Published online Oct 14, 2016. doi: 10.3748/wjg.v22.i38.8528
Peer-review started: May 20, 2016
First decision: July 12, 2016
Revised: July 29, 2016
Accepted: August 19, 2016
Article in press: August 19, 2016
Published online: October 14, 2016
Processing time: 145 Days and 22.1 Hours
To find the mechanisms by which special AT-rich sequence-binding protein 2 (SATB2) influences colorectal cancer (CRC) metastasis.
Cell growth assay, colony-forming assay, cell adhesion assay and cell migration assay were used to evaluate the biological characteristics of CRC cells with gain or loss of SATB2. Sphere formation assay was used to detect the self-renewal ability of CRC cells. The mRNA expression of stem cell markers in CRC cells with upregulated or downregulated SATB2 expression was detected by quantitative real-time polymerase chain reaction. Chromatin immunoprecipitation (ChIP) was used to verify the binding loci of SATB2 on genomic sequences of stem cell markers. The Cancer Genome Atlas (TCGA) database and our clinical samples were analyzed to find the correlation between SATB2 and some key stem cell markers.
Downregulation of SATB2 led to an aggressive phenotype in SW480 and DLD-1 cells, which was characterized by increased migration and invasion abilities. Overexpression of SATB2 suppressed the migration and invasion abilities in SW480 and SW620 cells. Using sequential sphere formation assay to detect the self-renewal abilities of CRC cells, we found more secondary sphere formation but not primary sphere formation in SW480 and DLD-1 cells after SATB2 expression was knocked down. Moreover, most markers for stem cells such as CD133, CD44, AXIN2, MEIS2 and NANOG were increased in cells with SATB2 knockdown and decreased in cells with SATB2 overexpression. ChIP assay showed that SATB2 bound to regulatory elements of CD133, CD44, MEIS2 and AXIN2 genes. Using TCGA database and our clinical samples, we found that SATB2 was correlated with some key stem cell markers including CD44 and CD24 in clinical tissues of CRC patients.
SATB2 can directly bind to the regulatory elements in the genetic loci of several stem cell markers and consequently inhibit the progression of CRC by negatively regulating stemness of CRC cells.
Core tip: We found that special AT-rich sequence-binding protein 2 (SATB2) had a suppressive effect on the tumor growth, adhesion and migration in vitro. Moreover, SATB2 could negatively regulate the stemness of colorectal cancer (CRC) cells by directly binding to the regulatory elements in the genetic loci of several stem cell markers. Our study provides a new mechanism for the involvement of SATB2 in CRC progression and helps us to better understand the metastasis traits of cancer stem cells.
- Citation: Li Y, Liu YH, Hu YY, Chen L, Li JM. Special AT-rich sequence-binding protein 2 acts as a negative regulator of stemness in colorectal cancer cells. World J Gastroenterol 2016; 22(38): 8528-8539
- URL: https://www.wjgnet.com/1007-9327/full/v22/i38/8528.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i38.8528
Special AT-rich sequence-binding protein 2 (SATB2) is an important DNA-binding protein involved in transcriptional regulation and chromatin remodeling. SATB2 plays key roles in osteoblastic differentiation, cortical neuron differentiation and skeletal development[1-4]. However, the role of SATB2 in cancer initiation and progression is still not well-understood. We previously found that SATB2 was a potential marker for metastasis of colorectal cancer (CRC) and low expression of SATB2 was correlated with tumor progression and poor prognosis in CRC patients[5]. Interestingly, more evidence shows that SATB2 is involved in progression of breast cancer, head and neck squamous cell carcinomas and osteosarcoma[6-8]. Importantly, SATB2 is strongly expressed in normal colorectal and appendiceal epithelium[9], demonstrating SATB2 as a diagnostic marker for CRC.
Recently, SATB2 and its analogue protein SATB1 are found to regulate embryonic stem cell differentiation by directly binding with NANOG genomic locus[10]. Renew and differentiation of trophoblast stem cells are also considered to be related with SATB proteins[11]. In the process of cancer development and progression, very few cancer cells with stem cell-like properties have greatly enhanced tumor-initiating potential within a tumor and these cells are termed as cancer stem cells (CSCs)[12,13]. However, the regulatory mechanisms of CSCs and relations between CSCs and cancer metastasis are still needed to be elucidated.
In this study, we found that SATB2 had a suppressive effect on the tumor growth, adhesion, and migration in vitro. Moreover, SATB2 could negatively regulate the stemness of CRC cells by directly binding to the regulatory elements in the genetic loci of several stem cell markers. Our study provides a new mechanism for the involvement of SATB2 in CRC progression and helps better understand the metastasis traits of CSCs.
The CRC cell lines used in our experiments were bought from the Cell Bank at the Chinese Academy of Sciences. The cells were cultured in RPMI-1640 medium in which fetal bovine serum (Hyclone, United States) was added to a final concentration of 10%. The cells were sustained in an incubator with 5% CO2 at 37 °C.
The information of pCAG-SATB2 vector and its control vector had been mentioned in our previous paper[5]. The pLKO.1-TRC vectors with different interference fragments targeting SATB2 were purchased from Thermo Scientific (United States, Item No. TRCN0000020684 to TRCN0000020688).
The pCAG-SATB2 was transfected into CRC cell lines to establish cells in which SATB2 expression was upregulated. The packaged virus with pLKO.1-TRC vectors were used to establish cells with stably downregulated expression of SATB2.
The mRNA expression levels of SATB2 and stem cell markers in CRC cell lines were measured by quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR Green (Takara, China) run in a 7500 real-time PCR system (ABI, United States). Expression levels of SATB2 and stem cell markers were evaluated using the ΔΔCt method and normalized according to the mRNA level of GAPDH. Primer sequences for qRT-PCR are listed in Table 1.
| Gene | Sequence |
| CD133-F | 5’ TTTGTCTTCTATTCTTGGCTTC 3’ |
| CD133-R | 5’ ACCTTGTCATAATCAATTTTGG 3’ |
| CD44-F | 5’ GGTTCATAGAAGGGCACGT 3’ |
| CD44-R | 5’ TGTCTTCGTCTGGGATGG 3’ |
| CD24-F | 5’ TCAAGTATTTGGGAAGTG 3’ |
| CD24-R | 5’ GTGTTCTAAATGTGGCTAT 3’ |
| OCT3/4-F | 5’ CGACCATCTGCCGCTTTGAG 3’ |
| OCT3/4-R | 5’ CCCCCTGTCCCCCATTCCTA 3’ |
| KLF4-F | 5’ TGGGTCTTGAGGAAGTGCTG 3’ |
| KLF4-R | 5’ TGTTTACGGTAGTGCCTGGTC 3’ |
| SOX2-F | 5’ CACCTACAGCATGTCCTACTC 3’ |
| SOX2-R | 5’ CATGCTGTTTCTTACTCTCCTC 3’ |
| PRL-1-F | 5’ GGCAAACTTCGAGTCTCCT 3’ |
| PRL-1-R | 5’ CCGGTTGATGAATGGCTAA 3’ |
| MEIS2-F | 5’ GTCCACGAACTGTGCGATAA 3’ |
| MEIS2-R | 5’ TTCGGAAGGGTACGGATG 3’ |
| AXIN2-F | 5’ AGCATTTTAATCAACAGCATCTA 3’ |
| AXIN2-R | 5’ TAACTAAGAATGTGATCCAAGAA 3’ |
| NANOG-F | 5’ CAACTGGCCGAAGAATAGCA 3’ |
| NANOG-R | 5’ GCAGGAGAATTTGGCTGGAA 3’ |
| GAPDH-F | 5’ GGAGCGAGATCCCTCCAAAAT 3’ |
| GAPDH-R | 5’ GGCTGTTGTCATACTTCTCATGG 3’ |
Protein extracts were obtained using the lysis buffer (KeyGen Biotech). After being quantified, equivalent amounts of protein extracts were separated using SDS-PAGE and transferred to the PVDF membrane (Roche Applied Sciences). The primary antibody was added onto each membrane and incubated at 4 °C overnight. The appropriate second antibody was used on the following day. Mouse monoclonal anti-SATB2 antibody (1:100, Abcam, United Kingdom), rabbit polyclonal anti-CD133 antibody (1:500, Abnova, China) and mouse monoclonal anti-α-tubulin antibody (1:1000, proteintech, United States) were used. The targeted bands were visualized and photographed with the FluorChem system (Alpha Innotech).
Cell aliquots (100 μL) were transferred into each well of 96-well microtiter plates at a concentration of 1 × 104 cells/mL. CCK-8 (Dojindo Laboratories, Japan) was used to test the cell proliferation ability every 24 h and would last 6 d. Each day we added 10 μL of CCK-8 reagents into each well and then incubated the plates at 37 °C for 2 h. After the incubation, the absorbance of each well was measured at 450 nm using the Vmax microplate spectrophotometer (Molecular Devices, CA).
Cells (1 × 102) were seeded into each well of 6-well culture plates and incubated at 37 °C for 14 d. Then the cells were stained with crystal violet solution and the pictures of stained cells were taken with a digital camera. Under a microscope, the colonies containing more than 50 cells were counted. The colony formation efficiency of each group was calculated as the colony number divided by inoculated cell number and then multiplied by 100%.
Fibronectin (Invitrogen, United States) was added into each well of 96-well plates at a concentration of 10 μg/mL and plates were incubated overnight at 4 °C. After that, 1% BSA was added and incubated at 37 °C for 1 h. Then diluted cells (1 × 105 cells/100 μL) were added to the coated wells and incubated at 37 °C for 1 h. After the non-adherent cells were washed out, we added CCK-8 reagent into each well and the plates were incubated at 37 °C for 2 h. The absorbance of each well was measured at 450 nm.
Transwells (BD Biosciences, United States) inserted with 8 μm pores were put into wells of 24-well plates. Cells were suspended with serum free medium and 2 × 105 cells were added inside the chamber. Below the matched chamber, 600 μL of RMPI-1640 medium containing 10% FBS was added. After incubation for 24 h, noninvasive cells on the membrane inside the transwell were removed. Invaded cells were fixed with methanol, stained with Giemsa or crystal violet and photographed.
CRC cells were stained with CD133 antibody as mentioned previously. Then the goat anti-rabbit secondary antibody conjugated with Alexa Fluor 594 (ZSGB-Bio, China) was used. DAPI was used to counterstain nuclei. The fluorescence was scanned and photographed with a confocal laser scanning microscope (Olympus, Japan). The average fluorescence intensity was calculated with Image J software.
The low attachment plates (Corning Incorporated, United States) were used to culture cells using serum-free medium according to a previous study[14]. We prepared the serum-free medium for sphere culturing by adding 10 μg of EGF, 5 μg of LIF and 10 μg of bFGF (Invitrogen, United States) into 500 mL of DMEM/F12 medium. Cells were cultured in the 24-well ULLA plates at a density of 5000 or 10000 cells/well for 1 wk. Spheres (> 50 μm) were counted using an immunofluorescent microscope (Olympus, Japan).
SW480 cells were cultured and harvested. Following procedures were provided by chromatin immunoprecipitation (CHIP-IT) Express Enzymatic and Enzymatic shearing Kit (Active Motif). Mouse monoclonal anti-SATB2 antibody (Abcam, Cambridge, United Kingdom) was used. The positive and negative control antibodies were provided in CHIP-IT control (Active Motif). The immunoprecipitated DNA was amplified by PCR. The primer sequences for PCR are listed in Table 2.
| Gene | Sequence |
| CD133-F | 5’ TTTGTCTTCTATTCTTGGCTTC 3’ |
| CD133-R | 5’ ACCTTGTCATAATCAATTTTGG 3’ |
| CD44(1)-F | 5’ CTCATGGCTCAGTCGCCCAATCA 3’ |
| CD44(1)-R | 5’ TTTGCTCCTGAGCTGTTGCGTGG 3’ |
| CD44(2)-F | 5’ AGATTAAGGAGCTAGGACTC 3’ |
| CD44(2)-R | 5’ AAGATCACTTGGCAAGAAAG 3’ |
| CD44(3)-F | 5’ GGCACGTGTGAAACCTTTCCATTC 3’ |
| CD44(3)-R | 5’ GCTGAGCTGGACGCCAAGCA 3’ |
| CD44(4)-F | 5’ GCCTTTCATCCCTCGGGTGTGC 3’ |
| CD44(4)-R | 5’ TTCCTCCCAGGGACCAGGCC 3’ |
| MEIS2(1)-F | 5’ GGATTCCTGGCCAAAGGACGC 3’ |
| MEIS2(1)-R | 5’ CTCCCCCTAAGAGCGGCTCCA 3’ |
| MEIS2(2)-F | 5’ ACTGCCCGCAAGGATTCCACAA 3’ |
| MEIS2(2)-R | 5’ GGACTGTGGACCAAATCCAGCACAG 3’ |
| AXIN2-F | 5’ TATTCAAGGCATCTTTTACTGGAC 3’ |
| AXIN2-R | 5’ AGCAAAGAACTAGCCAATAAGGAG 3’ |
RNA-Seq expression data (combining level 3 data from IlluminaGA_RNASeqV2 platforms) from CRC patients were downloaded from The Cancer Genome Atlas (TCGA), which had been analyzed in Cancer Browser (https://genome-cancer.ucsc.edu/). Correlations between SATB2 and stem cell markers were analyzed according to data from these clinical samples.
We collected 68 fresh samples from CRC patients operated from March to April in 2010 at Nanfang Hospital. Among them, there were 45 males and 23 females. The average age was 63.77 ± 16.22 years. We collected the tumor tissue and its adjacent normal tissue. Then the tissues were preserved in liquid nitrogen and RNA was extracted and analyzed subsequently.
SPSS V.13.0 statistical software package was used to perform all statistical analyses. The Student’s t-test was used to compare two groups of independent samples. One-way ANOVA was used to analyze differences among multiple groups and differences between groups were analyzed by LSD pairwise comparison. Pearson correlation analysis was used to calculate the correlation between SATB2 and stem cell markers. P < 0.05 was considered to be statistically significant for all the analyses.
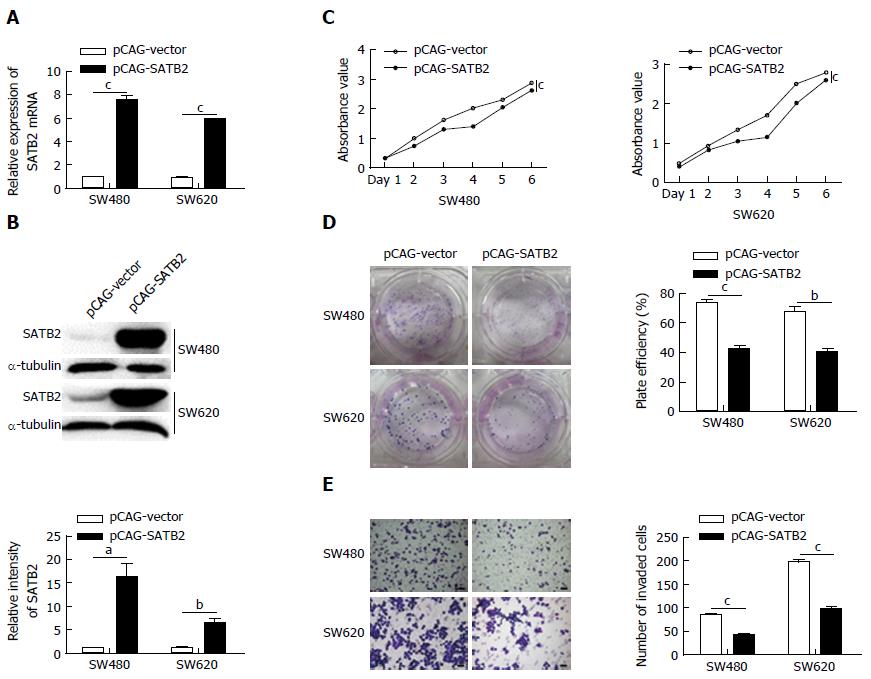
SATB2 was successfully overexpressed in SW480 and SW620 cells both at mRNA (SW480, P < 0.001; SW620, P < 0.001; Figure 1A) and protein (SW480, P < 0.05; SW620, P < 0.01; Figure 1B) levels. CCK-8 cell proliferation assay showed that overexpression of SATB2 inhibited cell proliferation in SW480 (P < 0.001) and SW620 (P < 0.001) cells (Figure 1C). Moreover, the colony formation assay indicated that cells with SATB2 overexpression had a deceased formation of colonies compared with control cells (SW480, P < 0.001; SW620, P < 0.01; Figure 1D). A significant decrease in cell migration was showed in CRC cells after the exogenous expression of SATB2 (SW480, P < 0.001; SW620, P < 0.001; Figure 1E).
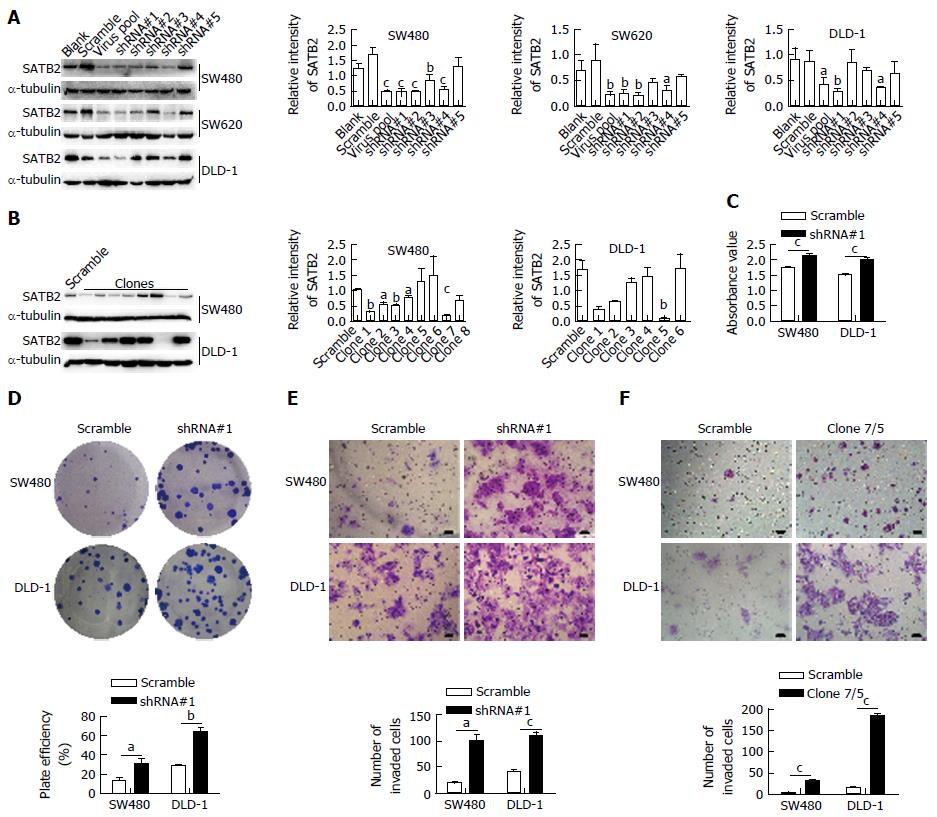
To further confirm the effect of SATB2 on the biological properties of CRC cells, we used the pLKO.1-TRC system with shRNA interference targeting SATB2 to produce virus to knock down SATB2 expression in CRC cells. The lentiviruses with different shRNAs targeting SATB2 were tested in SW480, SW620 and DLD-1 cells for optimal selection. The lentivirus with shRNA#1 targeting SATB2 had the optimal efficiency to knock down SATB2 expression in three tested CRC cell lines (SW480, P < 0.001; SW620, P < 0.01; DLD-1, P < 0.01) and was then used to establish the cell lines with SATB2 stable knockdown (Figure 2A). Single cells were isolated from the cells infected by the lentivirus with shRNA#1 targeting SATB2 and cultured for 2 wk to establish clones with SATB2 stable knockdown (Figure 2B). SW480/clone7 (P < 0.001) and DLD-1/clone5 (P < 0.01) were used in our next experiments. In contrast to our previous results, enhanced adhesion ability (SW480, P < 0.001; DLD-1, P < 0.001; Figure 2C), colony-forming capacity (SW480, P < 0.05; DLD-1, P < 0.01; Figure 2D) and migration ability (SW480/shRNA#1, P < 0.05; DLD-1/shRNA#1, P < 0.001; SW480/clone7, P < 0.001; DLD-1/clone5, P < 0.001; Figure 2E and F) were found in SW480 and DLD-1 cells after SATB2 was downregulated.
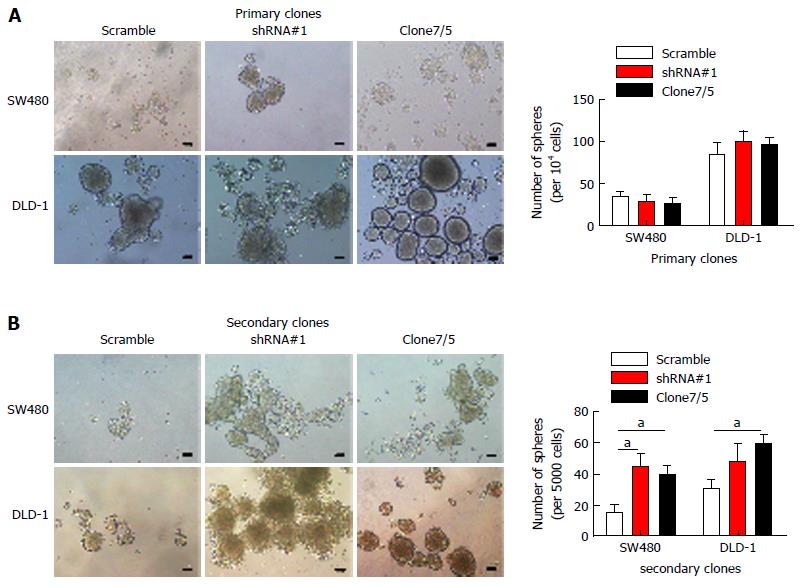
In our previous studies, we found that SATB2 expression was closely correlated with tumor invasion, lymph node metastasis, distant metastasis and Dukes’ classification in CRC patients[5]. Further, we found that SATB2 overexpression inhibited the proliferation and migration of CRC cells while knockdown of SATB2 promoted adhesion, colony-formation and migration of CRC cells in vitro. There is a subpopulation of CSCs that contributes to the biological traits of high-grade malignancy[15-17]. The CSCs were possibly the main cause of new tumor formation and tumor metastasis. We checked whether SATB2 could influence phenotype of stemness of CRC cells. As we know, self-renewal is one of the basic characteristics of stemness of CRC cells. So we observed the self-renewal of CRC cells using sequential sphere formation assay. Nevertheless, SATB2 knockdown had no effect on primary sphere formation in CRC cells (Figure 3A). Interestingly, more secondary sphere formation was found in SW480 and DLD-1 cells after SATB2 expression was knocked down (SW480/shRNA#1, P < 0.05; DLD-1/shRNA#1, P > 0.05; SW480/clone7, P < 0.05; DLD-1/clone5, P < 0.05; Figure 3B), indicating that SATB2 repressed the self-renewal ability of CRC cells.
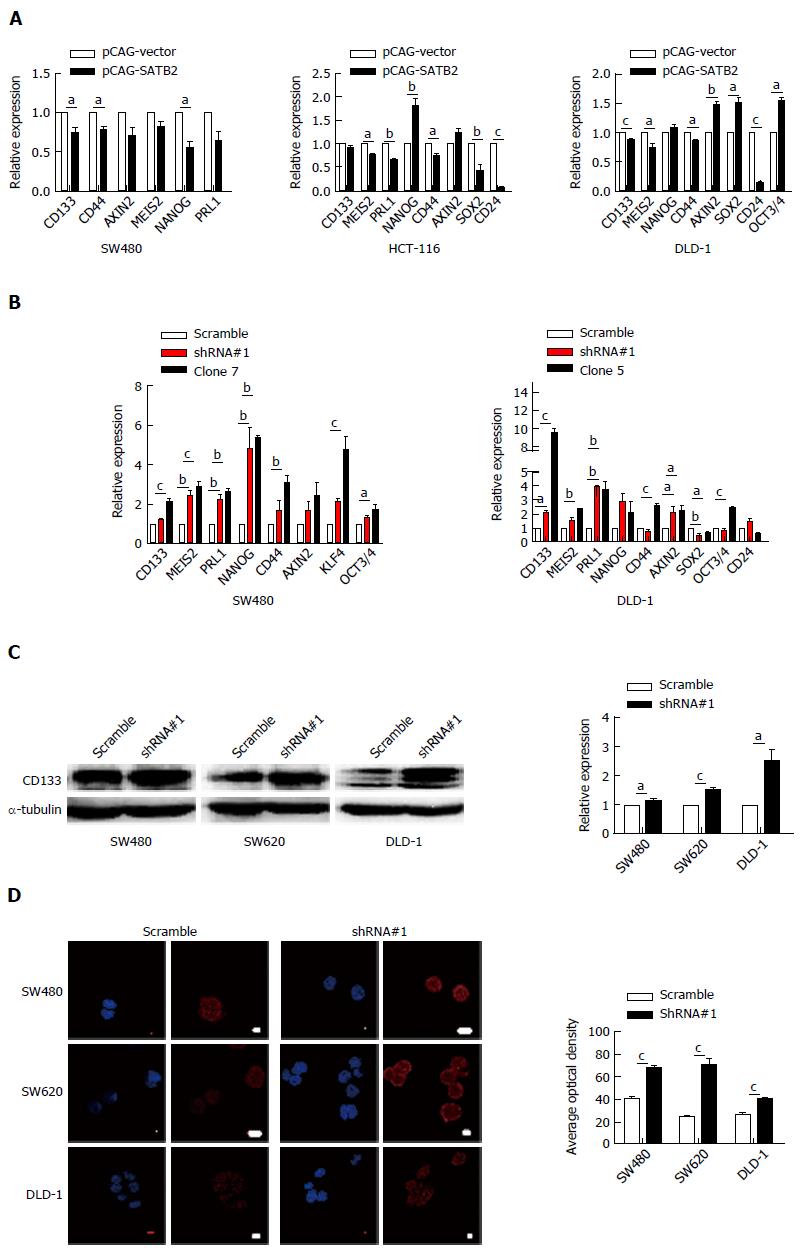
We found that SATB2 knockdown enhanced secondary sphere formation of CRC cells in vitro. It is logically supposed that SATB2 may affect the expression of markers of CSCs as a key transcriptional factor which controls gene expression. Therefore, we detected the mRNA expression of several key markers of CSCs, such as CD133, CD44, AXIN2, MEIS2 and NANOG, by qRT-PCR in CRC cells with gain or loss of SATB2 expression. Accordingly, most markers for stem cells were increased in cells with SATB2 knockdown and decreased in cells with SATB2 overexpression (Figure 4A and B), especially CD133 (SW480/pCAG-SATB2, P < 0.05; DLD-1/pCAG-SATB2, P < 0.001; SW480/clone7, P < 0.001; DLD-1/shRNA#1, P < 0.05; DLD-1/clone5, P < 0.001), CD44 (SW480/pCAG-SATB2, P < 0.05; HCT-116/pCAG-SATB2, P < 0.05; DLD-1/pCAG-SATB2, P < 0.05; SW480/clone7, P < 0.01; DLD-1/clone5, P < 0.001) and PRL1 (HCT-116/pCAG-SATB2, P < 0.01; SW480/shRNA#1, P < 0.01; SW480/clone7, P < 0.01; DLD-1/shRNA#1, P < 0.01; DLD-1/clone5, P < 0.01). Specifically, CD133 expression was further analyzed by Western blot (SW480, P < 0.05; SW620, P < 0.001; DLD-1, P < 0.05) and immunofluorescent staining (SW480, P < 0.001; SW620, P < 0.001; DLD-1, P < 0.001). And CD133 expression was increased in CRC cells after SATB2 was knocked down (Figure 4C and D).
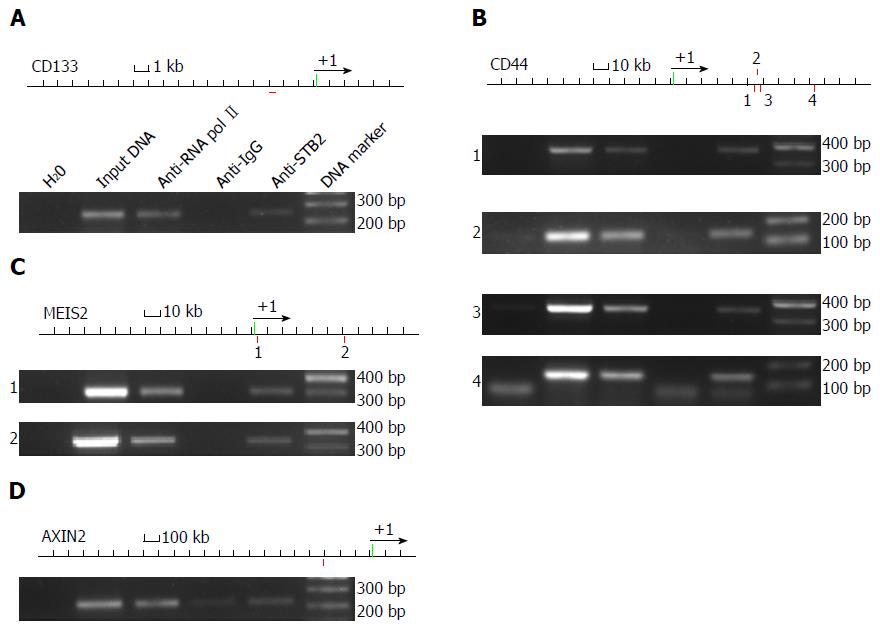
As a transcriptional factor, SATB2 may affect gene expression of stem cell markers by directly binding to regulatory elements of those genes. We used the Genomatix online software to find the possible SATB2 binding loci of those stem cell marker genes. We found that SATB2 may bind to regulatory elements of CD133 (Figure 5A), CD44 (Figure 5B), MEIS2 (Figure 5C) and AXIN2 (Figure 5D), at single or multiple sites. Then, we employed ChIP, followed by PCR, to test whether SATB2 could bind to regulatory elements of these genes. Chromatin fragments were prepared from SW480 cells. Mouse monoclonal anti-SATB2 antibody was used to precipitate the needed chromatin. Anti-RNA pol II and anti-IgG antibodies were used as positive and negative controls separately. Our results indicated that regulatory elements of CD133 (Figure 5A), CD44 (Figure 5B), MEIS2 (Figure 5C) and AXIN2 (Figure 5D) contained SATB2-binding sequences.
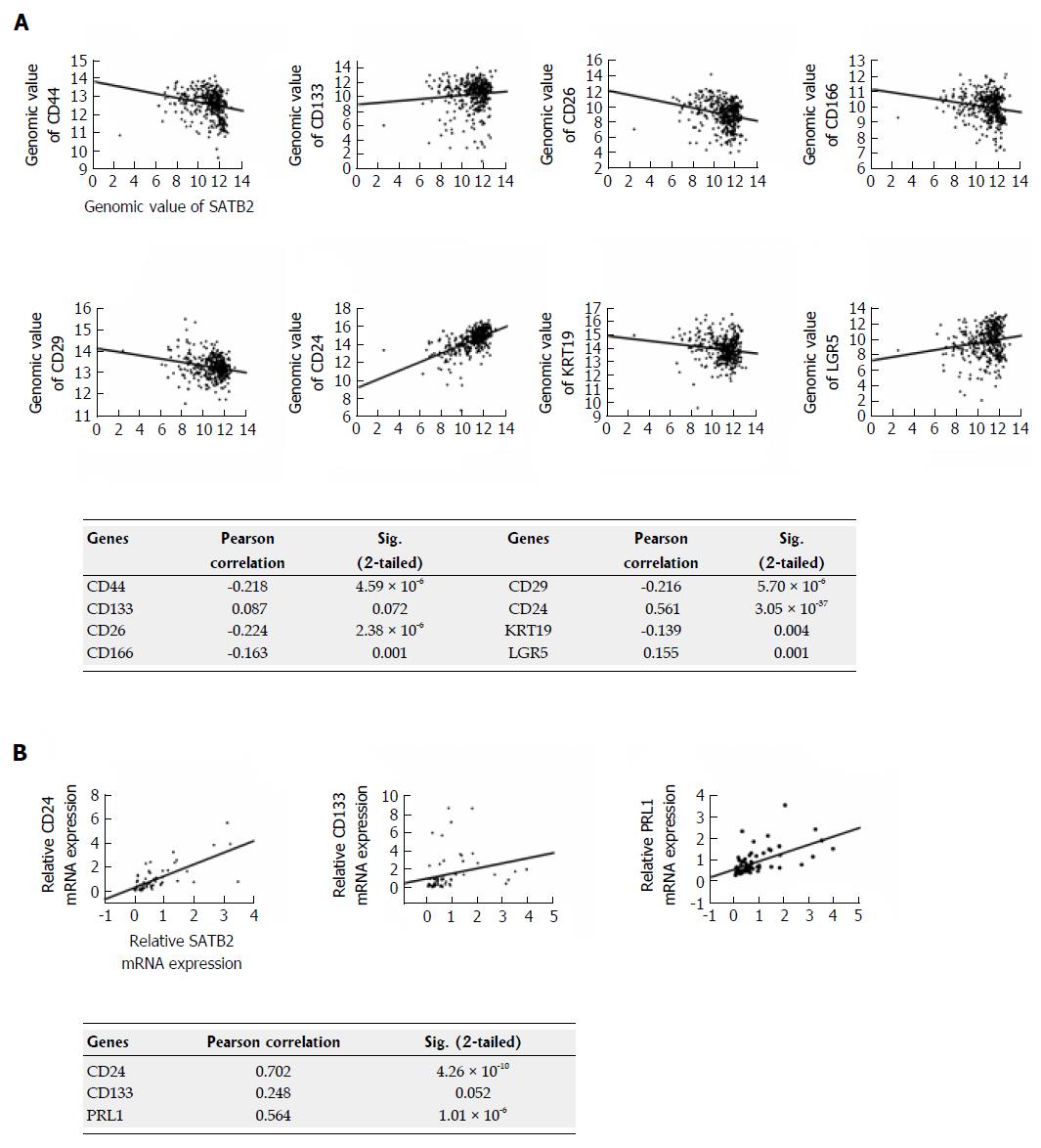
To further analyze the correlation between SATB2 and some key stem cell markers, RNA-Seq expression data in clinical samples of CRC from TCGA (http://cancergenome.nih.gov/) were used. Using data from TCGA, we found that SATB2 was negatively correlated with expression of CD44 (P < 0.001), CD26 (P < 0.001), CD166 (P < 0.01), CD29 (P < 0.001) and KRT19 (P < 0.01) and positively correlated with expression of CD24 (P < 0.001) and LGR5 (P < 0.01) (Figure 6A). Significantly, in our clinical CRC tissues, we further confirmed that SATB2 was positively correlated with CD24 (P < 0.001) expression (Figure 6B). However, the correlation between SATB2 and CD133 was marginal for significance analysis both in TCGA data (P = 0.072, Figure 6A) and our own clinical samples of CRC (P = 0.052, Figure 6B), suggesting that limited samples were included in both studies.
In our previous studies, SATB2 has been found to be a potential novel prognostic factor for CRC because of its strong correlation with local invasion, lymph node metastasis and distant metastasis in CRC[5]. After then, more evidence has confirmed SATB2 as a useful marker for CRC metastasis[18-21]. Even so, the mechanisms by which SATB2 is involved in CRC metastasis are still largely unclear.
Here, we found that SATB2 was a tumor suppressor in CRC. Gain-of-function studies showed that overexpression of SATB2 inhibited the proliferation and migration of CRC cells in vitro. Meanwhile, loss-of-function studies indicated that knockdown of SATB2 promoted adhesion, colony-formation and migration of CRC cells in vitro. These results are consistent with our clinical data, supporting the importance of SATB2 in tumor metastasis in CRC.
We also discovered that SATB2 was a negative regulator of stemness in CRC cells. At present, many solid tumors, including brain, colon, lung, breast, liver, prostate and bladder cancers, have been identified to have CSCs[13,22-26]. CSCs, commonly identified by their cell-surface-marker expression, have self-renewal and tumor-initiating ability and account for cancer relapse and metastasis[27]. Self-renewal ability of CSCs, also called stemness, is one of the basic characteristic of CSCs. Using primary and secondary sphere formation assay to detect self-renew of CRC cells, we found that SATB2 knockdown enhanced secondary sphere formation of CRC cells in vitro. Consistently, expression of CD133, NANOG and CD44, the key markers for CRC, was significantly increased when SATB2 was stably knocked down in CRC cells. Meanwhile, CD133 and CD44 were downregulated when SATB2 was overexpressed in CRC cells. As a transcriptional factor, SATB2 may regulate gene expression by directly binding to the regulatory elements of these stemness genes. NANOG was found to be directly regulated by SATB2 because of the binding to its promoter region[10]. In our study, Genomatix, an online web-based bioinformatic system, was first used to predict the potential genetic locus which might be recognized and bound by SATB2. We found that SATB2 could bind to one or more regulatory elements of CD133, CD44, MEIS2 and AXIN2. Importantly, ChIP assay confirmed that SATB2 could bind directly to the regulatory elements of CD133, CD44, MEIS2 and AXIN2. Interestingly, in TCGA clinical database and our clinical data of CRC, SATB2 was correlated with expression of several stem cell markers such as CD44 and CD24. Much more evidence is needed to explore the precise mechanisms by which SATB2 regulates stemness of cancer cells.
In conclusion, our studies found that SATB2 could directly bind to the regulatory elements in the genetic loci of several stem cell markers and consequently inhibit the progression of CRC by negatively regulating stemness of CRC cells.
Special AT-rich sequence-binding protein 2 (SATB2) is a key factor for transcriptional regulation and chromatin remodeling. Previously, the authors found that decreased expression of SATB2 was correlated with metastasis in colorectal cancer (CRC). Unfortunately, how SATB2 influences CRC metastasis is still unclear.
SATB2 and its analogue protein SATB1 are found to regulate embryonic stem cell differentiation by directly binding with NANOG genomic locus. Cancer stem cells (CSCs) with stem cell-like properties have been reported to enhance greatly tumor-initiating potential within a tumor. This implies the possible relationship between SATB2 and CSCs.
This is the first study to report the relationship between SATB2 and stemness of CRC cells. And our study provides a new mechanism for the involvement of SATB2 in CRC progression.
SATB2 inhibits the progression of CRC by negatively regulating stemness of CRC cells, which provides a new possible therapy for CRC patients.
SATB2 is a protein that binds AT sequence on the targeted genes to regulate their transcription. Low expression of SATB2 has been reported in CRC tissues. SATB2 expression was closely correlated with tumor invasion, lymph node metastasis, distant metastasis and Dukes’ classification in CRC patients. This predicts a possible role of SATB2 in regulating tumor metastasis.
This is a very interesting and may be a useful future technique. CRC is a leading cancerous disease affecting many people, therefore every method that could predict influencing factors on metastasis is very important for choosing the correct treatment or even follow-up schedule.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Furka A, Lakatos PT, Morris DLL, Perini MV S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Zhang FF
| 1. | Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Fariñas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 408] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 2. | Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, Tarabykin V. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am J Hum Genet. 2006;79:668-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 3. | Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 509] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 4. | Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 504] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 5. | Wang S, Zhou J, Wang XY, Hao JM, Chen JZ, Zhang XM, Jin H, Liu L, Zhang YF, Liu J. Down-regulated expression of SATB2 is associated with metastasis and poor prognosis in colorectal cancer. J Pathol. 2009;219:114-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 6. | Patani N, Jiang W, Mansel R, Newbold R, Mokbel K. The mRNA expression of SATB1 and SATB2 in human breast cancer. Cancer Cell Int. 2009;9:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | Chung J, Lau J, Cheng LS, Grant RI, Robinson F, Ketela T, Reis PP, Roche O, Kamel-Reid S, Moffat J. SATB2 augments ΔNp63α in head and neck squamous cell carcinoma. EMBO Rep. 2010;11:777-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Seong BK, Lau J, Adderley T, Kee L, Chaukos D, Pienkowska M, Malkin D, Thorner P, Irwin MS. SATB2 enhances migration and invasion in osteosarcoma by regulating genes involved in cytoskeletal organization. Oncogene. 2015;34:3582-3592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Zhao X, Qu Z, Tickner J, Xu J, Dai K, Zhang X. The role of SATB2 in skeletogenesis and human disease. Cytokine Growth Factor Rev. 2014;25:35-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 10. | Savarese F, Dávila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, Takahashi K, Jenuwein T, Kohwi-Shigematsu T, Fisher AG. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625-2638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Asanoma K, Kubota K, Chakraborty D, Renaud SJ, Wake N, Fukushima K, Soares MJ, Rumi MA. SATB homeobox proteins regulate trophoblast stem cell renewal and differentiation. J Biol Chem. 2012;287:2257-2268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 12. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7712] [Article Influence: 350.5] [Reference Citation Analysis (0)] |
| 13. | Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821-5828. [PubMed] |
| 14. | Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367-2382. [PubMed] |
| 15. | Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302-1313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 923] [Cited by in RCA: 907] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 16. | Pang R, Law WL, Chu AC, Poon JT, Lam CS, Chow AK, Ng L, Cheung LW, Lan XR, Lan HY. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 2010;6:603-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 427] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 17. | Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29:32-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 353] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 18. | Eberhard J, Gaber A, Wangefjord S, Nodin B, Uhlén M, Ericson Lindquist K, Jirström K. A cohort study of the prognostic and treatment predictive value of SATB2 expression in colorectal cancer. Br J Cancer. 2012;106:931-938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 19. | Yang MH, Yu J, Chen N, Wang XY, Liu XY, Wang S, Ding YQ. Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2. PLoS One. 2013;8:e85353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 20. | Yang MH, Yu J, Jiang DM, Li WL, Wang S, Ding YQ. microRNA-182 targets special AT-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J Transl Med. 2014;12:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Dragomir A, de Wit M, Johansson C, Uhlen M, Pontén F. The role of SATB2 as a diagnostic marker for tumors of colorectal origin: Results of a pathology-based clinical prospective study. Am J Clin Pathol. 2014;141:630-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 106] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 22. | Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM, Akita H, Takiuchi D, Hatano H, Nagano H. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120:3326-3339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 490] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 23. | Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C, De Maria R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1158] [Cited by in RCA: 1253] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 24. | Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 460] [Cited by in RCA: 453] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 25. | O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2977] [Cited by in RCA: 3045] [Article Influence: 160.3] [Reference Citation Analysis (0)] |
| 26. | Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946-10951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1998] [Cited by in RCA: 2033] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 27. | Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 718] [Cited by in RCA: 715] [Article Influence: 44.7] [Reference Citation Analysis (0)] |