Published online Oct 7, 2016. doi: 10.3748/wjg.v22.i37.8361
Peer-review started: May 23, 2016
First decision: June 20, 2016
Revised: July 4, 2016
Accepted: August 1, 2016
Article in press: August 1, 2016
Published online: October 7, 2016
Processing time: 132 Days and 15 Hours
To investigate autophagy-related genes, particularly ATG12, in apoptosis and cell cycle in hepatitis B virus (HBV)-associated hepatocellular carcinoma (HCC) and non-HBV-HCC cell lines.
The expression of autophagy-related genes in HBV-associated hepatocellular carcinoma and non-HBV-HCC cell lines and human liver tissues was examined by quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR) and western blotting. The silencing of target genes was used to examine the function of various genes in apoptosis and cell cycle progression.
The expression of autophagy related genes ATG5, ATG12, ATG9A and ATG4B expression was analyzed in HepG2.2.15 cells and compared with HepG2 and THLE cells. We found that ATG5 and ATG12 mRNA expression was significantly increased in HepG2.2.15 cells compared to HepG2 cells (P < 0.005). Moreover, ATG5-ATG12 protein levels were increased in tumor liver tissues compared to adjacent non-tumor tissues mainly from HCC patients with HBV infection. We also analyzed the function of ATG12 in cell apoptosis and cell cycle progression. The percentage of apoptotic cells increased by 11.4% in ATG12-silenced HepG2.2.15 cells (P < 0.005) but did not change in ATG12-silenced HepG2 cells under starvation with Earle’s balanced salt solution. However, the combination blockade of Notch signaling and ATG12 decreased the apoptotic rate of HepG2.2.15 cells from 55.6% to 50.4% (P < 0.05).
ATG12 is important for HBV-associated apoptosis and a potential drug target for HBV-HCC. Combination inhibition of ATG12/Notch signaling had no additional effect on HepG2.2.15 apoptosis.
Core tip: ATG5-ATG12 protein expression was increased in hepatitis B virus (HBV)-transfected HepG2.2.15 cells when they were compared with HepG2 cells and was increased in tumor liver tissues compared to adjacent non-tumor tissues from hepatocellular carcinoma (HCC) patients with HBV infection. We showed that silencing of ATG12 increased cell apoptosis of HepG2.2.15 cells but not HepG2 cells under starvation conditions. These results suggest that ATG5-ATG12 proteins are important for the survival of HBV-associated HCC during states of limited tumor nutrients. The inhibition of ATG12 might be a good target for HBV-associated HCC treatment.
- Citation: Kunanopparat A, Kimkong I, Palaga T, Tangkijvanich P, Sirichindakul B, Hirankarn N. Increased ATG5-ATG12 in hepatitis B virus-associated hepatocellular carcinoma and their role in apoptosis. World J Gastroenterol 2016; 22(37): 8361-8374
- URL: https://www.wjgnet.com/1007-9327/full/v22/i37/8361.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i37.8361
Previous studies have reported that HBV expression is correlated with autophagy induction[1-3]. HBV enhances and uses autophagy for its replication via the HBx protein, which binds and activates phosphatidylinositol-3-kinase class 3 (PIK3C3), an enzyme important for the initiation of autophagy. Autophagy inhibitors or the silencing of enzymes essential for the formation of autophagosomes suppresses HBV DNA synthesis with a minimal effect on the HBV mRNA levels[2]. The role of autophagy in the production of HBV virions was demonstrated in HBV transgenic mice with a liver-specific deficiency of Atg5[4]. We recently confirmed that ATG12 knock down reduced HBV DNA levels in HepG2.2.15 cells and induced the interferon signaling pathway, suggesting that autophagy machinery may aid HBV survival by reducing antiviral innate immunity[5].
Many studies have provided evidence to support the role of autophagy in human cancer. Beclin-1 was the first mammalian autophagy gene to be identified. The monoallelic deletion of Beclin-1 at chromosome 17q21 is sporadically observed in approximately 75% of ovarian cancers[6,7], 50% of breast cancers[8], and 40% of prostate cancers[9]. Other mutations in autophagy genes such as Atg5, Atg12, Atg9B are frequent in gastric and colon cancers[10]. UVRAG, a Beclin1-interacting protein[10,11] and Atg4C were also shown to suppress tumor gene activity[12]. Liver-specific Beclin-1 knockout heterozygous mice showed increased rates of hepatocellular carcinoma in old aged mice[13,14]. Furthermore, mosaic Atg5-/- mice developed benign liver tumors at 6-mo of age[15] and Atg7 hepatocyte-specific knockout mice also developed liver tumors later in life[15]. These data support the idea that autophagy defects contribute to tumorigenesis.
However, autophagy deficient cells can occur via cellular damage caused by dysfunctional mitochondria, oxidative stress, endoplasmic reticulum stress, necrosis and p62 accumulation[16]. The accumulation of cell damage can lead to chromosome instability[17] and inflammatory responses[18], resulting in tumor development. Although, autophagy functions as a tumor suppressor in primary cells, it is important for cancer cell survival. Interestingly, spontaneously occurring liver tumors did not progress in chimeric mice with Atg5 or Atg7 loss[15]. This finding implies that autophagy is required for tumor progression. Additionally, autophagy is required for cancer progression of other types of cancers. For example, an Atg3 deletion in hematopoietic cells prevents BCR-Abl-mediated leukemia[19]. Some tumor cells are susceptible to growth inhibition or death when autophagy is inhibited. Guo et al[20], found that Ras-driven tumors required autophagy for tumor cell survival upon starvation. Therefore, autophagy has a dual-function in cancer. It functions as a tumor suppressor during cancer initiation, but also functions to promote tumor progression and metastasis later in the development process.
Because HBV infection is associated with hepatocellular carcinoma (HCC) and requires the induction of autophagy for its survival, we investigated the involvement of autophagic genes in cancer cell survival using HBV-associated HCC and non HBV-HCC cell lines and liver tissues.
The immortalized human liver epithelial cell line (THLE-2; ATCC® CRL-2706™, Manassas, VA, United States) was cultured in BEGM medium (CC3170 Bullet Kit; Lonza, Walkersville, MD, United States) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 5 ng/mL epidermal growth factor and 70 ng/mL phosphoethanolamine and maintained at 37 °C in 5% CO2. The flasks were precoated with a mixture of 0.01 mg/mL fibronectin, 0.03 mg/mL bovine collagen type I and 0.01 mg/mL bovine serum albumin dissolved in BEBM medium. The human hepatocellular carcinoma (HepG2) and HBV-transfected HepG2.2.15 cell lines were obtained from Professor Antonio Bertoletti (Singapore Institute for Clinical Sciences, A*Star). These two cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, United States) supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and streptomycin at 37 °C in 5% CO2. For HepG2.2.15 cells, the medium was supplemented with 150 μg/mL of G418 (Geneticin; Gibco, United States) to maintain HBV plasmids.
To induce starvation conditions, the cells were incubated in serum-free Earle’s balanced salt solution (EBSS; starvation medium; Invitrogen, United States) for the indicated number of hours (between 4-8 h).
Human liver tissues were obtained from King Chulalongkorn Memorial Hospital. The use of tissue biopsy was approved by the Institutional Review Board (IRB No. 396/55), Faculty of Medicine, Chulalongkorn University. The sections of tumor tissue and adjacent non-tumor tissue were performed at the Department of Pathology, Chulalongkorn University. The tissue samples were divided into two groups: HBV-infected and non-HBV infected groups. The diagnosis of HBV infection from HCC patients was established by seropositivity for HBsAg, HBV viral DNA, anti-HBcAg and seronegativity to anti-HBsAg. All fresh tissue samples were snap frozen in liquid nitrogen and stored at -80 °C prior to use.
Total cellular RNA was extracted using Real Genomics Total RNA Extraction kit (RBC Bioscience, Taiwan). The quantity of RNA was measured using a spectrophotometer. RNA concentrations in a solution were read and a 260 nm absorbance reading of 1.0 was equivalent to about 40 μg/mL of RNA and the ratio of absorbance at 260 and 280 nm was used to assess the RNA purity of RNA preparations. Then 1 μg of total RNA was converted to cDNA using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, United States).
PCR reactions were performed in a 20 μL reaction volume in a 96-well plate (Applied Biosystems, United States). The PCR mixture contained 2 μL of cDNA template, 10 μL of commercial (2 ×) Power SYBR® Green PCR Master Mix (Applied Biosystems), and each primer at a final concentration of 0.5 μmol/L. Real-time PCR runs were carried out using an ABI Thermal cycler 7500 real-time PCR instrument (Applied Biosystems). The conditions for thermal cycling were as follows: initial denaturation at 95 °C for 10 min, followed by 40 amplification cycles at 95 °C for 15 s and then 60 °C for 1 min. For each PCR run, a negative (no-template) control was used to test for false-positive results or contamination. The absence of nonspecific amplification was confirmed by generating a melt curve using the Applied Biosystems real-time PCR system software. The primers utilized in this study are summarized in Table 1.
| Gene names | Primer sequences (5’-3’) | |
| Beclin-1 | BECN1-F | GGATCAGGAGGAAGC |
| BECN1-R | GATGTGGAAGGTTGC | |
| ATG5 | ATG5-F | GCTTCGAGATGTGTGGTTTGG |
| ATG5-R | ACTTTGTCAGTTACCAACGTCA | |
| ATG12 | ATG12-F | TTGTGGCCTCAGAACAGTTG |
| ATG12-R | GAGAGTTCCAACTTCTTGGTCTG | |
| ATG9A | ATG9A-F | CGTGTGGGAAGGACAG |
| ATG9A-R | GGCGCTTTCTCCACTC | |
| ATG4B | ATG4B-F | TCCATAGGCCAGTGGTACG |
| ATG4B-R | TGCACAACCTTCTGATTTCC | |
| B-actin | B-actin-F | ACCAACTGGGACGACATGGAGAA |
| B-actin-R | GTGGTGGTGAAGCTGTAGCC | |
To analyze the relative gene expression data, we used real-time quantitative PCR and the 2-ΔΔCT method[21]. The CT values were obtained from real-time PCR instrumentation. The quantity of mRNA relative to a reference gene was calculated using the formula 2-ΔCT, where ΔCT = (CT target RNA - CT reference RNA). Comparison of gene expression was based on a comparative CT method (ΔΔCT), and the relative RNA expression was quantified according to the formula of 2-ΔΔCT, where ΔΔCT = (CT target RNA (experimental group) - CT reference RNA (experimental group)) - (CT target RNA (control group) - CT reference RNA (control group)). Genes with high expression levels were selected for the functional study.
All cell lines and human liver tissues samples were lysed with RIPA buffer (25 mmol/L Tris-HCl, pH 7.6, 150 mmol/L NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate). After sonication, protein concentrations of cell lysates were measured using BCA assay (Thermo Scientific, United States). Then 20 μg of protein from each cell line was loaded onto a 12% polyacrylamide gel and run at 130 V for 80 min. The proteins were transferred onto nitrocellulose membranes and were immunoblotted with primary mAb against Atg9a, Atg12, cNotch1, GAPDH (dilution, 1:1000; rabbit mAb; Cell Signaling Technologies, United States) and Atg4b (dilution, 1:1000; mouse mAb; Abcam, United States). After washing, the membranes were incubated with secondary antibodies conjugated with horseradish peroxidase (dilution 1:2000) and were analyzed with enhanced chemiluminescence (ECL) substrate (SuperSignal West Femto Chemiluminescent Substrate; Thermo Scientific). Images were quantified using a Licor-odyssey image system (LI-COR®, United States).
HepG2 cells were grown in DMEM supplemented with 10% FBS, antibiotic-free at 37 °C in 5% CO2 for 24 h. HepG2 cells were transfected with 500-1000 ng of pGFP-HBx plasmids (Addgene, MA, United States) or pGFP (negative control) using 1-3 μL of Lipofectamine® 2000 Transfection Reagent (Invitrogen, United States). Opti-MEM®I Reduced Serum Medium (Invitrogen) was used to dilute Lipofectamine® 2000 and plasmids, which were then gently mixed at a 1:1 ratio, incubated for 40 min at room temperature. HepG2 cells were diluted in complete growth medium without antibiotics to a final concentration of 4 × 105 cells/mL, then the cells were added to culture plates with plasmid-Lipofectamine® 2000 complexes and gently mixed by rocking the plate. The cells were incubated for 24-72 h at 37 °C in a 5% CO2 incubator. The relative autophagic gene expression was detected by real-time quantitative PCR.
HepG2 and HepG2.2.15 cells were transiently transfected with 500 ng of short hairpin RNA (shRNA) plasmid specific to ATG12 or a negative control (SureSilencing shRNA Plasmid; Qiagen, United States). The shATG12 plasmids contained following sequence: 5’-GCAAATCCTCTATGCCTTCTT-3’. Negative controls were inserted with a mock target sequence as follows: 5’-GGAATCTCATTCGATGCATAC-3’. Transfection of shRNA plasmids was carried out using Lipofectamine® 2000 (Invitrogen) according to the manufacturer’s instructions. At 72 h post-transfection, cells were collected and the silencing efficiency of the shRNA was detected by western blotting.
Specific autophagic silenced cells (shATG12) were assayed for apoptosis using PE-labeled annexin V staining (BioLegend, San Diego, CA, United States) and Fixable Viability Dye eFluor® 780 staining (eBioscience, CA, United States). Total apoptotic cells and early apoptotic cells were defined as annexin V-positive and Fixable Viability Dye eFluor® 780-negative and late apoptotic cells were defined as annexin V-positive and Fixable Viability Dye eFluor® 780-positive. For cell cycle analysis, cells were fixed with 70% ice-cold ethanol for at least 2 h on ice. The supernatant was removed, and the pellet was washed and treated with RNaseA to resuspend cells. Cells were then stained with propidium iodide (PI) solution at room temperature in the dark for 30 min. Cells were acquired on a FACScan (BD Biosciences, United States) and analyzed with FlowJo 10.0 software (FlowJo LLC., United States).
Significant differences were determined by unpaired t test with GraphPad Prism software, version 5.0 (San Diego, CA, United States). Statistical significance was set at a P value of < 0.05.
To investigate the involvement of autophagy in HCC, we used Beclin-1 as a marker for HBV-induced autophagy under starvation conditions. We analyzed Beclin-1 mRNA expression in HBV-transfected HepG2.2.15 cells and the parental cell line, HepG2. At 4 h post-starvation, Beclin-1 was up regulated in both cell lines and was then down regulated at 8 h (data not shown). Thus, we selected autophagy induction with starvation in EBSS for 4 h for all subsequent experiments.
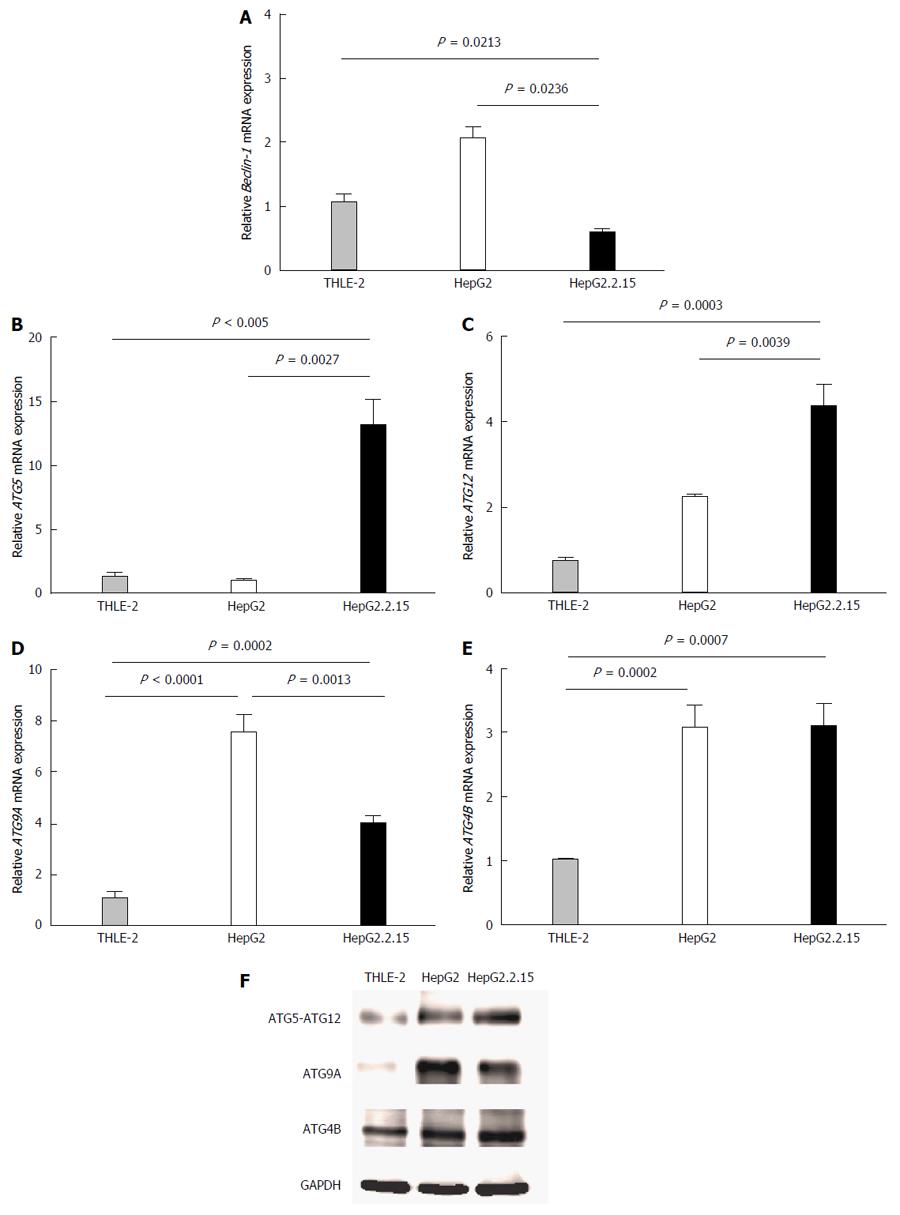
Autophagy related genes that represent each major part of the autophagy machinery including (1) induction step (Beclin1); (2) the first ubiquitin-like conjugation molecules (ATG5 and ATG12); (3) the cysteine protease that catalyzes the second ubiquitin-like conjugation molecules (ATG4B); and (4) the transporter membrane required for autophagosome formation (ATG9A) were tested. The mRNA expression levels of these selected molecules were determined at baseline and at 4 h post-culture in EBSS. There were no significant differences at baseline (data not shown). At 4 h post starvation, Beclin1 mRNA was down regulated in HepG2.2.15 compared to THLE2 and HepG2 (P = 0.0213 and P = 0.0236, respectively) (Figure 1A). No significant difference was observed for ATG4B expression between HBV-transfected HepG2.2.15 and HepG2, but it was significantly increased in HepG2 and HepG2.2.15 compared with THLE-2 (P = 0.0002 and P = 0.0007, respectively) (Figure 1E). ATG9A mRNA expression was higher in HepG2 and HepG2.2.15 compared to THLE2 but significantly down regulated in HBV-transfected HepG2.2.15 compared with HepG2 (P = 0.0013) (Figure 1D). Only the mRNA expression of ATG5 and ATG12 was significantly increased in HBV-transfected HepG2.2.15 compared with HepG2 cells (P = 0.0027 (Figure 1B) and P = 0.0039 (Figure 1C), respectively). The results of mRNA levels were confirmed by western blot analysis. ATG5-ATG12 proteins were up regulated in HBV-transfected HepG2.2.15 whereas ATG9A were down regulated compared to HepG2 cells. There was no difference in ATG4B protein expression between HBV-transfected HepG2.2.15 and HepG2 (Figure 1F).
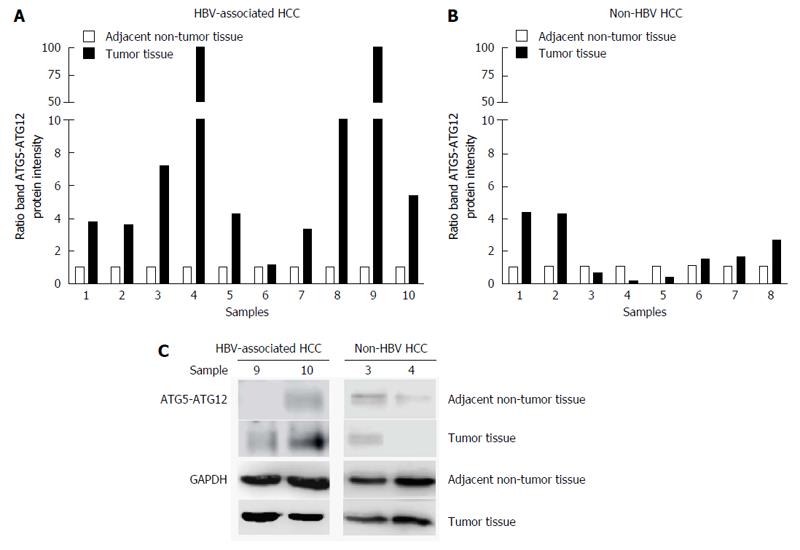
Next, we analyzed ATG5-ATG12 levels in tumor tissues compared to adjacent non-tumor tissues from HCC patients with or without HBV infection. In HBV-associated HCC tissues, ATG5-ATG12 protein levels were increased in the majority of tumor liver tissues compared to adjacent non-tumor tissues (9/10 sample pairs) (Figure 2A and C). In non-HBV HCC, ATG5-ATG12 protein levels were only increased in 3/8 tumor liver tissue samples (Figure 2B and C). These results suggested that ATG5-ATG12 proteins might have a selective advantage in HBV-associated HCC compared to non-HBV HCC.
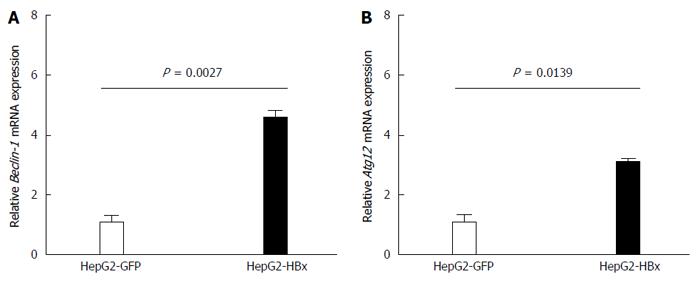
The X protein was shown to induce HBV replication by increasing Beclin-1 transcription leading to the induction of autophagy[1]. In this study, we analyzed the effect of HBx on Beclin-1 and on ATG12 by transfection of HepG2 cells with the pGFP-HBx plasmid. Then, we analyzed Beclin-1 and ATG12 mRNA expressions using quantitative real-time RT-PCR. We found that Beclin-1 (P = 0.0027) (Figure 3A) and ATG12 (P = 0.0139) (Figure 3B) mRNA expression were significantly increased in HepG2-GFP-HBx compared with HepG2-GFP after 48 h of transfection. These results suggested that HBX plays a role in ATG12 induction, either directly or indirectly through Beclin1.
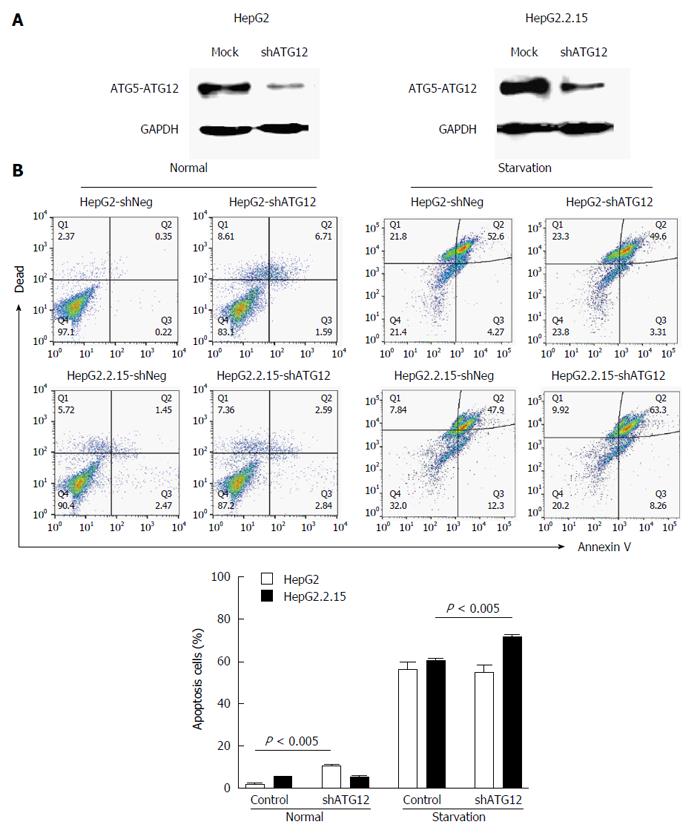
Next, we studied the functional role of the ATG12 autophagic gene by monitoring the biological effect after knockdown of its expression. Cells containing plasmids with the greatest silencing efficiency were subjected to western blot analysis for protein expression. The transfection efficiency was estimated by monitoring GFP expression under inverted fluorescence microscope (data not shown). The protein level of ATG12 in transfected HepG2 and HepG2.2.15 cells was decreased compared with mock controls (Figure 4A).
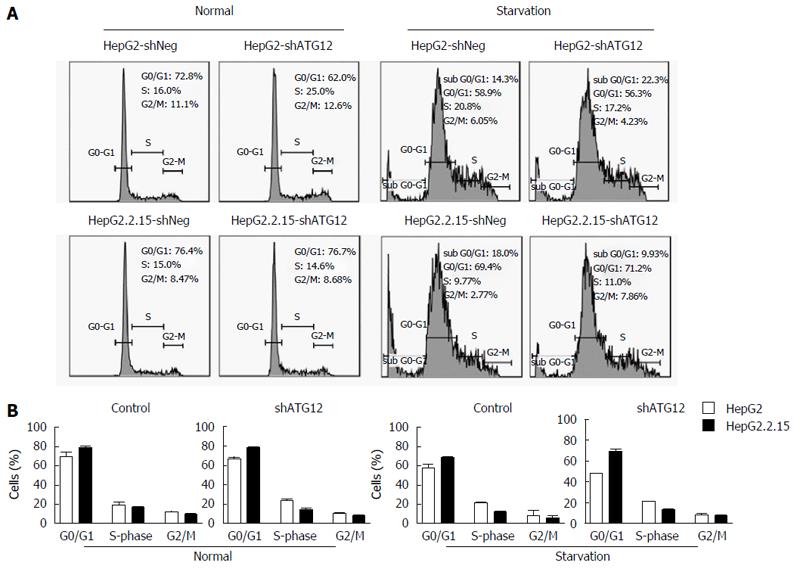
We investigated apoptosis and cell cycle progression in HepG2.2.15 cells compare to HepG2 cells after the silencing of ATG12. The percentage of apoptotic cells was slightly increased (8.3%) in ATG12-silenced HepG2 but unchanged in HepG2.2.15 under normal conditions, which is similar to what we have previously reported[5]. Under starvation conditions with EBSS, the percentage of apoptotic cells increased 11.4% in ATG12-silenced HepG2.2.15 but did not change in ATG12-silenced HepG2 (Figure 4B). No significant changes in cell cycle progression were observed in either cell line (Figure 5). These results suggest that ATG5-ATG12 proteins are important for the survival of HBV-associated HCC during states of limited tumor nutrients.
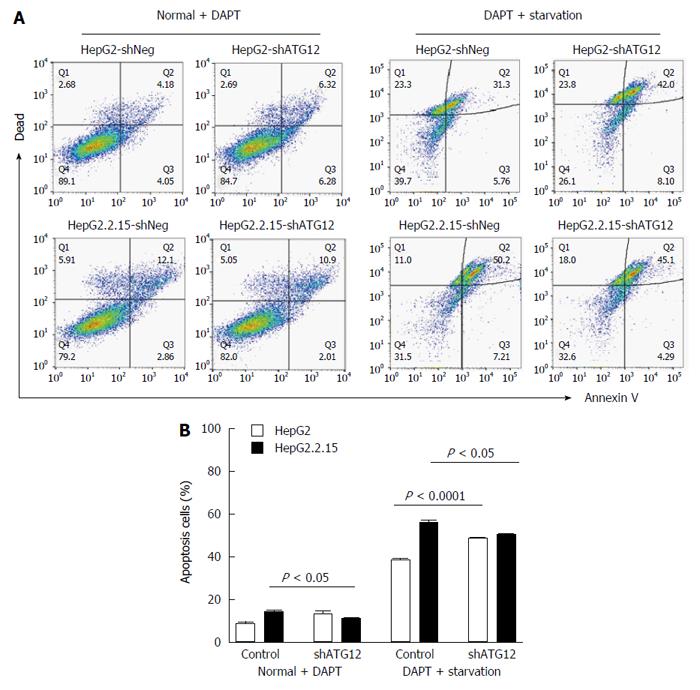
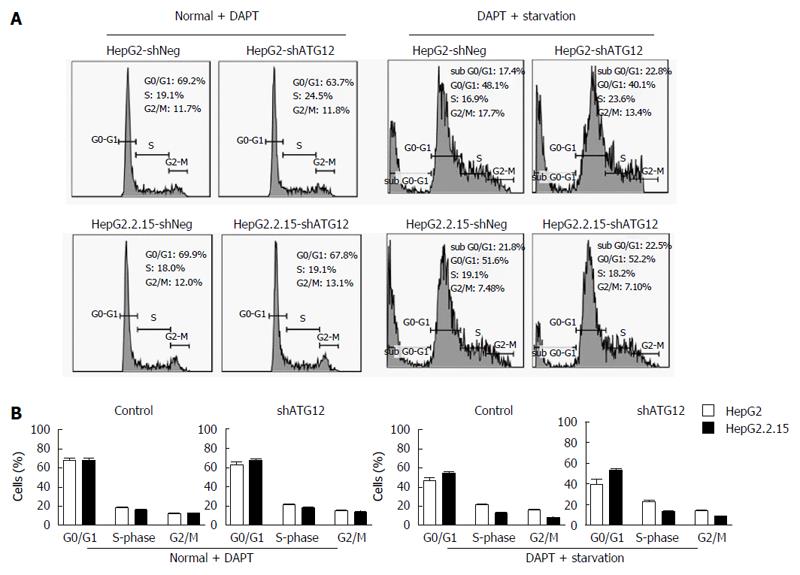
We recently reported that Notch signaling played a role in cell cycle progression and apoptosis, particularly in HBV-associated HCC[22]. Because ATG12 was up regulated in HepG2.2.15 compared to HepG2, and plays a role in cell apoptosis, we assessed the effect of ATG12 silencing in combination with a Notch inhibitor in the HepG2.2.15 cell line. Gamma-secretase inhibitors, also known as N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; DAPT, is used to block the Notch pathway[23]. When using DAPT combined with gene silencing of ATG12 under starvation conditions, HepG2 cell apoptosis was increased from 38.5% to 48.3%, compared to DAPT treatment alone, although there was no synergistic effect (Figure 6). Interestingly, the combination blockade of DAPT and ATG12 gave the opposite result by decreasing the apoptosis rate in HepG2.2.15 from 55.6% to 50.4% (Figure 6). No significant changes in cell cycle progression were observed for the combination treatment of ATG12 siRNA and DAPT (Figure 7). These results suggested that autophagy and Notch signaling are independent from each other in HBV-expressing cells compared to non-HBV HCC.
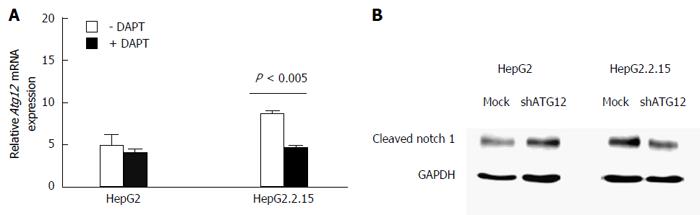
Finally, we analyzed ATG12 expression after treatment with DAPT and detected cleaved Notch1 expression after silencing ATG12 genes at baseline. ATG12 mRNA was decreased by DAPT treatment in HepG2.2.15 (Figure 8A). Moreover, cleaved Notch1 was diminished in ATG12-silenced HepG2.2.15 under normal conditions (Figure 8B).
Autophagy has been reported to be associated with HBV and HCC[13,15,24]. However, the mechanism is still poorly understood. Moreover, recent evidence suggests that specific ATG genes might contribute differently to viral infection and cancer development[25-28]. Some ATG genes have autophagy independent functions[29-32]. In this study, we focused on the ATG5-ATG12 complex that was up regulated in HepG2.2.15 and were highly expressed in HBV-associated HCC compared to non-HBV cell lines and HCC.
HBX appears to play a role in ATG12 induction, probably indirectly through the stimulation of Beclin-1 and PIK3C3[1,2]. We showed that silencing of ATG12, which represented the elimination of the ATG5-ATG12 complex, increased cell apoptosis under starvation conditions in HepG2.2.15 but not in HepG2. These results suggested that ATG5-ATG12 is important for the survival of HBV-associated HCC during states of limited tumor nutrients. A previous study by Mao et al[33] showed that HepG2.2.15 cells have a higher survival rate than HepG2 cells after culturing in a starvation medium for 48 h. Cell death was reduced through activation of the autophagy pathway. However, our study did not show any significant difference between the rate of cell death for HepG2 and HepG2.2.15 after starvation for 4 h. This might be explained by our shorter starvation time. The inhibition of autophagy-induced liver cancer cell apoptosis under starvation conditions was reported by Chang et al[34] They showed that HepG2 and HuH6 induced autophagy (Beclin-1, ATG5 and LC3B expression) for cell survival under starvation conditions with serum-free medium or chemotherapy. The inhibition of autophagy via the silencing of autophagic genes (Beclin-1 and ATG5) or 3-MA treatment induced HuH6 cell apoptosis under starvation conditions[34]. A study by Liu et al[35] demonstrated that starvation with serum-free medium for 48 h induced autophagy and apoptosis in 7702, HepG2, Hep3B and Huh7 cell lines; however, the inhibition of autophagy via Bafilomycin A1 was not affected in HepG2 apoptosis under starvation conditions similar to our result. These results imply that targeting the inhibition of autophagy might have different effects depending on the cell type and various metabolic stress conditions present.
Although ATG12 has been demonstrated to have pro-apoptotic activity through the ATG12-ATG3 conjugate[36], ATG12 also has anti-apoptotic activity. In mammalian HeLa cells, disruption of autophagy by silencing ATG12 promotes cells to die via apoptosis under starvation conditions[37]. ATG12 and other autophagic proteins (LC3 and ATG5) are associated with mitochondrial quality control of human umbilical vein endothelial cells. ATG12, ATG5 and LC3B were up-regulated after mitochondrial damage, leading to increased anti-apoptotic effects and increased life span in an in vitro aging model[38]. The combination of trastuzumab with silencing of ATG12 reduces cell viability and tumor growth in nude mice[39]. Therefore, we suggest that the inhibition of ATG12 might be a good target for the treatment of HBV-associated HCC.
Many cancer drug treatments increase tumor cell autophagy to protect cancer cells from apoptosis. Autophagy inhibitors in combination with chemotherapy are designed to induce apoptosis in human cancers. For example, autophagy inhibition was previously shown to enhance the growth inhibitory effects of sorafenib[40] as well as the combination of vorinostat with sorafenib in HCC cell lines[41]. In this study, we examined apoptosis cell death after the inhibition of ATG12 in combination with Notch signaling in HepG2 and HepG2.2.15 cell lines. The combination treatment increased cell apoptosis in HepG2 cells. However, when we blocked both Notch signaling and ATG12 under starvation conditions, cell apoptosis did not increase. There appears to be some interaction between ATG12 and Notch signaling, specifically in HepG2.2.12, because the Notch inhibitor DAPT caused the down-regulation of ATG12 mRNA but this was not observed in HepG2. In addition, the inhibition of ATG12 impaired Notch activation in HepG2.2.15 but increased Notch activation in HepG2.
This observation regarding the regulation between autophagy and notch signaling in HepG2.2.15 was unexpected. Both autophagy and Notch signaling are highly conserved signaling pathways in eukaryotic cells. The core Notch signaling pathway is through the binding of Notch ligand to Notch receptor which induces two proteolytic cleavages, metalloprotease and γ-secretase, to release the Notch intracellular domain, which translocates to the nucleus and binds to DNA to regulate the transcription of target genes[42]. The Notch pathway is associated with both tumor suppression and tumorigenesis in HCC[43,44]. The connection of autophagy and Notch signaling was demonstrated in Drosophila where the ATG4 mutation enhanced the notched-wing phenotype resulting in defective Notch signaling[45]. In contrast, the overexpression of ATG1 enhanced the notched-wing phenotype[46], suggesting that some autophagic proteins have additional functions independent of autophagy that are related to the enhancement or suppression of Notch signaling.
Mammalian target of rapamycin (mTOR) is a positive regulator of Notch signaling[47] and a negative regulator of autophagy[48]. Previous reports have shown that silencing of Notch1 activated phosphorylated Akt and decreased mTOR to induce glioblastoma cell apoptosis[49]. The inhibition of Notch signaling induced autophagy via PTEN-PI3K/Akt/mTOR pathway to promote the adipogenic differentiation of BM-MSCs[50]. Other studies have demonstrated that autophagy regulates Notch signaling by reducing the impact of Notch1 on stem cell differentiation. The silencing of autophagy (ATG7 or ATG16L1) induced Notch1 and cleaved Notch1 in HEK cells[51].
The unexpected relationship between autophagy and Notch signaling in HepG2.2.15 was different from HepG2 and might be explained by the HBV genome. The HBV X protein activates the NF-κB pathway (p65 and p50) via Notch signaling through a specific ligand and receptor to promote cell survival[52]. After treatment with DAPT, NF-κB expression was decreased[53]. Thus, the activation of NF-κB can either stimulate or inhibit autophagy via the upregulation of Beclin-1 in T-cells[54], whereas prolonged NF-κB activation suppresses ATG5 and Beclin-1 expression in macrophages[55]. However, the connection between HBV and Notch related autophagy needs further study.
In conclusion, our previous study demonstrated the role of autophagy machinery in HBV replication. We found that ATG12-knockdown reduced HBV DNA levels in HepG2.2.15 and induced the interferon signaling pathway. Thus, the ATG12 machinery of autophagy may aid HBV survival by reducing antiviral innate immunity[5]. Moreover, this study demonstrated that autophagy is related to HBV-associated cell death responses. Atg12 silenced HepG2.2.15 showed increased apoptosis under starvation conditions. The function of Atg12 seems to be dominated in HBV-associated HCC. These results suggested that ATG5-ATG12 is important for the survival of HBV-associated HCC during states of limited tumor nutrients. However, as mentioned previously that autophagy has a dual-function in cancer. Despite its role as tumor promotion in the later phase, it can act as a tumor suppressor during cancer initiation. Therefore, the therapeutic intervention that target autophagy has to take this information into consideration too. Moreover, our result showed that the use of autophagy inhibition in combination with other anti-tumor therapies such as Notch inhibitor, might not be a good strategy. Further characterization of HBV and Notch related autophagy is needed.
Hepatitis B virus (HBV) infection is associated with hepatocellular carcinoma (HCC) and has been proved to induce autophagy for its replication and survival. The involvement of some autophagic genes on cancer cell survival is still unknown. Therefore, we investigated the role of ATG genes in HBV-associated HCC and non HBV-HCC in this study.
Autophagy is a catabolic process for cell survival under nutrient limitation or stress conditions. However, autophagy can act as either an oncogene or tumor suppressor gene. A recent report showed that HBV used the autophagic pathway for its own benefit; however the molecular mechanism is still unclear.
ATG5-ATG12 protein was up regulated in HepG2.2.15 and expressed in a high percentage of HBV-associated HCC compared to non-HBV cell lines and HCC. The Atg12 silenced HepG2.2.15 cell line had increased apoptosis under starvation conditions. This is the first study to show a function of Atg12 in apoptosis in HBV-associated HCC.
These findings raise the possibility of targeting the autophagic pathway for the treatment of HBV-associated HCC patients.
This paper reported that ATG5-ATG12 protein expression was increased in HBV-transfected HepG2.2.15 cells compared to HepG2 cells and was increased in tumor liver tissues compared to adjacent non-tumor tissues from HCC patients with HBV infection. The silencing of ATG12 increased cell apoptosis under starvation conditions in HepG2.2.15 cells but not in HepG2 cells. Their results suggest that ATG5-ATG12 is important for the survival of HBV-associated HCC in the state of tumor nutrient limitation. The inhibition of ATG12 might be a good target for HBV-associated HCC.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Thailand
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Kim K, Rodriguez-Frias F S- Editor: Ma YJ L- Editor: A E- Editor: Wang CH
| 1. | Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 2. | Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383-4388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 254] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 3. | Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319-6333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 231] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 4. | Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol. 2011;85:13453-13456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 5. | Kunanopparat A, Hirankarn N, Kittigul C, Tangkijvanich P, Kimkong I. Autophagy machinery impaired interferon signalling pathways to benefit hepatitis B virus replication. Asian Pac J Allergy Immunol. 2016;34:77-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59-65. [PubMed] |
| 7. | Eccles DM, Russell SE, Haites NE, Atkinson R, Bell DW, Gruber L, Hickey I, Kelly K, Kitchener H, Leonard R. Early loss of heterozygosity on 17q in ovarian cancer. The Abe Ovarian Cancer Genetics Group. Oncogene. 1992;7:2069-2072. [PubMed] |
| 8. | Saito H, Inazawa J, Saito S, Kasumi F, Koi S, Sagae S, Kudo R, Saito J, Noda K, Nakamura Y. Detailed deletion mapping of chromosome 17q in ovarian and breast cancers: 2-cM region on 17q21.3 often and commonly deleted in tumors. Cancer Res. 1993;53:3382-3385. [PubMed] |
| 9. | Gao X, Zacharek A, Salkowski A, Grignon DJ, Sakr W, Porter AT, Honn KV. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995;55:1002-1005. [PubMed] |
| 10. | Lebovitz CB, Bortnik SB, Gorski SM. Here, there be dragons: charting autophagy-related alterations in human tumors. Clin Cancer Res. 2012;18:1214-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639-645. [PubMed] |
| 12. | Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573-18583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 316] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 13. | Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-1820. [PubMed] |
| 14. | Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077-15082. [PubMed] |
| 15. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1059] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 16. | Roy S, Debnath J. Autophagy and tumorigenesis. Semin Immunopathol. 2010;32:383-396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367-1381. [PubMed] |
| 18. | Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237-248. [PubMed] |
| 19. | Altman BJ, Jacobs SR, Mason EF, Michalek RD, MacIntyre AN, Coloff JL, Ilkayeva O, Jia W, He YW, Rathmell JC. Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis. Oncogene. 2011;30:1855-1867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 106] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 20. | Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1090] [Cited by in RCA: 1052] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 21. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133635] [Article Influence: 5568.1] [Reference Citation Analysis (1)] |
| 22. | Kongkavitoon P, Tangkijvanich P, Hirankarn N, Palaga T. Hepatitis B Virus HBx Activates Notch Signaling via Delta-Like 4/Notch1 in Hepatocellular Carcinoma. PLoS One. 2016;11:e0146696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 23. | Purow B. Notch inhibition as a promising new approach to cancer therapy. Adv Exp Med Biol. 2012;727:305-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 24. | Kotsafti A, Farinati F, Cardin R, Cillo U, Nitti D, Bortolami M. Autophagy and apoptosis-related genes in chronic liver disease and hepatocellular carcinoma. BMC Gastroenterol. 2012;12:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 25. | Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314-9325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 286] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 26. | Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26:1401-1410. [PubMed] |
| 27. | Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500-508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 326] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 28. | Dreux M, Chisari FV. Impact of the autophagy machinery on hepatitis C virus infection. Viruses. 2011;3:1342-1357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050-14055. [PubMed] |
| 30. | Takeshita F, Kobiyama K, Miyawaki A, Jounai N, Okuda K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy. 2008;4:67-69. [PubMed] |
| 31. | Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397-409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 209] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 32. | Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, Sharma AK, Amin S, Hu CD, Zhang J. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455-12468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 298] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 33. | Mao Y, Da L, Tang H, Yang J, Lei Y, Tiollais P, Li T, Zhao M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun. 2011;415:68-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 34. | Chang Y, Chen L, Liu Y, Hu L, Li L, Tu Q, Wang R, Wu M, Yang J, Wang H. Inhibition of autophagy may suppress the development of hepatoblastoma. FEBS J. 2011;278:4811-4823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 35. | Liu K, Shi Y, Guo XH, Ouyang YB, Wang SS, Liu DJ, Wang AN, Li N, Chen DX. Phosphorylated AKT inhibits the apoptosis induced by DRAM-mediated mitophagy in hepatocellular carcinoma by preventing the translocation of DRAM to mitochondria. Cell Death Dis. 2014;5:e1078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 36. | Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590-600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 37. | Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025-1040. [PubMed] |
| 38. | Mai S, Muster B, Bereiter-Hahn J, Jendrach M. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy. 2012;8:47-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 39. | Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Urruticoechea A, Martin-Castillo B, Menendez JA. Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies: utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment. Oncotarget. 2012;3:1600-1614. [PubMed] |
| 40. | Shi YH, Ding ZB, Zhou J, Hui B, Shi GM, Ke AW, Wang XY, Dai Z, Peng YF, Gu CY. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 276] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 41. | Yuan H, Li AJ, Ma SL, Cui LJ, Wu B, Yin L, Wu MC. Inhibition of autophagy significantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J Gastroenterol. 2014;20:4953-4962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 42. | Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678-689. [PubMed] |
| 43. | Viatour P, Ehmer U, Saddic LA, Dorrell C, Andersen JB, Lin C, Zmoos AF, Mazur PK, Schaffer BE, Ostermeier A. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med. 2011;208:1963-1976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 44. | Strazzabosco M, Fabris L. Notch signaling in hepatocellular carcinoma: guilty in association! Gastroenterology. 2012;143:1430-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | Thumm M, Kadowaki T. The loss of Drosophila APG4/AUT2 function modifies the phenotypes of cut and Notch signaling pathway mutants. Mol Genet Genomics. 2001;266:657-663. [PubMed] |
| 46. | Kwon MH, Callaway H, Zhong J, Yedvobnick B. A targeted genetic modifier screen links the SWI2/SNF2 protein domino to growth and autophagy genes in Drosophila melanogaster. G3 (Bethesda). 2013;3:815-825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Ma J, Meng Y, Kwiatkowski DJ, Chen X, Peng H, Sun Q, Zha X, Wang F, Wang Y, Jing Y. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Invest. 2010;120:103-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 198] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 48. | Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870-34879. [PubMed] |
| 49. | Zhao N, Guo Y, Zhang M, Lin L, Zheng Z. Akt-mTOR signaling is involved in Notch-1-mediated glioma cell survival and proliferation. Oncol Rep. 2010;23:1443-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 50. | Song BQ, Chi Y, Li X, Du WJ, Han ZB, Tian JJ, Li JJ, Chen F, Wu HH, Han LX. Inhibition of Notch Signaling Promotes the Adipogenic Differentiation of Mesenchymal Stem Cells Through Autophagy Activation and PTEN-PI3K/AKT/mTOR Pathway. Cell Physiol Biochem. 2015;36:1991-2002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 51. | Wu X, Fleming A, Ricketts T, Pavel M, Virgin H, Menzies FM, Rubinsztein DC. Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat Commun. 2016;7:10533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 146] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 52. | Wang F, Zhou H, Yang Y, Xia X, Sun Q, Luo J, Cheng B. Hepatitis B virus X protein promotes the growth of hepatocellular carcinoma by modulation of the Notch signaling pathway. Oncol Rep. 2012;27:1170-1176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 53. | Luo J, Zhou H, Wang F, Xia X, Sun Q, Wang R, Cheng B. The hepatitis B virus X protein downregulates NF-κB signaling pathways through decreasing the Notch signaling pathway in HBx-transformed L02 cells. Int J Oncol. 2013;42:1636-1643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 54. | Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594-2608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 211] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 55. | Schlottmann S, Buback F, Stahl B, Meierhenrich R, Walter P, Georgieff M, Senftleben U. Prolonged classical NF-kappaB activation prevents autophagy upon E. coli stimulation in vitro: a potential resolving mechanism of inflammation. Mediators Inflamm. 2008;2008:725854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |