Published online Sep 21, 2016. doi: 10.3748/wjg.v22.i35.7882
Peer-review started: April 21, 2016
First decision: May 27, 2016
Revised: June 30, 2016
Accepted: August 1, 2016
Article in press: August 1, 2016
Published online: September 21, 2016
Processing time: 147 Days and 16.5 Hours
Patients with extensive ulcerative colitis (UC) of more than eight years duration have an increased risk of colorectal cancer. Molecular biomarkers for dysplasia and cancer could have a great clinical value in managing cancer risk in these UC patients. Using a wide range of molecular techniques - including cutting-edge OMICS technologies - recent studies have identified clinically relevant biomarker candidates from a variety of biosamples, including colonic biopsies, blood, stool, and urine. While the challenge remains to validate these candidate biomarkers in multi-center studies and with larger patient cohorts, it is certain that accurate biomarkers of colitis-associated neoplasia would improve clinical management of neoplastic risk in UC patients. This review highlights the ongoing avenues of research in biomarker development for colitis-associated colorectal cancer.
Core tip: With the incidence of ulcerative colitis on the rise, there is a need to develop clinically useful biomarkers capable of identifying and monitoring the subset of ulcerative colitis (UC) patients at highest risk for developing colon cancer. Recent studies have reported the efforts in developing clinically relevant biomarkers, laying a foundation for further clinical biomarker development. While the challenge remains to validate these candidate biomarkers in multi-center studies using larger patient cohorts, it is certain that accurate biomarkers of colitis-associated neoplasia would improve current clinical management of neoplastic risk in UC patients.
- Citation: Chen R, Lai LA, Brentnall TA, Pan S. Biomarkers for colitis-associated colorectal cancer. World J Gastroenterol 2016; 22(35): 7882-7891
- URL: https://www.wjgnet.com/1007-9327/full/v22/i35/7882.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i35.7882
Ulcerative colitis (UC) is an inflammatory bowel disease, characterized by chronic, recurrent inflammation of the colon. Patients with extensive UC of more than eight years duration have an increased risk of colorectal cancer. These patients are usually advised to undergo lifelong colonoscopic surveillance to detect the presence of dysplasia (pre-cancer) or cancer in the colon. Colitis-associated cancer (CAC) is distinct from sporadic colorectal cancer, both in disease mechanism and presentation. Progression in UC is usually dependent upon several factors. For patients with disease duration in excess of 8-10 years, the risk of developing cancer increases at a rate of 0.5%-1% per year. For example, a patient with 20 years of UC will have a neoplastic risk of approximately 10%-20%[1,2]. While the patient’s age at disease onset factors into cancer risk[3], disease duration has a greater impact on cancer risk than chronological age. In addition, risk for cancer development increases with severity of inflammation[4] and extent of disease[5]. Patients with pancolitis are at higher risk compared to those with distal disease.
While current colonoscopic surveillance programs are cost-effective and able to reduce cancer incidence and mortality rate in UC patients[2,6], current methods of surveillance are expensive and invasive. Moreover, pathologic evaluation of colitis-associated neoplasia is subjective; even experienced gastrointestinal pathologists who developed the histologic standards for UC neoplasia could only agree on the diagnosis of indefinite for dysplasia half of the time[7]. Clearly, an objective molecular biomarker for dysplasia and cancer would have great clinical value in the management of cancer risk in UC patients.
In UC, the colon epithelium undergoes repeated cycles of inflammation and tissue repair, resulting in oxidative stress and accumulation of reactive oxidative species (ROS)[8-11]. Excessive ROS causes oxidative stress and damage to DNA, proteins and lipids, leading to tumor initiation. Colitis-associated colorectal cancer progresses in a step-wise fashion from negative for dysplasia (NEG) → indefinite for dysplasia (IND) → low-grade dysplasia (LGD) → high-grade dysplasia (HGD) → cancer[12]. Neoplasia in UC is facilitated by several molecular alterations, some of which are detectable prior to dysplasia. These alterations include: telomere shortening, chromosomal and microsatellite instability, aneuploidy, and loss of p53[13-21]. In fact, alterations to the p53 tumor suppressor gene are important early events and occur in 47%-85% of colitis-associated cancers[22,23]. The second mechanism by which inflammation could contribute to carcinogenesis is through the release of proinflammatory cytokines[24-26]. Cytokines released from chronic inflammation participate in all stages of cancer development, including tumor initiation, tumor promotion, angiogenesis and metastasis. NF-κB is thought to be a key molecular link between inflammation and carcinogenesis[23]. When activated, it upregulates the expression of many pro-inflammatory mediators, such as adhesion molecules, cytokines, TNF-α and IL-6, which play a critical role in tumor development and progression of colitis-associated cancer[23].
The molecular alterations which occur in the pathogenesis of CAC are distinct from those in sporadic colon carcinogenesis. For example, the loss of APC occurs early in development of sporadic colorectal cancer, whereas it is usually a late event in UC associated disease progression if it occurs at all. In addition, p53 mutations appear as an early event in CAC, even prior to dysplasia yet p53 mutations are a late event in the sporadic disease. This evidence suggests that there are some unique pathways associated with progression of colitis-associated cancer. Studies show that the non-dysplastic mucosa is genomically abnormal in UC patients who have neoplasia elsewhere in their colon (Progressors)[15,27], i.e., there is a colon-wide field defect in UC Progressors. This field defect involves the abnormal expression of proteins, genomic instability in repetitive DNA, and mitochondrial dysfunction in both the non-dysplastic and dysplastic mucosa from UC Progressors[15,16,28-30]. Such molecular changes tend to be absent in UC patients who are dysplasia-free (UC Non-progressors). These molecular alterations present in UC Progressors provide targets for the development of biomarkers for diagnostics and therapeutic applications in CAC.
The current standard of care for surveillance of chronic UC patients involves colonoscopy starting after 8-10 years of disease duration. Surveillance is recommended every 1-2 years with frequency increasing over time to match the elevating risk with years of disease duration. Surveillance involves chromoendoscopy, where a solution is sprayed throughout the colon to highlight areas of dysplasia or, alternatively, high-definition colonoscopy with 4 quadrant random biopsy sampling at 10 cm intervals throughout the colon for subsequent histological determination of neoplasia. Moreover, UC-associated neoplastic lesions can be multi-focal and can appear either flat or raised[12] and may be difficult to diagnose amongst chronically inflamed epithelium. Overall, this translates to collection of roughly 30 to 45 biopsies in order to get a reasonable sampling of the colon. The histologic diagnosis of dysplasia[12], especially at early stages, can be complicated by the presence of inflammation in the biopsy. Patients who receive a diagnosis of dysplasia are deemed high risk for developing cancer and subsequently undergo a more frequent surveillance regimen or a colectomy. However, the proportion of UC patients who develop cancer is low, such that identification of low risk patients and subsequent use of alternate non-colonoscopy based surveillance could ultimately reduce health care costs associated with unnecessary colonoscopy.
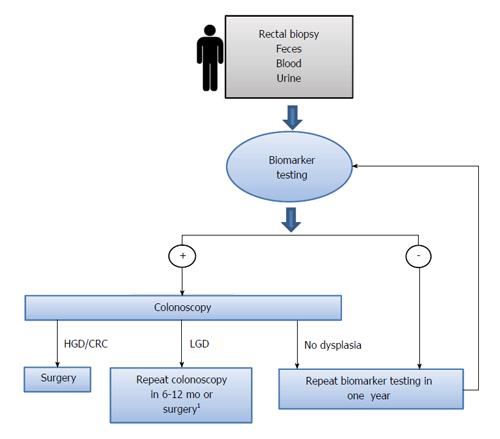
It is therefore important to identify and develop biomarkers with high sensitivity that could be obtained through non- or less invasive methods (i.e., blood sample, single rectal biopsy, urine) in order (1) to stratify the UC patients which require a more intense colonoscopic surveillance regimen; and (2) to provide earlier diagnostic and better prognostic information. Diagnostic biomarkers could be integrated with existing endoscopy-based UC cancer surveillance to improve the current clinical paradigm, as illustrated in Figure 1. Chronic UC Patients could be screened using a biomarker first using biosamples such as blood, urine or rectal biopsy. Only patients with a positive biomarker result would undergo further evaluation with a full colonoscopy. Patients with a negative biomarker result would be followed by biomarker testing on an annual basis. Therefore, biomarker testing could improve clinical management by identifying those UC patients who have a very low risk of dysplasia and thus reducing the need for colonoscopy.
Unlike sporadic colorectal cancer, colitis-associated cancer does not usually follow a linear pathway of gene loss or disruption but rather appears to be driven largely by inflammation-associated damage. Combined with the unclear etiology of this disease, researchers are pursuing multiple independent avenues to identify sensitive and specific biomarkers for early UC cancer. While there are currently many biomarkers in development to discriminate subtypes of patients with inflammatory bowel disease (IBD-ulcerative colitis vs Crohn’s disease), there are few biomarkers for differentiating UC non-Progressors (patients who do not develop dysplasia) from UC Progressors (patients who develop dysplasia or cancer). We will discuss some of the major efforts here. Due to limited space of the review, there will be a primary focus on markers with the most promising clinical potential. Table 1 summarizes the biomarker candidates from recent studies according to the types of biospecimens that have been used including colonic tissue, blood, urine and stool. Since many of these studies were performed on limited numbers of patient samples, they will need to be independently validated in multi-center larger cohort studies.
| Type of specimen | Analyte | Biomarker | Ref. |
| Blood | Protein | p53 | [58] |
| Colon tissue | DNA | Chromosomal instability | [15,16,18-20,30-35] |
| IBD specific mutation: SOX9, EP300, NRG1, and IL16 | [36] | ||
| DNA methylation | ER, MYOD, p16, and CSPG2 | [40] | |
| FOXE1, SYNE1 | [41] | ||
| EYA4 | [27,42] | ||
| RUNX3, MINT1, and COX-2 | [43] | ||
| p14 | [44] | ||
| BVES | [45] | ||
| microRNA | miR-143 and miR-145 | [46] | |
| Decrease of 3 microRNAs (miR-192, miR-375, and miR-422b), upregulation of 8 microRNAs (miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, miR-195, and Let-7f) | [47] | ||
| miR-21 and miR-155 | [48] | ||
| Protein | miR-26b | [50] | |
| p53 and CHGA | [51] | ||
| p53 and AMACR | [52] | ||
| Bcl-xl | [53] | ||
| PDCD4 | [54] | ||
| COX | [55] | ||
| 8-NG and 8-oxodG | [56] | ||
| RNA | TRAP1 | [29,57] | |
| CCND1, SERPINB6, PAP, IL8, and NOS2A | [37] | ||
| Stool | ACSL1, BIRC3, CLC, CREM, ELTD1, FGG, S100A9, THBD, and TPD52L1 | [38] | |
| A panel of 20 genes (including cancer genes CYP27B1, RUNX3, SAMSN1, EDIL3, NOL3, CXCL9, ITGB2, and LYN) | [39] | ||
| DNA methylation | VIM, EYA4, BMP3, and NDRG4 | [63] | |
| Metabolites | Buryrate, acetate, methylamine, and trimethylamine | [62] | |
| Protein | Calprotectin and lactoferrin | [61] | |
| Urine | Metabolites | Prostaglandin-E | [59] |
| Protein | MMP-2 and MMP-2/NGAL | [60] |
Biomarker development for UC-associated cancer has, for the most part, focused on the use of colonic biopsies obtained during colonoscopy or colectomy due to (1) the known pancolonic abnormalities that precede CAC; (2) colonic biopsies are a readily available source of material; and (3) retrospective material can be acquired. Immunohistochemical staining of formalin-fixed paraffin embedded (FFPE) material has provided information about disease etiology and clues to possible biomarker candidates. Isolation of genetic material (i.e., DNA, RNA, miRNA) as well as proteins for proteomic analysis from FFPE sections or frozen material has taken this to the next level. The hope is to identify alterations associated with UC cancerous or dysplastic regions that are also present in non-neoplastic and remote regions, ideally rectum. Rectal biopsies could be obtained with any laxative preparation, without sedation, and using a quick and minimally invasive procedure (proctoscopy) that could be performed in any clinic.
DNA: DNA-based assays using genetic material isolated from purified colonic epithelium has shown promise as a biomarker source. However, since UC doesn’t follow the hallmark loss of gene pathways like sporadic colorectal cancer, several studies have focused on more general genome-wide (rather than locus specific) instability. Pan-colonic chromosomal instability has been shown to be detectable relatively early in UC disease progression by BAC array[18,31], fluorescence in-situ hybridization (FISH)[19,20] and arbitrarily primed PCR (AP-PCR)[15,16], yet none of these assays are amenable to high throughput analysis that would be required for a clinical UC neoplastic biomarker. Clonal expansions of poly glutamine repeats (PolyG) are detectable in DNA isolated from colonic epithelium relative to adjacent stromal or muscle DNA controls. The poly G assay was validated in two independent cohorts which confirmed discrimination between UC Progressors and Non-progressors, and demonstrated its utility for UC patients with early disease onset (i.e., diagnosis before 50 years of age)[30,32]. A more recent study examined chromosomal instability in UC neoplastic progression using copy number variation microarrays, and found increased numbers of copy number variations with progression of UC to UC-associated colorectal cancer[33].
UC can also be thought of as an aging disease of the colon; qFISH and qPCR of colonic epithelium from UC patients showed accelerated telomere shortening compared to leukocyte or adjacent stromal controls[34,35]. Shortened telomeres result in chromosomal instability due to higher frequencies of anaphase bridges and subsequent chromosomal breakage, losses, and gains[19]. These techniques lack either the very high specificity demanded by a CAC biomarker (qPCR) or capability for high throughput (qFISH) to create a clinically useable test.
With the development of whole-genome sequencing, comprehensive investigation of the genomic landscape has identified an increasing array of mutational signatures associated with specific diseases. In CAC, the chronic oxidative injury can cause mounting epithelial cell DNA damage which over time, overwhelms the G1 cell cycle checkpoint and results in p53 mutation. A recent study investigated somatic mutation patterns from inflammatory bowel disease (IBD, including ulcerative colitis and Crohn’s disease) associated with colorectal tumor using whole-exome sequencing[36]. Not surprisingly, TP53 was the most commonly mutated gene, with mutational prevalence similar to that of sporadic colorectal tumors (63% of cases). However, APC and KRAS were mutated at significantly lower rates in tumors from patients with IBD than in sporadic colorectal tumors (13% and 20% of cases, respectively). Several IBD-specific gene mutations, including SOX9, EP300, NRG1, and IL16 were identified, confirming the notion that IBD association colorectal cancer (CRC) has its unique genetic composition compared to sporadic CRC.
RNA: Gene expression analysis using RT-PCR of materials isolated from colonic biopsy has resulted in identification of genes which are involved in or associated with UC neoplasia. Microarrays have been used to investigate gene expressional changes in UC neoplastic progression by comparing the mucosa of non-dysplastic, dysplastic and cancerous colonic tissues[37]. The study identified genes associated with UC dysplasia and UC cancer. Five genes were further verified to be common to UC dysplasia and adenocarcinoma relative to non-dysplastic UC (CCND1, SERPINB6, PAP, IL8, and NOS2A).
Because the dysplasia or cancer in UC patients can appear as flat mucosa and/or is multi-foci, there is an interest in identifying neoplastic biomarkers that are present in both dysplastic and non-dysplastic tissues. Such biomarkers could then be tested in random biopsies, including rectal which could be obtained with a minimally invasive procedure. Numerous studies have identified gene expression changes in non-dysplastic tissues from UC Progressors. In one study, a gene signature which included ACSL1, BIRC3, CLC, CREM, ELTD1, FGG, S100A9, THBD, and TPD52L1 were progressively increased with neoplastic progression[38]. These findings support the concept of a field defect phenomena in CAC. Two markers (S100A9 and REG1α) were further validated by IHC showing increased staining in the dysplastic and non-dysplastic tissues from UC Progressors compared to UC Non-progressors and normal controls[38]. In a separate study, by surveying the expressions of 189 carcinogenesis related genes in non-dysplastic rectal mucosa from UC Progressors and Non-progressors, researchers identified a panel of 20 genes (including cancer genes such as CYP27B1, RUNX3, SAMSN1, EDIL3, NOL3, CXCL9, ITGB2, and LYN) in rectal tissues as Progressor associated genes[39]. Using non-dysplastic rectal tissues, the 20-gene panel was able to predict UC-cancer patients from UC-non cancer patients with 83% accuracy and a negative predictive value of 100%.
DNA methylation: Promotor methylation plays an important role in tumorigenesis though transcriptional silencing of critical genes. The early study of DNA methylation in UC dysplasia could be traced back to a study investigating methylation status of five age or cancer related genes in UC dysplasia[40]. Hypermethylation of ER, MYOD, p16, and CSPG2 were detectable in the high-grade dysplasia and cancer tissues from UC Progressors. Moreover, hypermethylation of ER, MYOD and p16 could be detected in the non-dysplastic tissues from Progressors compared to Non-progressors[40]. In another study using methylation-specific PCR, hypermethylation of the tumor growth genes, FOXE1 and SYNE1, was detected in approximately 60%-80% of cancer samples (whereas it was undetectable in controls), suggesting an increase in methylation with disease progression[41]. Methylation of the eyes absent homolog 4 (EYA4) gene was present both in neoplastic and remote non-neoplastic tissue of UC patients with cancer but absent in UC control patients without neoplasia[27,42]. Another study found altered methylation status of RUNX3, MINT1, and COX-2 in both the non-neoplastic regions and neoplastic regions of UC -CRC colons as compared with that in the UC controls[43]. Either RUNX3 or MINT1 showed interaction with COX-2 with an additive effect[43]. Further study is needed to evaluate whether this three-gene panel can predict the likelihood of patients to progress to CRC and/or to identify patients with dysplasia. While CpG island hypermethylation of p14 (ARF) but not p16 (INK4) was detectable in 100% of UC dysplasia tested, only 20% (2/10) of patients with dysplasia showed hypermethylation in DNA extracted from non-dysplastic rectum[44]. In a recent study, the reduced expression of a tight junction-associated protein, BVES in UC neoplasia was shown to be the result of promotor hypermethylation of this gene[45]. The BVES promoter hypermethylation could be detected in dysplastic colonic tissues as well as distant non-malignant-appearing mucosa from UC Progressors in comparison to UC Non-progressor and non-UC controls. The study further suggests that BVES interacts with PR61α to promote inflammatory tumorigenesis through c-Myc destruction. Based on the results, BVES promoter hypermethylation status could be a potential biomarker to identify patients with UC at risk of cancer[45].
MicroRNA: MicroRNAs are small non-coding 20-25 nucleotide single-stranded RNA molecules which usually bind to the 3’ untranslated region of target mRNA transcripts, effectively silencing them by inhibition of translation or through degradation. MicroRNAs can function as oncogenes to enhance cellular proliferation and survival or as tumor suppressors. Downregulation of miR-143 and miR-145, as well as concomitant upregulation of their predicted targets, IRS-1, K-Ras, API5, and MEK-2 was found in colon biopsies of UC patients, suggesting that some microRNAs could contribute directly to transformation of UC colonic epithelium[46]. In colitis, the inflammatory microenvironment can modulate microRNA expression and further influence target gene expressions. Currently, there is much focus on the investigation of microRNAs that affect immune response in order to maintain intestinal homeostasis. MicroRNA analysis of colonic biopsies revealed decreased expression of 3 microRNAs (including miR-192, miR-375, and miR-422b) in active UC whereas upregulation of 8 microRNAs (miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, miR-195, and Let-7f) was noted in active UC compared to normal controls[47]. Upregulation of miR-21 and miR-155 in inflamed colon from UC patients compared to controls was confirmed in an independent study[48]. Mouse studies suggest that miR-155 is involved in the proinflammatory cellular response due to decreased numbers of CD4+T, Th1/Th17, CD11b+, and CD11c+ cells in miR-155-/- mice[49]. Dysregulation of some microRNAs could be related to inflammation and thus could be used to discriminate colitis associated cancers from sporadic colorectal cancers. For example, miR-26b was shown upregulated in CAC, and down-regulated in sporadic colon cancer[50].
Protein: Protein-based research has revealed some interesting CAC biomarker candidates. Protein participants in major known pathways involved in UC tumorigenesis have been further scrutinized to evaluate their potential as biomarkers. While p53 mutations have been shown to occur as an early event in UC neoplasia[13], the value of the mutated p53 protein as a clinical marker by IHC is controversial. The results of IHC studies are variable and limited due to the use of different antibodies and small sample sizes per study. Protein expression of p53 and chromogranin A provides moderate sensitivity (66.7%) and specificity (80%) for HGD detection[51]. Other protein expression data shows that co-detection of p53 and α-methylacyl coenzyme A racemase (AMACR), using IHC, could discriminate UC patients who had dysplasia or cancer from those who did not; and further, the co-expression of p53 and AMACR was detectable in biopsies taken as early as 10-14 months prior to dysplasia[52]. In a different study, expression of the apoptosis gene, Bcl-xl, is absent in non-dysplastic UC epithelium, but highly positive in dysplasia or cancer samples by IHC[53]. Nuclear IHC staining of the programmed cell death 4 (PDCD4) tumor suppressor showed a reduction in colonic UC dysplasia and cancer tissue samples, and could be useful as a biomarker in the histological assessment of IBD-associated dysplastic lesions[54]. Mitochondrial alterations as defined by patchy loss of cytochrome C oxidase (COX) IHC staining within colonic crypts preceded dysplasia and the staining was lowest in regions of dysplasia and adjacent regions[55]. Chronic inflammation in UC results in the generation of reactive oxygen and nitrogen species, leading to the accumulation of DNA damage, which can be measured by 8-nitroguanine (8-NG) and 8-oxo-7,8-dyhydro-2’-deoxyguanosine (8-oxodG). IHC of rectal mucosa from patients with UC-associated dysplasia showed statistically significant increases in 8-NG and decreases in 8-oxodG compared to UC non-neoplastic controls[56].
Proteomics has also been applied in colitis-associated colon cancer studies with research interests ranging from investigation of disease mechanism to biomarker discovery. To investigate proteomic alterations linked to UC-associated dysplasia and invasive cancer, one study applied stable isotope label based quantitative proteomics to examine the protein expression in the colonic mucosa[28]. The study identified a roster of proteins that displayed at least a 2-fold expression change in random non-dysplastic colon biopsies from Progressors compared to random biopsies from Non-progressors. Among the differentially expressed proteins in the random non-dysplastic biopsies, almost 60% of them were also concurrently expressed in the dysplastic tissues from the same Progressors. These findings suggest that changes in protein expression occur very early in the neoplastic process, before the histologic changes become evident in epithelial cells[28]. Protein activities associated with neoplastic progression included proteins related to mitochondrial function, oxidative activity, and calcium-binding. Protein network analysis suggested that SP1 and c-MYC may play key roles in UC early and late stages of neoplastic progression, respectively.
In a follow-up quantitative proteomics study, individual random rectal samples from UC Progressors were profiled compared to random rectal samples from UC Non-progressors[29]. The study identified over 60 proteins that were differentially expressed in both non-dysplastic rectal tissue and the corresponding dysplastic colonic tissue from Progressors. Mitochondrial proteins, cytoskeletal proteins, RAS superfamily and proteins related to apoptosis were the important protein classes differentially associated with Progressors. One of the mitochondrial proteins, TRAP1, was further validated by IHC in an independent UC cohort, and showed up-regulation in the colon tissues of UC Progressors, but not in the colon tissues of UC Non-progressors[57]. Moreover, up-regulation of TRAP1 preceded the neoplastic changes: it was present in both the dysplastic and non-dysplastic tissue of UC Progressors. TRAP1 staining in dysplastic tissue could achieve 94% sensitivity and 80% specificity in separating Progressors from Non-progressors. In random non-dysplastic rectal biopsies, TRAP1 staining could separate Progressors from Non-progressors with 59% sensitivity and 80% specificity[57].
Blood represents an ideal diagnostic specimen for clinical tests due to its low cost and easy accessibility. It provides abundant material (e.g., DNAs, RNAs, proteins, cells, exosomes etc.) for analysis by a variety of molecular approaches. The gold standard for blood testing of protein levels has traditionally been enzyme linked immunosorbent assays (ELISA) while new technologies such as modified ELISAs, targeted proteomics, and nanotechnology are emerging, making it possible to more thoroughly investigate cancer associated protein abnormalities, particularly for proteins with low abundance. One study reported serum p53 levels detected by ELISA was higher in UC patients compared to normal controls, however elevated levels were detectable in only 8/13 of UC patients with CRC or p53[58]. While p53 is an early event in UC-related tumorigenesis, detection of p53 protein in serum showed only moderate sensitivity-limiting its utility as a neoplastic biomarker for high-risk patients.
Urine is one of the most convenient sources of material for UC biomarker discovery due to the ease of collection. Most of the current urine-based biomarker studies are focused on discriminating UC patients from those with other types of inflammatory bowel disease (i.e., Crohn’s Disease) or to monitor treatment non-adherence. Only a few urine studies reported data that is relevant to neoplastic biomarker discovery in CAC. In a small cross-sectional study, the urinary metabolite, prostaglandin-E, (PGE-MUM assay) was reported to have some potential as a biomarker for UC activity (compared to C-reactive protein), which may assist in staging disease inflammatory activity[59]. Another report suggested that urine MMP-2 and MMP-2/NGAL levels could discriminate UC patients from normal controls[60]. The urinary biomarkers from the current studies may be useful as non-invasive biomarkers for disease activity measurements in IBD patients. However, it is unclear whether these markers would be of value in discriminating UC with and without dysplasia.
Due to the close proximity to intestinal mucosa, stool samples are a rich resource of material for UC biomarker development. It is estimated that approximately one half of stool is comprised of gut microflora and that upwards of one million colon epithelial cells can be isolated from one gram of stool, which would allow for the study of changes in the genetic material or proteome of a patient’s colonocytes, as well as their gut microbiome. Given the complexity of stool samples, it is critical to distinguish changes caused by dysplasia or disease progression, chronic inflammation or consequences of inflammation (i.e., DNA damage, reactive oxygen species, etc.), microbial changes, and/or diet. While biomarkers for UC-associated cancer from stool have not yet been adapted for clinical use, fecal biomarkers are being used to diagnose IBD and irritable bowel syndrome (IBS) patients, to distinguish between UC and CD patients, as well as to assess active inflammation. Currently, the two most widely used stool biomarkers, calprotectin and lactoferrin, are derived from neutrophils, which penetrate the intestinal mucosa in areas of active inflammation and are subsequently shed into the lumen. Both biomarkers may be valuable in assessing the active inflammation as a way to gauge therapeutic response in UC patients and as a non-invasive method to monitor relapses[61]. Use of NMR-based spectroscopy to analyze stool is also gaining appeal due to minimal sample preparation, the ability to detect multiple metabolites at once, and the high reproducibility. Mass resonance metabolomics studies have revealed reductions in buryrate, acetate, methylamine, and trimethylamine as well as differences in the levels of several amino acids when stool samples of UC patients are compared to controls[62]. In addition to protein and metabolite markers, DNA isolated from stool offers a promise as a UC biomarker source. Although mutational analysis failed to discriminate UC-associated cancer samples from cancer-free patients, the methylation changes of four genes - including vimentin, eyes absent homolog 4 (EYA4), bone morphogenetic protein 3 (BMP3), and N-myc downstream regulated gene 4 (NDRG4) - showed AUC (areas under the ROC curve) ranging from 0.84-0.91 in distinguishing UC patients with or without neoplasia, suggesting both high specificity and sensitivity for dysplasia detection[63].
With the incidence of ulcerative colitis on the rise, there is a need to develop clinically useful biomarkers capable of identifying and monitoring the subset of UC patients at highest risk for developing colon cancer. Cologuard is a newly FDA approved stool biomarker test for sporadic colorectal cancer screening. The most recent study suggests that it can detect 100% colorectal cancer in people between the ages of 40-85[64]. However, this test has about 52% sensitivity in detecting premalignant lesions, such as adenomas, and its utility for screening dysplasia in UC patients has not been tested. Therefore, a new objective molecular test for detection of dysplasia and cancer could be helpful for the clinical management of cancer risk in UC patients. Recent studies have reported the efforts in developing clinically relevant biomarkers for early detection of UC associated cancer using a variety of biosamples, including colonic biopsies, blood, stool, and urine. These studies have revealed a roster of biomarker candidates and laid a foundation for further clinical biomarker development. While the challenge remains to validate these candidate biomarkers in multi-center studies and with larger patient cohorts, it is certain that accurate biomarkers of colitis-associated neoplasia would improve clinical management of neoplastic risk in UC patients.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chiang HMT, Gazouli M S- Editor: Ma YJ L- Editor: A E- Editor: Wang CH
| 1. | Ekbom A, Helmick C, Zack M, Adami HO. Ulcerative colitis and colorectal cancer. A population-based study. N Engl J Med. 1990;323:1228-1233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1294] [Cited by in RCA: 1198] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 2. | Prior P, Gyde SN, Macartney JC, Thompson H, Waterhouse JA, Allan RN. Cancer morbidity in ulcerative colitis. Gut. 1982;23:490-497. [PubMed] |
| 3. | Baars JE, Kuipers EJ, van Haastert M, Nicolaï JJ, Poen AC, van der Woude CJ. Age at diagnosis of inflammatory bowel disease influences early development of colorectal cancer in inflammatory bowel disease patients: a nationwide, long-term survey. J Gastroenterol. 2012;47:1308-1322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 4. | Rutter M, Saunders B, Wilkinson K, Rumbles S, Schofield G, Kamm M, Williams C, Price A, Talbot I, Forbes A. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126:451-459. [PubMed] |
| 5. | Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526-535. [PubMed] |
| 6. | Risques RA, Rabinovitch PS, Brentnall TA. Cancer surveillance in inflammatory bowel disease: new molecular approaches. Curr Opin Gastroenterol. 2006;22:382-390. [PubMed] |
| 7. | Riddell RH, Goldman H, Ransohoff DF, Appelman HD, Fenoglio CM, Haggitt RC, Ahren C, Correa P, Hamilton SR, Morson BC. Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Hum Pathol. 1983;14:931-968. [PubMed] |
| 8. | Ellis EM. Reactive carbonyls and oxidative stress: potential for therapeutic intervention. Pharmacol Ther. 2007;115:13-24. [PubMed] |
| 9. | Keshavarzian A, Sedghi S, Kanofsky J, List T, Robinson C, Ibrahim C, Winship D. Excessive production of reactive oxygen metabolites by inflamed colon: analysis by chemiluminescence probe. Gastroenterology. 1992;103:177-185. [PubMed] |
| 10. | Keshavarzian A, Banan A, Farhadi A, Komanduri S, Mutlu E, Zhang Y, Fields JZ. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut. 2003;52:720-728. [PubMed] |
| 11. | Roessner A, Kuester D, Malfertheiner P, Schneider-Stock R. Oxidative stress in ulcerative colitis-associated carcinogenesis. Pathol Res Pract. 2008;204:511-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 202] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 12. | Bressenot A, Cahn V, Danese S, Peyrin-Biroulet L. Microscopic features of colorectal neoplasia in inflammatory bowel diseases. World J Gastroenterol. 2014;20:3164-3172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 13. | Brentnall TA, Crispin DA, Rabinovitch PS, Haggitt RC, Rubin CE, Stevens AC, Burmer GC. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107:369-378. [PubMed] |
| 14. | Brentnall TA, Crispin DA, Bronner MP, Cherian SP, Hueffed M, Rabinovitch PS, Rubin CE, Haggitt RC, Boland CR. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res. 1996;56:1237-1240. [PubMed] |
| 15. | Chen R, Rabinovitch PS, Crispin DA, Emond MJ, Koprowicz KM, Bronner MP, Brentnall TA. DNA fingerprinting abnormalities can distinguish ulcerative colitis patients with dysplasia and cancer from those who are dysplasia/cancer-free. Am J Pathol. 2003;162:665-672. [PubMed] |
| 16. | Chen R, Rabinovitch PS, Crispin DA, Emond MJ, Bronner MP, Brentnall TA. The initiation of colon cancer in a chronic inflammatory setting. Carcinogenesis. 2005;26:1513-1519. [PubMed] |
| 17. | Chen R, Bronner MP, Crispin DA, Rabinovitch PS, Brentnall TA. Characterization of genomic instability in ulcerative colitis neoplasia leads to discovery of putative tumor suppressor regions. Cancer Genet Cytogenet. 2005;162:99-106. [PubMed] |
| 18. | Lai LA, Risques RA, Bronner MP, Rabinovitch PS, Crispin D, Chen R, Brentnall TA. Pan-colonic field defects are detected by CGH in the colons of UC patients with dysplasia/cancer. Cancer Lett. 2012;320:180-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | O’Sullivan JN, Bronner MP, Brentnall TA, Finley JC, Shen WT, Emerson S, Emond MJ, Gollahon KA, Moskovitz AH, Crispin DA. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet. 2002;32:280-284. [PubMed] |
| 20. | Rabinovitch PS, Dziadon S, Brentnall TA, Emond MJ, Crispin DA, Haggitt RC, Bronner MP. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res. 1999;59:5148-5153. [PubMed] |
| 21. | Risques RA, Lai LA, Himmetoglu C, Ebaee A, Li L, Feng Z, Bronner MP, Al-Lahham B, Kowdley KV, Lindor KD. Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res. 2011;71:1669-1679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Itzkowitz SH. Molecular biology of dysplasia and cancer in inflammatory bowel disease. Gastroenterol Clin North Am. 2006;35:553-571. [PubMed] |
| 23. | O’Connor PM, Lapointe TK, Beck PL, Buret AG. Mechanisms by which inflammation may increase intestinal cancer risk in inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1411-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 24. | Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7-17. [PubMed] |
| 25. | Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539-545. [PubMed] |
| 26. | Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101-2114.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1333] [Cited by in RCA: 1511] [Article Influence: 100.7] [Reference Citation Analysis (0)] |
| 27. | Kisiel JB, Garrity-Park MM, Taylor WR, Smyrk TC, Ahlquist DA. Methylated eyes absent 4 (EYA4) gene promotor in non-neoplastic mucosa of ulcerative colitis patients with colorectal cancer: evidence for a field effect. Inflamm Bowel Dis. 2013;19:2079-2083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Brentnall TA, Pan S, Bronner MP, Crispin DA, Mirzaei H, Cooke K, Tamura Y, Nikolskaya T, Jebailey L, Goodlett DR. Proteins That Underlie Neoplastic Progression of Ulcerative Colitis. Proteomics Clin Appl. 2009;3:1326. [PubMed] |
| 29. | May D, Pan S, Crispin DA, Lai K, Bronner MP, Hogan J, Hockenbery DM, McIntosh M, Brentnall TA, Chen R. Investigating neoplastic progression of ulcerative colitis with label-free comparative proteomics. J Proteome Res. 2011;10:200-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 30. | Salk JJ, Salipante SJ, Risques RA, Crispin DA, Li L, Bronner MP, Brentnall TA, Rabinovitch PS, Horwitz MS, Loeb LA. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc Natl Acad Sci USA. 2009;106:20871-20876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 31. | Bronner MP, Skacel M, Crispin DA, Hoff PD, Emond MJ, Lai LA, Tubbs RR, O’Sullivan JN, Rabinovitch PS, Brentnall TA. Array-based comparative genomic hybridization in ulcerative colitis neoplasia: single non-dysplastic biopsies distinguish progressors from non-progressors. Mod Pathol. 2010;23:1624-1633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 32. | Salk JJ, Bansal A, Lai LA, Crispin DA, Ussakli CH, Horwitz MS, Bronner MP, Brentnall TA, Loeb LA, Rabinovitch PS. Clonal expansions and short telomeres are associated with neoplasia in early-onset, but not late-onset, ulcerative colitis. Inflamm Bowel Dis. 2013;19:2593-2602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 33. | Shivakumar BM, Rotti H, Vasudevan TG, Balakrishnan A, Chakrabarty S, Bhat G, Rao L, Pai CG, Satyamoorthy K. Copy number variations are progressively associated with the pathogenesis of colorectal cancer in ulcerative colitis. World J Gastroenterol. 2015;21:616-622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (2)] |
| 34. | O’Sullivan JN, Finley JC, Risques RA, Shen WT, Gollahon KA, Rabinovitch PS. Quantitative fluorescence in situ hybridization (QFISH) of telomere lengths in tissue and cells. Curr Protoc Cytom. 2005;Chapter 12:Unit 12.6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 35. | Risques RA, Lai LA, Brentnall TA, Li L, Feng Z, Gallaher J, Mandelson MT, Potter JD, Bronner MP, Rabinovitch PS. Ulcerative colitis is a disease of accelerated colon aging: evidence from telomere attrition and DNA damage. Gastroenterology. 2008;135:410-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 140] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 36. | Robles AI, Traverso G, Zhang M, Roberts NJ, Khan MA, Joseph C, Lauwers GY, Selaru FM, Popoli M, Pittman ME. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology. 2016;150:931-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 212] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 37. | Colliver DW, Crawford NP, Eichenberger MR, Zacharius W, Petras RE, Stromberg AJ, Galandiuk S. Molecular profiling of ulcerative colitis-associated neoplastic progression. Exp Mol Pathol. 2006;80:1-10. [PubMed] |
| 38. | Pekow J, Dougherty U, Huang Y, Gometz E, Nathanson J, Cohen G, Levy S, Kocherginsky M, Venu N, Westerhoff M. Gene signature distinguishes patients with chronic ulcerative colitis harboring remote neoplastic lesions. Inflamm Bowel Dis. 2013;19:461-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Watanabe T, Kobunai T, Yamamoto Y, Ikeuchi H, Matsuda K, Ishihara S, Nozawa K, Iinuma H, Kanazawa T, Tanaka T. Predicting ulcerative colitis-associated colorectal cancer using reverse-transcription polymerase chain reaction analysis. Clin Colorectal Cancer. 2011;10:134-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573-3577. [PubMed] |
| 41. | Papadia C, Louwagie J, Del Rio P, Grooteclaes M, Coruzzi A, Montana C, Novelli M, Bordi C, de’ Angelis GL, Bassett P. FOXE1 and SYNE1 genes hypermethylation panel as promising biomarker in colitis-associated colorectal neoplasia. Inflamm Bowel Dis. 2014;20:271-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Osborn NK, Zou H, Molina JR, Lesche R, Lewin J, Lofton-Day C, Klatt KK, Harrington JJ, Burgart LJ, Ahlquist DA. Aberrant methylation of the eyes absent 4 gene in ulcerative colitis-associated dysplasia. Clin Gastroenterol Hepatol. 2006;4:212-218. [PubMed] |
| 43. | Garrity-Park MM, Loftus EV, Sandborn WJ, Bryant SC, Smyrk TC. Methylation status of genes in non-neoplastic mucosa from patients with ulcerative colitis-associated colorectal cancer. Am J Gastroenterol. 2010;105:1610-1619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 44. | Moriyama T, Matsumoto T, Nakamura S, Jo Y, Mibu R, Yao T, Iida M. Hypermethylation of p14 (ARF) may be predictive of colitic cancer in patients with ulcerative colitis. Dis Colon Rectum. 2007;50:1384-1392. [PubMed] |
| 45. | Parang B, Kaz A, Barrett CW, Short SNW, Keating C, Mittal M, Naik R, Washington M, Revetta F, Smith J. BVES regulates c-Myc stability via PP2A and suppresses colitis-induced tumourigenesis. Gut. 2016;Online First. [RCA] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 46. | Pekow JR, Dougherty U, Mustafi R, Zhu H, Kocherginsky M, Rubin DT, Hanauer SB, Hart J, Chang EB, Fichera A. miR-143 and miR-145 are downregulated in ulcerative colitis: putative regulators of inflammation and protooncogenes. Inflamm Bowel Dis. 2012;18:94-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 47. | Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624-1635.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 404] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 48. | Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, Ando T, Oyamada Y, Isozaki Y, Hongo H. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol. 2010;25 Suppl 1:S129-S133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 49. | Singh UP, Murphy AE, Enos RT, Shamran HA, Singh NP, Guan H, Hegde VL, Fan D, Price RL, Taub DD. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143:478-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 50. | Benderska N, Dittrich AL, Knaup S, Rau TT, Neufert C, Wach S, Fahlbusch FB, Rauh M, Wirtz RM, Agaimy A. miRNA-26b Overexpression in Ulcerative Colitis-associated Carcinogenesis. Inflamm Bowel Dis. 2015;21:2039-2051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Shigaki K, Mitomi H, Fujimori T, Ichikawa K, Tomita S, Imura J, Fujii S, Itabashi M, Kameoka S, Sahara R. Immunohistochemical analysis of chromogranin A and p53 expressions in ulcerative colitis-associated neoplasia: neuroendocrine differentiation as an early event in the colitis-neoplasia sequence. Hum Pathol. 2013;44:2393-2399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | van Schaik FD, Oldenburg B, Offerhaus GJ, Schipper ME, Vleggaar FP, Siersema PD, van Oijen MG, Ten Kate FJ. Role of immunohistochemical markers in predicting progression of dysplasia to advanced neoplasia in patients with ulcerative colitis. Inflamm Bowel Dis. 2012;18:480-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 53. | van der Woude CJ, Moshage H, Homan M, Kleibeuker JH, Jansen PL, van Dekken H. Expression of apoptosis related proteins during malignant progression in chronic ulcerative colitis. J Clin Pathol. 2005;58:811-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 54. | Ludwig K, Fassan M, Mescoli C, Pizzi M, Balistreri M, Albertoni L, Pucciarelli S, Scarpa M, Sturniolo GC, Angriman I. PDCD4/miR-21 dysregulation in inflammatory bowel disease-associated carcinogenesis. Virchows Arch. 2013;462:57-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 55. | Ussakli CH, Ebaee A, Binkley J, Brentnall TA, Emond MJ, Rabinovitch PS, Risques RA. Mitochondria and tumor progression in ulcerative colitis. J Natl Cancer Inst. 2013;105:1239-1248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 56. | Saigusa S, Araki T, Tanaka K, Hashimoto K, Okita Y, Fujikawa H, Okugawa Y, Toiyama Y, Inoue Y, Uchida K. Identification of patients with developing ulcerative colitis-associated neoplasia by nitrative DNA damage marker 8-nitroguanin expression in rectal mucosa. J Clin Gastroenterol. 2013;47:e80-e86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 57. | Chen R, Pan S, Lai K, Lai LA, Crispin DA, Bronner MP, Brentnall TA. Up-regulation of mitochondrial chaperone TRAP1 in ulcerative colitis associated colorectal cancer. World J Gastroenterol. 2014;20:17037-17048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 58. | Yoshizawa S, Matsuoka K, Inoue N, Takaishi H, Ogata H, Iwao Y, Mukai M, Fujita T, Kawakami Y, Hibi T. Clinical significance of serum p53 antibodies in patients with ulcerative colitis and its carcinogenesis. Inflamm Bowel Dis. 2007;13:865-873. [PubMed] |
| 59. | Arai Y, Arihiro S, Matsuura T, Kato T, Matsuoka M, Saruta M, Mitsunaga M, Matsuura M, Fujiwara M, Okayasu I. Prostaglandin E-major urinary metabolite as a reliable surrogate marker for mucosal inflammation in ulcerative colitis. Inflamm Bowel Dis. 2014;20:1208-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 60. | Manfredi MA, Zurakowski D, Rufo PA, Walker TR, Fox VL, Moses MA. Increased incidence of urinary matrix metalloproteinases as predictors of disease in pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1091-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 61. | Wang S, Wang Z, Shi H, Heng L, Juan W, Yuan B, Wu X, Wang F. Faecal calprotectin concentrations in gastrointestinal diseases. J Int Med Res. 2013;41:1357-1361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 62. | Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546-551. [PubMed] |
| 63. | Kisiel JB, Yab TC, Nazer Hussain FT, Taylor WR, Garrity-Park MM, Sandborn WJ, Loftus EV, Wolff BG, Smyrk TC, Itzkowitz SH. Stool DNA testing for the detection of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2013;37:546-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 64. | Redwood DG, Asay ED, Blake ID, Sacco PE, Christensen CM, Sacco FD, Tiesinga JJ, Devens ME, Alberts SR, Mahoney DW. Stool DNA Testing for Screening Detection of Colorectal Neoplasia in Alaska Native People. Mayo Clin Proc. 2016;91:61-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |