA aberrations. Biogenesis and maturation of miRNA occurs under the control of various enzymes or co-regulators such as exportin 5, Drosha, Dicer, TRBP, Ago2 and RISC. However, they are frequently deregulated under various pathological conditions. For example partial deletion of Drosha and/or Dicer1[36,37] and mutation in TRBP or Exportin[38,39] leads to aberrant expression of miRNAs in different cancers[36,40,41].
MiRNA deregulation has been well established in most cancer types[42,43]. Various studies have suggested that miRNA expression is commonly down-regulated in human tumors, while some studies also reported miRNA over-expression in cancer[44]. In general, miRNAs are involved in transcriptional regulation of important genes that control key signaling pathways involved in tumorigenesis and tumor development such as apoptosis, cellular proliferation, angiogenesis and regulation of the microenvironment. Here, we discuss the roles of miRNAs in regulation of common steps in carcinogenesis.
MiRNAs control cell cycle directly by targeting cell cycle regulatory genes and indirectly by targeting various signaling pathways[45-48]. Cyclins (cyclin A, B, D, E) and cyclin dependent kinases (CDK 2, 4, 6) promote the cell cycle with subsequent inactivation of Rb protein and activation of E2F transcription factor[49-51]. Conversely, the INK4 (p16, p15, p18 and p19) and Cip/Kip (p21, p27, and p57) families suppress the cell cycle by activating CDK inhibitor[52-55]. These entire genes are well known targets of miRNAs (Supplementary Table 1).
Tumor suppressive miRNAs inhibit the cell cycle through repression of a wide spectrum of positive regulators[56,57]; however, oncogenic miRNAs such as miR-17-92, promote cell cycle progression by targeting its negative regulators[58]. Upregulation of miR-221/222 and miR-21 has been shown to promote G1/S transition[59], while over-expression of miR-16 and mir-34 family miRNAs may lead to G0/G1 arrest[60]. Further, alteration of miRNA levels is associated with cell cycle dysregulation contributing to carcinogenesis[61]. The let-7 and miR-15 families which are major tumor-suppressor miRNAs are frequently lost or downregulated in various cancers[46,62]. Additionally, the mir-17-92 and mir-221/222 are often upregulated in cancer inducing cell proliferation through activation of the cell cycle and Akt pathway[63,64]. Further, recent studies have demonstrated a complex interaction between miRNAs and several transcription factors (p53, cMYC) governing the cell-cycle[65,66].
Diminished apoptosis is considered a hallmark of cancer progression[67]. Apoptosis may occur via either an intrinsic or extrinsic pathway. miRNAs dysregulation has been shown to control both the extrinsic and intrinsic apoptotic pathway, especially in cancer cells, by controlling the expression of pro-apoptotic and anti-apoptotic protein (Supplementary Table 2). In general, pro-apoptotic miRNAs (mir-15, mir-16, let-7f, mir-34, mir-1, mir-101, mir-29) target anti-apoptotic genes or negative regulators of apoptosis while anti-apoptotic miRNAs (mir-21, mir-133, mir-17-92, mir-206, mir-143, mir-145, mir-155, mir-221/222) target pro-apoptotic genes or positive regulators[68-70]. In cancer cells, the expression of pro-apoptotic miRNAs were demonstrated to decrease, while several anti-apoptotic miRNAs were frequently up-regulated, thereby inhibiting apoptosis[63,71].
P53, an important player of apoptosis, is negatively regulated by mir-125b and mir-380-5p while miR-29 family members were identified as positive regulators by targeting upstream CDC42 and p85α[72]. Further, miR-24 modulates XIAP expression level, while mir-203 and mir-218 regulate survivin expression and consequently regulate the apoptosis threshold in cancer cells[73-75]. In addition, mir-10a, let-7a, mir-144, mir-133, mir-24a and mir-155 were shown to affect caspases activation resulting diminished apoptosis[70,76].
MiRNAs, collectively termed as “metastamir”, play significant roles in metastasis by regulating the expression of different genes involved in various steps of metastasis such as EMT, cancer cell detachment, invasion and migration (Supplementary Table 3)[77-80]. mir-200f and mir-203 are well-known epithelial markers that are associated with suppression of EMT and metastasis when over-expressed by targeting ZEB1/2 and Snail1/2 expression[81,82]. mir-221/222, mir-103/107, mir-27, mir-9, mir-155, mir-81a and mir216a/217 are EMT inducer while, mir-30a, mir-34a/b/c, mir-124, mir-203, mir-145, mir-204/211, mir-138, mir-215, mir-708 and mir-205 are EMT inhibitors[83]. Further, miR-143, miR-29b, miR-206, mir-340, miR-218, mir-491-5p, miR-338-3p, let-7, miR-31, mir-21, mir-181 and mir-22/222 regulate extracellular matrix remodeling through modulation of matrix metallo-proteinases in cancer[84].
MiR-10b, mir-21, mir-520c and mir-373 were reported as pro-metastatic miRNAs[85] while let-7, mir-126, mir-335, mir-206 and mir-31 were found to be anti-metastatic miRNAs[86]. Down-regulation of the miR-200f, miR-148a miR-148b and miR-9 family, and upregulation miR-210 is believed to be a metastasis-specific feature[78].
Angiogenesis is mediated by cross-talk between pro and anti-angiogenic signaling pathways[87]. Downregulation/depletion of the Dicer and Drosha were shown to impair angiogenesis demonstrating the importance of miRNA in angiogenesis[88]. Several angiomirs targeting angiogenesis have been also identified (Supplementary Table 4)[89]. Specifically, miR-17-92 cluster, miR-27b, miR-126, miR-130a, miR-210, miR-296, mir-21, mir-31, let-7f and mir-378 have pro-angiogenic function and promote tumor angiogenesis, while miR-221 /miR-222, miR-320, mir-26a, miR-15, miR-16, miR-20a and miR-20b are anti-angiomiRs[90-93].
MiRNAs have been also shown to regulate endothelial cell (EC) function, vascular development, physiology and disease[90,94-97]. ECs demonstrated high expression of miR-21, let-7f, miRNA-23-24 cluster, mir-15b, mir-16, mir-100, miR-126, miR-221/222 and miR-17-92 cluster[92,98]. miR-126 was suggested to be an EC specific miRNA that promotes angiogenesis response to VEGF and bFGF[99].
Metabolic reprogramming constitutes the unique biochemical characteristic and the very origin of cancer[100]. Recently, miRNAs were established to serve as master supervisors of energy metabolism such as carbohydrate, lipid, insulin, protein and nucleic acid metabolism, directly by influencing the metabolic machinery (transporters), or indirectly by modulating the expression of metabolic enzymes/kinases or production of certain metabolites via targeting the genes encoding them (Supplementary Table 5)[101-103]. The miRNA regulation of carbohydrate metabolism involving glucose transporters (GLUTs) and the key enzymes (HKs, GAPDH, PFK1 etc.) has been extensively investigated[104-106]. miRNAs dregulations has also been found to affect key molecules associated with various signaling pathways leading defective metabolism in cancer such as p53, HIF-1alpha, Ras, AKT, AMPK and cMyc[107-109].
Emerging evidence has shown a clear link between miRNA alterations and tumor progression from normal phenotype[110-113]. Although, miRNA profile has been implicated as a potential biomarker and therapeutic target for cancer[114-118], the characterization of miRNA alterations in GBC, its relationship with GBC pathogenesis, and prognosis has been only poorly deciphered based on a limited number of studies, to date (Table 1)[119].
| Ref. | Source | miRNA | Level | Target gene | Functional characterization | Clinical association with | Function |
| Kitamura et al[121], 2012 | GBCs from BK5.erbb2 mice vs GBC from wild-type mice | mir-106a, mir-96 mir-223 mir-27a mir-17 mir-15b mir-142-5p mir-142-3p mir-21 | ↑ | - | - | - | - |
| mir-665 mir-714 mir-763 mir-466f-3p mir-145 mir-193 mir-467e mir-143 mir-881 mir-720 mir-706 -mir-122 mir-378 | ↓ | - | - | - | - | ||
| Li et al[133], 2015 | Paired GBC, normal tissue and blood samples | mir-21, mir-370, mir-187, mir-122, mir-202 | ↑ | - | - | mir-187, mir-143, mir-202 were associated with metastasis, TNM | - |
| let-7a, mir-200b, mir-143, mir-31, mir-335, mir-551 | ↓ | ||||||
| Peng et al[125], 2013 | Paired GBC, normal tissue | mir-335 | ↓ | - | - | Histologic grade, stage, metastasis, poor survival | - |
| Zhou et al[127], 2014 | Paired GBC, paracancerous tissue | mir-106a, mir-96 mir-223 mir-27a mir-17 mir-15b mir-142-5p mir-142-3p mir-21 | ↑ | - | - | - | - |
| mir-665 mir-714 mir-763 mir-466f-3p mir-145 mir-193 mir-467e mir-143 mir-881 mir-720 mir-706 -mir-122 mir-378 | ↓ | - | - | - | - | ||
| hsa-mir-135a-5p | ↓ | VLDLR | Cell proliferation, cell cycle distribution | Histologic grade | TSG | ||
| Jin et al[126], 2014 | Paired GBC tissue, tissue | mir-34a | ↓ | PNUTS | Cell proliferation | Poor prognosis | TSG |
| Zhou et al[128], 2014 | Paired GBC, paracancerous tissue | mir-26a | ↓ | HMGA2 | Cell proliferation | Histological grade | TSG |
| Letelier et al[129], 2014 | Normal GB, GBC tissue | mir-133a/b, mir-143-3p/5p, mir-991-5p, mir-125b-5p, mir-29c-3p, mir-195-5p, mir-139-5p, mir-29c-5p, mir-100-5p, mir-148a-3p, mir-376c, mir-187-3p, mir-365a-3p, mir-29b-3p, mir-497-5p, mir-654-3p, mir-411-5p, mir-125a-5p, mir-26a-5p, mir-101-3p, mir-495, mir-381-3p, mir-154-5p, mir-99a-3p, mir-328, mir-299-5p, mir-30e-3p, mir-29b-2-5p, mir-379-5p, mir-140-5p, mir-24-1-5p, mir-101-5p | ↓ | Distinguish GBC from normal samples | - | - | |
| - | TSG | ||||||
| mir-145-3p/5p | ↓ | - | Cell growth, cell viability, apoptosis | - | TSG | ||
| mir-1 | ↓ | VEGF-A , AXL | Cell growth, cell viability, apoptosis | - | TSG | ||
| Ma et al[131], 2014 | Paired GBC, normal tissue | mir-130a | ↓ | - | Cell proliferation, invasion | - | TSG |
| Cai et al[130], 2015 | Paired GBC, normal tissue | mir-146b-5p | ↓ | EGFR | Apoptosis, G1 phase arrest | Carcinoma size, progression | TSG |
| Ma et al[132], 2015 | Paired GBC, normal tissue | mir-218-5p | ↓ | BMI-1 | Cell proliferation, invasion | - | TSG |
| Chang et al[123], 2013 | Paired GBC, normal GB tissue | mir-20a | ↑ | SMAD-7 | EMT, metastasis | Worse overall survival | OG |
| Kono et al[122], 2014 | Paired GBC, normal GB tissue | mir-155 | ↑ | - | Cell proliferation, invasion | Lymph node metastasis, invasion, poor prognosis | OG |
| Qiu et al[124], 2014 | GBC, normal tissue | mir-182 | ↑ | CADM1 | Cell migration, invasion | Metastasis | OG |
Initially, Shu et al[120] (2012), used immunohistochemistry to investigated the expression of Dicer and Drosha in GBC patients and non-dysplastic gallbladder epithelia. They reported significant downregulation of Dicer or Drosha in GBC than in non-dysplastic gallbladder epithelia, and that the loss of Dicer and Drosha expression was closely related to the metastasis, invasion, decreased overall survival and poor-prognosis of GBC patients. Since Dicer and Drosha are two key enzymes involved in miRNA production, the loss of expression of these two enzymes is considered to partially account for the down regulation of miRNA expression followed by overexpression of oncogenes or survival and proliferation associated genes in GBC. Thus, they provided the first evidence of a role of miRNA in GBC pathogenesis.
Kitamura et al[121] (2012) compared the expression of miRNA in GBC from BK5.erbB2 mice (animal model for GBC) to that in control gallbladders from wild-type mice using a microarray. They reported significant up-regulation of miR-106a, miR-96, miR-223, miR-27a miR-17, miR-15b, miR-142-5p, miR-142-3p and miR-21 as well as down-regulation of miR-665, miR-714, miR-763, miR-466f-3p, miR-145, miR-193, miR-467e, miR-143, miR-881, miR-720, miR-706, miR-122 and miR-378 in BK5.erbB2 mice compared to normal gallbladder. Furthermore, BK5.erbB2 mice treated with histone deacetylase inhibitor PCI-24781 showed significant downregulation of miR-21, miR-142-3p, miR-142-5p and miR-223 and upregulation of miR-122.
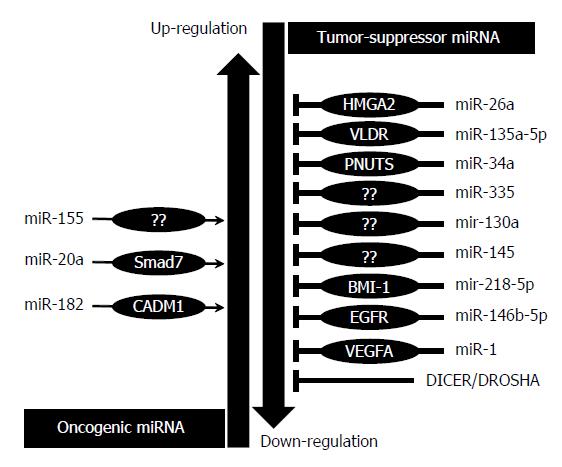
MiRNAs over-expression in GBC - Oncogenic miRNAs: Mir-155, mir-20a and mir-182 have been reported as oncomiRNAs that promote progression of gallbladder carcinoma when overexpressed (Figure 2).
MiR-155 has been reported to be up-regulated in GBCs as compared with gallbladders with pancreaticobiliary maljunction (P = 0.007) and normal gallbladders (P = 0.04). The higher expression of miR-155 was found to be significantly correlated with aggressive behavior of GBCs such as the presence of lymph nodes, metastasis and vessel invasion, indicating a poor prognosis. Further, GBC cell lines transfected with miR-155 inhibitors showed significant decrease in cell proliferation and invasion. Conversely, cells transfected with miR-155 showed significant increase in cell proliferation relative to negative controls, confirming the role of miR-155 as a potent regulator of proliferation and invasion in GBC. These findings suggest that modulation of the miR-155 level may be applied to the treatment of GBCs, particularly for inhibition of cancer progression such as lymph node invasion. Thus, mir-155 may represent a prognostic marker and therapeutic target for GBC[122].
Chang et al[123] (2013) identified 17 prominent metastatic inducers of GBCs. Among them, mir-20a was found to be highly expressed in tumor tissues as compared to normal gallbladder and was closely associated with local invasion, distant metastasis, and poor prognosis of GBC patients. Patients with higher miR-20a expression exhibited a worse overall survival than those with lower expression. The ectopic expression of miR-20a was shown to induce EMT and enhance metastasis of GBC cells in vitro and in vivo by directly targeting the 3’ UTR of Smad7 and subsequently promoting nuclear translocation of β-catenin, suggesting that the TGF-β1/miR-20a/SMAD7 axis plays an important role in the progression of GBC.
Another study reported significant upregulation of mir-182 in GBC relative to normal control and in the metastatic tumor when compared with primary tumor that did not metastasize. Upregulation of miR-182 expression in GBC cells was found to promote cell migration and invasion while downregulation of miR-182 inhibited TGF-β-induced cell migration and invasion. They also identified CADM1 as a new target gene of miR-182 that is negatively regulated by mir-182 in vitro and in vivo. Importantly, re-expression of CADM1 in GBC cells partially abrogates mir-182-induced cell invasion[124].
MiRNAs downregulation in GBC - Tumor suppressive miRNAs: Tumor suppressive miRNAs in GBC include mir-34a, miR-335, miR-135-5p, miR-26a, miR-1, miR-145 and mir-146b-5p which were frequently found to be down-regulated in GBC (Figure 2).
Peng et al[125] (2013) demonstrated downregulation of miR-335 in the majority of GBC patients that is associated with aggressive tumor behaviors such as high histologic grade, advanced pathologic T stage, clinical stage, lymph node metastasis and shorter overall survival. Reduced miR-335 expression represents an independent predictor of poor prognosis for overall survival in GBC patients. Later, Jin et al[126] (2014) identified mir-34a as a tumor suppressor in GBC. Moreover, they demonstrated significantly lower miR-34a expression in GBC as compared to peritumoral tissue, which correlated with poor prognosis of GBC patients. Forced overexpression of miR-34a reduced the colony-forming ability of CD44+CD133+ GBC tumor stem-like cells in vitro and inhibited tumor growth in a xenograft animal model. This inhibitory effect was associated with downregulation of phosphatase 1 nuclear targeting subunit (PNUTS) expression and a decrease in telomerase length.
By using the miRNA expression profiling chip in four paired GBC and paracancerous tissues, Zhou et al[127], 2014 identified aberrant expression of 23 miRNAs in GBC tissue as compared with their paired noncancerous tissue. Among them, miR-135a-5p and miR-26a were considered to significantly influence GBC cell proliferation. miR-135-5p expression was correlated with the neoplasm histologic grade. Ectopic mir-135a expression inhibited GBC cell proliferation in vitro and in vivo and disrupted cell cycle distribution leading to arrest the cells in the G 1⁄S phase. miR-135a exerted this function by directly targeting very low density lipoprotein receptor (VLDLR), leading to activation of the p38 MAPK pathway. In a subsequent study, they reported miR-26a downregulation in GBC associated with neoplasm histological grade. miR-26a significantly inhibited the proliferation of GBC cells by targeting high mobility group AT-hook 2 (HMGA2) via G1 arrest. Furthermore, they demonstrated that HMGA2 mRNA levels and miR-26a levels were negatively correlated and reintroduction of HMGA2 antagonized inhibition of the GBC cell proliferation by miR-26a[128].
A comprehensive miRNA profiling study showed consistent downregulation of 36 miRNAs separating the GBC and normal samples. Among them, expression of hsa-miR-133a, hsa-miR-133b, hsa-miR-143, hsa-miR-145, hsa-miR-1, and hsa-miR-29c were further validated by qRT-PCR. These miRNAs collectively targeted a number of genes including TGF-β, ErbB3, WNT, MAPK and VEGF, Notch as well as those participating in various biological processes associated with human cancer such as transcription regulation, signal transduction, positive regulation of cell proliferation, cell adhesion and angiogenesis. miR-1 and miR-145 expression were significantly decreased in GBC cell lines. Ectopic expression of miR-1 and miR-145 was shown to decrease cell viability, inhibit colony formation or clonogenic survival and increase apoptosis. mir-1 may act via reduced gene expression of VEGF-A and AXL, resulting in inhibition of gallbladder cancer cell growth, in vitro[129].
Decreased expression of mir-146b-5p was also reported in GBC tissue, which is significantly correlated with carcinoma size and development[130]. miR-146b-5p overexpression was found to inhibit cell growth through enhanced apoptosis and G1 phase arrest. Enforced expression of epidermal growth factor receptor (EGFR) reversed the ability of miR-146b-5p to inhibit proliferation suggesting that EGFR is a mediator of the biological effects of miR-146b-5p in GBC[130].
HOTAIR’s, which are long non-coding RNAs with oncogenic activity, have been found to negatively regulate miRNA-130a expression in GBC, promoting the invasiveness and proliferation of cancer cells. miRNA-130a has tumor-suppressive activity, and has been previously reported to be downregulated in a variety of carcinomas[131]. Later, Ma et al[132] (2015) demonstrated that downregulation of miR-218-5p in GBC occurred via Colon Cancer-Associated Transcript-1 (CCAT1). Specifically, CCAT1 was found to promote the proliferation and invasiveness of GBC cells by upregulating the miRNA-218-5p target gene, Bmi1, by competitively “spongeing” miRNA-218-5p. Recently, Li et al[133] (2015) demonstrated deregulation of 11 miRNAs (upregulated- miR-21, miR-370, miR-187, miR-122, miR-202; and down-regulated- let-7a, miR-200b, miR-143, miR-31, miR-335, and miR-551) in GBC tissue samples relative to neighboring noncancerous tissue.
Li et al[133] (2015) demonstrated upregulation of miR-21, miR-200b, miR-187, miR-122, and miR-202 while down-regulation of let-7a, miR-370, miR-143, miR-31, miR-335, and miR-551 in blood samples of GBC patients relative to control blood samples. However, only the level of let-7a, miR-21, miR-187, miR-143, miR-202, and miR-335 was reported to differ significantly between GBC patients and the control. Furthermore, only miR-187, miR-143, and miR-122 were associated with lymphatic metastasis, suggesting that these three miRNAs can be used for GBC prognosis and therapeutic efficacy.
Envisioning the important role of miRNA in carcinogenesis, as well as the role of genetic variants to modulate the miRNA processing and expression, thereby resulting in diverse functional consequences, Srivastava et al[134], investigated the association of common potentially functional polymorphisms in pre-miRNA (hsa-miR-146a, hsa-mir-196a2 and hsa-mir-499) with GBC susceptibility in a North Indian population. However, they found no association suggesting that these miRNA variants may not contribute to GBC susceptibility. Recently, we evaluated the association of genetic variants of miR-27a (rs895819), miR-570 (rs4143815) and miR-181a (rs12537), which alter miRNA processing and expression, with genetic susceptibility, treatment response and toxicity (personal communication). None of the studied SNPs were found to be associated with overall GBC susceptibility, survival outcomes of GBC patients (locally advanced, metastatic) and response to chemo-radiotherapy. However, in Generalized Multifactor Dimensionality Reduction(GMDR) analysis, miR-27ars895819, miR-570rs4143815, and miR-181ars12537 combination was found to be associated with GBC susceptibility and treatment response while miR-27ars895819-miR-181ars12537 combination was associated with neutropenia toxicity in patients undergoing chemo-radiotherapy suggesting the the cumulatively influence of miRNA variants on GBC susceptibility and treatment outcomes[135].
Currently, the use of miRNA as a viable therapeutic target is the most fascinating area of cancer research that has gain overwhelming amount of importance because (1) miRNA are small molecule comprising of a known and evolutionary conserved sequences; (2) it can target multiple genes and regulate wide range of biological process; (3) the potential targets of a particular miRNAs can be predicted by using various bioinformatical tools; such as miRanda (http://www.microRNA.org), microCosm (previously known as miRBase targets, http://www.mirbase.org), Targetscan (http://www.targetscan.org), or PicTar (http://pictar.mdc-berlin.de); (4) miRNA expression is frequently dysregulated in various cancer; and (5) the cancer phenotype can be changed by targeting miRNA expression[17,136-138]. Consequently, growing evidences have confirmed that restoration of miRNAs signature i.e., inhibition of over-expressed and oncogenic miRNAs by using chemically modified anti-miR oligonucleotides as well as restoration of down-regulated or tumor suppressive miRNAs by using synthetic miRNA mimics or viral expression constructs, constitute a novel treatment strategy for cancer. In addition several miRNAs modulation has been found to reverse the drug resistance in various cancer enhancing the therapeutic implication of miRNAs[139-141].
Till date, several studies have successfully investigated the therapeutic effects of miRNA in preclinical models and xenografts, e.g., reconstitution of miR-34 and let-7 was found to reduce the tumour growth in a murine model of non-small cell lung cancer (NSCLC). On the other hand, depletion of miR-21 in combination therap has been shown to control pancreatic ductal adenocarcinoma in a mouse model. MRX34 (mir-34 mimic) is the first microRNA mimic to enter clinical development for cancer and is currently under phase I clinical trial (NCT01829971) involving primary hepatocellular carcinoma (HCC metastatic liver cancer) patients. Since, mir-34 and let-7 was shown to function like TSG while mir-21 was found to be oncogenic in GBC, similar miRNA therapy could be also applied for GBC like other cancers. Thus, miRNA holds promising perspectives for GBC management where treatment options are already limited, however, there is still limited study exploring the role of miRNA in GBC and we still need to identify key miRNAs having therapeutic importance in GBC. Moreover, though the effectiveness and safety of miRNA-derived drugs depends on cellular context and pre-existing genetic and epigenetic lesions due to heterogeneous nature of disease, this area of research needs to be carefully appraised in GBC.
miRNAs constitute an important regulator of carcinogenesis. Frequent deregulation of various miRNAs in GBC has been found to affect tumor growth and survival, suggesting miRNAs as promising biomarkers for GBC diagnosis and prognosis. Further, miRNA signature in blood sample represents a future non-invasive diagnostic biomarker for GBC. miRNAs also represent wide-ranging applications as new targets for treatment by inhibition of overexpressed oncogenic miRNAs or inhibition of tumor suppressive miRNAs. The high stability of miRNAs when compared with mRNA molecules in both blood samples and formalin-fixed, paraffin-embedded tissue samples offers a great advantage suggesting miRNAs as a promising approach to ameliorate GBC. However, the number of studies investigating the miRNAs signature in GBC is limited. Therefore, larger well-designed studies with clinical outcome are needed to investigate the complex network of miRNAs in the pathogenesis of GBC.
P- Reviewer: Kai K S- Editor: Yu J L- Editor: A E- Editor: Wang CH
| 1. | Rai R, Sharma KL, Misra S, Kumar A, Mittal B. CYP17 polymorphism (rs743572) is associated with increased risk of gallbladder cancer in tobacco users. Tumour Biol. 2014;35:6531-6537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 2. | Bizama C, García P, Espinoza JA, Weber H, Leal P, Nervi B, Roa JC. Targeting specific molecular pathways holds promise for advanced gallbladder cancer therapy. Cancer Treat Rev. 2015;41:222-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 3. | Rai R, Tewari M, Kumar M, Singh TB, Shukla HS. Expression profile of cholecystokinin type-A receptor in gallbladder cancer and gallstone disease. Hepatobiliary Pancreat Dis Int. 2011;10:408-414. [PubMed] |
| 4. | Rai R, Tewari M, Kumar M, Singh AK, Shukla HS. p53: its alteration and gallbladder cancer. Eur J Cancer Prev. 2011;20:77-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Zhu AX, Hong TS, Hezel AF, Kooby DA. Current management of gallbladder carcinoma. Oncologist. 2010;15:168-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 241] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 6. | Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol. 2014;6:99-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 488] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 7. | Feig JL, Giles KM, Osman I, Franks AG. How microRNAs modify protein production. J Invest Dermatol. 2015;135:e32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Ross SA, Davis CD. MicroRNA, nutrition, and cancer prevention. Adv Nutr. 2011;2:472-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 9. | Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 755] [Cited by in RCA: 860] [Article Influence: 61.4] [Reference Citation Analysis (0)] |
| 10. | Ke XS, Liu CM, Liu DP, Liang CC. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7:516-523. [PubMed] |
| 11. | Shah AA, Meese E, Blin N. Profiling of regulatory microRNA transcriptomes in various biological processes: a review. J Appl Genet. 2010;51:501-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Panera N, Gnani D, Crudele A, Ceccarelli S, Nobili V, Alisi A. MicroRNAs as controlled systems and controllers in non-alcoholic fatty liver disease. World J Gastroenterol. 2014;20:15079-15086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Ha TY. MicroRNAs in Human Diseases: From Cancer to Cardiovascular Disease. Immune Netw. 2011;11:135-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 257] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 14. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5384] [Cited by in RCA: 5607] [Article Influence: 295.1] [Reference Citation Analysis (0)] |
| 15. | Stahlhut Espinosa CE, Slack FJ. The role of microRNAs in cancer. Yale J Biol Med. 2006;79:131-140. [PubMed] |
| 16. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1980] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 17. | Price C, Chen J. MicroRNAs in Cancer Biology and Therapy: Current Status and Perspectives. Genes Dis. 2014;1:53-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 18. | Megiorni F, Pizzuti A, Frati L. Clinical Significance of MicroRNA Expression Profiles and Polymorphisms in Lung Cancer Development and Management. Patholog Res Int. 2011;2011:780652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Xue Z, Wen J, Chu X, Xue X. A microRNA gene signature for identification of lung cancer. Surg Oncol. 2014;23:126-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Chen M, Calin GA, Meng QH. Circulating microRNAs as Promising Tumor Biomarkers. Adv Clin Chem. 2014;67:189-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432-10439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 223] [Cited by in RCA: 282] [Article Influence: 25.6] [Reference Citation Analysis (1)] |
| 22. | Gayral M, Jo S, Hanoun N, Vignolle-Vidoni A, Lulka H, Delpu Y, Meulle A, Dufresne M, Humeau M, Chalret du Rieu M. MicroRNAs as emerging biomarkers and therapeutic targets for pancreatic cancer. World J Gastroenterol. 2014;20:11199-11209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 23. | Ferracin M, Pedriali M, Veronese A, Zagatti B, Gafà R, Magri E, Lunardi M, Munerato G, Querzoli G, Maestri I. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225:43-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 24. | Søkilde R, Vincent M, Møller AK, Hansen A, Høiby PE, Blondal T, Nielsen BS, Daugaard G, Møller S, Litman T. Efficient identification of miRNAs for classification of tumor origin. J Mol Diagn. 2014;16:106-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Guo Z, Maki M, Ding R, Yang Y, Zhang B, Xiong L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep. 2014;4:5150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 26. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8596] [Article Influence: 409.3] [Reference Citation Analysis (0)] |
| 27. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 28. | Weber C. MicroRNAs: from basic mechanisms to clinical application in cardiovascular medicine. Arterioscler Thromb Vasc Biol. 2013;33:168-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Leonardi GC, Candido S, Carbone M, Colaianni V, Garozzo SF, Cinà D, Libra M. microRNAs and thyroid cancer: biological and clinical significance (Review). Int J Mol Med. 2012;30:991-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 30. | Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-4060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2902] [Cited by in RCA: 2997] [Article Influence: 142.7] [Reference Citation Analysis (0)] |
| 31. | Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622-4629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 196] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 32. | Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1964] [Cited by in RCA: 1986] [Article Influence: 94.6] [Reference Citation Analysis (0)] |
| 33. | Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209-216. [PubMed] |
| 34. | Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1109] [Cited by in RCA: 1107] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 35. | Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 395] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 36. | Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1101] [Cited by in RCA: 1127] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 37. | Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700-2704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 38. | Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 249] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 39. | Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 282] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 40. | Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345-2350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 188] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 41. | Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 274] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 42. | Mraz M, Pospisilova S. MicroRNAs in chronic lymphocytic leukemia: from causality to associations and back. Expert Rev Hematol. 2012;5:579-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 43. | Kusenda B, Mraz M, Mayer J, Pospisilova S. MicroRNA biogenesis, functionality and cancer relevance. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:205-215. [PubMed] |
| 44. | Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643-2646. [PubMed] |
| 45. | Yu Z, Baserga R, Chen L, Wang C, Lisanti MP, Pestell RG. microRNA, cell cycle, and human breast cancer. Am J Pathol. 2010;176:1058-1064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 46. | Liang LH, He XH. Macro-management of microRNAs in cell cycle progression of tumor cells and its implications in anti-cancer therapy. Acta Pharmacol Sin. 2011;32:1311-1320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 47. | Cui W, Zhang S, Shan C, Zhou L, Zhou Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280:3962-3974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 48. | Giglio S, Cirombella R, Amodeo R, Portaro L, Lavra L, Vecchione A. MicroRNA miR-24 promotes cell proliferation by targeting the CDKs inhibitors p27Kip1 and p16INK4a. J Cell Physiol. 2013;228:2015-2023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 49. | Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079-3093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 1307] [Article Influence: 108.9] [Reference Citation Analysis (0)] |
| 50. | Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220-5227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 870] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 51. | Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol. 2013;5:a008904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 260] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 52. | Denicourt C, Dowdy SF. Cip/Kip proteins: more than just CDKs inhibitors. Genes Dev. 2004;18:851-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 150] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 53. | Seo YH, Joo YE, Choi SK, Rew JS, Park CS, Kim SJ. Prognostic significance of p21 and p53 expression in gastric cancer. Korean J Intern Med. 2003;18:98-103. [PubMed] |
| 54. | Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 775] [Cited by in RCA: 855] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 55. | Cánepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life. 2007;59:419-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 216] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 56. | Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391-5404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 369] [Cited by in RCA: 390] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 57. | Wang F, Fu XD, Zhou Y, Zhang Y. Down-regulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep. 2009;42:725-730. [PubMed] |
| 58. | Ouyang Q, Xu L, Cui H, Xu M, Yi L. MicroRNAs and cell cycle of malignant glioma. Int J Neurosci. 2016;126:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 59. | Zhou JY, Ma WL, Liang S, Zeng Y, Shi R, Yu HL, Xiao WW, Zheng WL. Analysis of microRNA expression profiles during the cell cycle in synchronized HeLa cells. BMB Rep. 2009;42:593-598. [PubMed] |
| 60. | Bandi N, Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer. 2011;10:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 61. | Sharma G, Dua P, Agarwal SM. A Comprehensive Review of Dysregulated miRNAs Involved in Cervical Cancer. Curr Genomics. 2014;15:310-323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 62. | Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 316] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 63. | Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27-33. [PubMed] |
| 64. | Olive V, Li Q, He L. mir-17-92: a polycistronic oncomir with pleiotropic functions. Immunol Rev. 2013;253:158-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 65. | Takwi A, Li Y. The p53 Pathway Encounters the MicroRNA World. Curr Genomics. 2009;10:194-197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 66. | Tao J, Zhao X, Tao J. c-MYC-miRNA circuitry: a central regulator of aggressive B-cell malignancies. Cell Cycle. 2014;13:191-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 67. | Fulda S. Tumor resistance to apoptosis. Int J Cancer. 2009;124:511-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 451] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 68. | Alanazi I, Hoffmann P, Adelson DL. MicroRNAs are part of the regulatory network that controls EGF induced apoptosis, including elements of the JAK/STAT pathway, in A431 cells. PLoS One. 2015;10:e0120337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 69. | Wang Y, Lee CG. MicroRNA and cancer--focus on apoptosis. J Cell Mol Med. 2009;13:12-23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 272] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 70. | Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer. 2011;47:163-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 212] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 71. | Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6:8474-8490. [PubMed] |
| 72. | Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 505] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 73. | Xie Y, Tobin LA, Camps J, Wangsa D, Yang J, Rao M, Witasp E, Awad KS, Yoo N, Ried T. MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 2013;32:2442-2451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 74. | Xu D, Wang Q, An Y, Xu L. MiR-203 regulates the proliferation, apoptosis and cell cycle progression of pancreatic cancer cells by targeting Survivin. Mol Med Rep. 2013;8:379-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 75. | Hu Y, Xu K, Yagüe E. miR-218 targets survivin and regulates resistance to chemotherapeutics in breast cancer. Breast Cancer Res Treat. 2015;151:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 76. | Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol. 2010;223:289-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 77. | Lindsey S, Langhans SA. Crosstalk of Oncogenic Signaling Pathways during Epithelial-Mesenchymal Transition. Front Oncol. 2014;4:358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 78. | Le XF, Merchant O, Bast RC, Calin GA. The Roles of MicroRNAs in the Cancer Invasion-Metastasis Cascade. Cancer Microenviron. 2010;3:137-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 79. | Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci. 2015;22:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 125] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 80. | Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283-1290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 156] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 81. | Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1717] [Cited by in RCA: 1849] [Article Influence: 108.8] [Reference Citation Analysis (0)] |
| 82. | Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z, Dong JT. Epigenetic Silencing of miR-203 Upregulates SNAI2 and Contributes to the Invasiveness of Malignant Breast Cancer Cells. Genes Cancer. 2011;2:782-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 136] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 83. | Zaravinos A. The Regulatory Role of MicroRNAs in EMT and Cancer. J Oncol. 2015;2015:865816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 219] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 84. | Rutnam ZJ, Wight TN, Yang BB. miRNAs regulate expression and function of extracellular matrix molecules. Matrix Biol. 2013;32:74-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 85. | Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115-119. [PubMed] |
| 86. | Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31:653-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 217] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 87. | Gordon MS, Mendelson DS, Kato G. Tumor angiogenesis and novel antiangiogenic strategies. Int J Cancer. 2010;126:1777-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 88. | Chen S, Xue Y, Wu X, Le C, Bhutkar A, Bell EL, Zhang F, Langer R, Sharp PA. Global microRNA depletion suppresses tumor angiogenesis. Genes Dev. 2014;28:1054-1067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 89. | Wang P, Luo Y, Duan H, Xing S, Zhang J, Lu D, Feng J, Yang D, Song L, Yan X. MicroRNA 329 suppresses angiogenesis by targeting CD146. Mol Cell Biol. 2013;33:3689-3699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 90. | Gallach S, Calabuig-Fariñas S, Jantus-Lewintre E, Camps C. MicroRNAs: promising new antiangiogenic targets in cancer. Biomed Res Int. 2014;2014:878450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 91. | Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2011;17:1359-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1212] [Cited by in RCA: 1322] [Article Influence: 94.4] [Reference Citation Analysis (0)] |
| 92. | Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 376] [Cited by in RCA: 360] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 93. | Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. MicroRNAs: molecular features and role in cancer. Front Biosci (Landmark Ed). 2012;17:2508-2540. [PubMed] |
| 94. | Costa A, Afonso J, Osório C, Gomes AL, Caiado F, Valente J, Aguiar SI, Pinto F, Ramirez M, Dias S. miR-363-5p regulates endothelial cell properties and their communication with hematopoietic precursor cells. J Hematol Oncol. 2013;6:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 95. | Marcelo KL, Goldie LC, Hirschi KK. Regulation of endothelial cell differentiation and specification. Circ Res. 2013;112:1272-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 219] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 96. | Su Z, Si W, Li L, Zhou B, Li X, Xu Y, Xu C, Jia H, Wang QK. MiR-144 regulates hematopoiesis and vascular development by targeting meis1 during zebrafish development. Int J Biochem Cell Biol. 2014;49:53-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 97. | Jamaluddin MS, Weakley SM, Zhang L, Kougias P, Lin PH, Yao Q, Chen C. miRNAs: roles and clinical applications in vascular disease. Expert Rev Mol Diagn. 2011;11:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 98. | Heusschen R, van Gink M, Griffioen AW, Thijssen VL. MicroRNAs in the tumor endothelium: novel controls on the angioregulatory switchboard. Biochim Biophys Acta. 2010;1805:87-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 99. | Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1380] [Cited by in RCA: 1336] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 100. | Chandra V, Hong KM. Effects of deranged metabolism on epigenetic changes in cancer. Arch Pharm Res. 2015;38:321-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 101. | Chen B, Li H, Zeng X, Yang P, Liu X, Zhao X, Liang S. Roles of microRNA on cancer cell metabolism. J Transl Med. 2012;10:228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 152] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 102. | Lynn FC. Meta-regulation: microRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 144] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 103. | Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA. 2012;109:8983-8988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 383] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 104. | Yoshino H, Enokida H, Itesako T, Kojima S, Kinoshita T, Tatarano S, Chiyomaru T, Nakagawa M, Seki N. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci. 2013;104:1567-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 105. | Fei X, Qi M, Wu B, Song Y, Wang Y, Li T. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 2012;586:392-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 128] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 106. | Yamasaki T, Seki N, Yoshino H, Itesako T, Yamada Y, Tatarano S, Hidaka H, Yonezawa T, Nakagawa M, Enokida H. Tumor-suppressive microRNA-1291 directly regulates glucose transporter 1 in renal cell carcinoma. Cancer Sci. 2013;104:1411-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 107. | Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 432] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 108. | Dumortier O, Hinault C, Van Obberghen E. MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 2013;18:312-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 176] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 109. | Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356:301-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 110. | Ge YZ, Xin H, Lu TZ, Xu Z, Yu P, Zhao YC, Li MH, Zhao Y, Zhong B, Xu X. MicroRNA expression profiles predict clinical phenotypes and prognosis in chromophobe renal cell carcinoma. Sci Rep. 2015;5:10328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 111. | Braicu C, Cojocneanu-Petric R, Chira S, Truta A, Floares A, Petrut B, Achimas-Cadariu P, Berindan-Neagoe I. Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomedicine. 2015;10:791-800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 112. | Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B, Amsellem-Ouazana D. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013;132:2479-2491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 113. | Brase JC, Johannes M, Schlomm T, Fälth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sültmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 458] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 114. | Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review). Int J Oncol. 2012;41:1897-1912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 269] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 115. | Wang J, Zhang KY, Liu SM, Sen S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules. 2014;19:1912-1938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 116. | Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 224] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 117. | Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61:56-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 358] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 118. | Takahashi RU, Miyazaki H, Ochiya T. The Roles of MicroRNAs in Breast Cancer. Cancers (Basel). 2015;7:598-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 119. | Li Z, Yu X, Shen J, Law PT, Chan MT, Wu WK. MicroRNA expression and its implications for diagnosis and therapy of gallbladder cancer. Oncotarget. 2015;6:13914-13921. [PubMed] |
| 120. | Shu GS, Yang ZL, Liu DC. Immunohistochemical study of Dicer and Drosha expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Pathol Res Pract. 2012;208:392-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 121. | Kitamura T, Connolly K, Ruffino L, Ajiki T, Lueckgen A, DiGiovanni J, Kiguchi K. The therapeutic effect of histone deacetylase inhibitor PCI-24781 on gallbladder carcinoma in BK5.erbB2 mice. J Hepatol. 2012;57:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 122. | Kono H, Nakamura M, Ohtsuka T, Nagayoshi Y, Mori Y, Takahata S, Aishima S, Tanaka M. High expression of microRNA-155 is associated with the aggressive malignant behavior of gallbladder carcinoma. Oncol Rep. 2013;30:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 123. | Chang Y, Liu C, Yang J, Liu G, Feng F, Tang J, Hu L, Li L, Jiang F, Chen C. MiR-20a triggers metastasis of gallbladder carcinoma. J Hepatol. 2013;59:518-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 124. | Qiu Y, Luo X, Kan T, Zhang Y, Yu W, Wei Y, Shen N, Yi B, Jiang X. TGF-β upregulates miR-182 expression to promote gallbladder cancer metastasis by targeting CADM1. Mol Biosyst. 2014;10:679-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 125. | Peng HH, Zhang YD, Gong LS, Liu WD, Zhang Y. Increased expression of microRNA-335 predicts a favorable prognosis in primary gallbladder carcinoma. Onco Targets Ther. 2013;6:1625-1630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 126. | Jin K, Xiang Y, Tang J, Wu G, Li J, Xiao H, Li C, Chen Y, Zhao J. miR-34 is associated with poor prognosis of patients with gallbladder cancer through regulating telomere length in tumor stem cells. Tumour Biol. 2014;35:1503-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 127. | Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding J, Liang L, Yang G, Chen Z, Ma B. MicroRNA-135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci. 2014;105:956-965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 128. | Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding J, Liang L, Hu J, Shen H, Chen Z. MicroRNA-26a acts as a tumor suppressor inhibiting gallbladder cancer cell proliferation by directly targeting HMGA2. Int J Oncol. 2014;44:2050-2058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 129. | Letelier P, García P, Leal P, Álvarez H, Ili C, López J, Castillo J, Brebi P, Roa JC. miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int J Clin Exp Pathol. 2014;7:1849-1867. [PubMed] |
| 130. | Cai J, Xu L, Cai Z, Wang J, Zhou B, Hu H. MicroRNA-146b-5p inhibits the growth of gallbladder carcinoma by targeting epidermal growth factor receptor. Mol Med Rep. 2015;12:1549-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 131. | Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD, Qin YY, Gong W, Quan ZW. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer. 2014;13:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 132. | Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 297] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 133. | Li G, Pu Y. MicroRNA signatures in total peripheral blood of gallbladder cancer patients. Tumour Biol. 2015;36:6985-6990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 134. | Srivastava K, Srivastava A, Mittal B. Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet. 2010;55:495-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 135. | Gupta A, Sharma A, Yadav A, Rastogi N, Agrawal S, Kumar A, Kumar V, Misra S, Mittal B. Evaluation of miR-27a, miR-181a, and miR-570 genetic variants with gallbladder cancer susceptibility and treatment outcome in a North Indian population. Mol Diagn Ther. 2015;19:317-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 136. | Shah MY, Calin GA. MicroRNAs as therapeutic targets in human cancers. Wiley Interdiscip Rev RNA. 2014;5:537-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 137. | Rothschild SI. microRNA therapies in cancer. Mol Cell Ther. 2014;2:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 138. | Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 581] [Cited by in RCA: 666] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 139. | Li JH, Luo N, Zhong MZ, Xiao ZQ, Wang JX, Yao XY, Peng Y, Cao J. Inhibition of microRNA-196a might reverse cisplatin resistance of A549/DDP non-small-cell lung cancer cell line. Tumour Biol. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 140. | Fujita Y, Kojima T, Kawakami K, Mizutani K, Kato T, Deguchi T, Ito M. miR-130a activates apoptotic signaling through activation of caspase-8 in taxane-resistant prostate cancer cells. Prostate. 2015;75:1568-1578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 141. | Liu L, Zheng W, Song Y, Du X, Tang Y, Nie J, Han W. miRNA-497 Enhances the Sensitivity of Colorectal Cancer Cells to Neoadjuvant Chemotherapeutic Drug. Curr Protein Pept Sci. 2015;16:310-315. [PubMed] |