Published online Sep 14, 2016. doi: 10.3748/wjg.v22.i34.7824
Peer-review started: April 18, 2016
First decision: May 12, 2016
Revised: June 28, 2016
Accepted: August 8, 2016
Article in press: August 8, 2016
Published online: September 14, 2016
Processing time: 146 Days and 10.1 Hours
To review Hepatitis C virus (HCV) prevalence and genotypes distribution worldwide.
We conducted a systematic study which represents one of the most comprehensive effort to quantify global HCV epidemiology, using the best available published data between 2000 and 2015 from 138 countries (about 90% of the global population), grouped in 20 geographical areas (with the exclusion of Oceania), as defined by the Global Burden of Diseases project (GBD). Countries for which we were unable to obtain HCV genotype prevalence data were excluded from calculations of regional proportions, although their populations were included in the total population size of each region when generating regional genotype prevalence estimates.
Total global HCV prevalence is estimated at 2.5% (177.5 million of HCV infected adults), ranging from 2.9% in Africa and 1.3% in Americas, with a global viraemic rate of 67% (118.9 million of HCV RNA positive cases), varying from 64.4% in Asia to 74.8% in Australasia. HCV genotype 1 is the most prevalent worldwide (49.1%), followed by genotype 3 (17.9%), 4 (16.8%) and 2 (11.0%). Genotypes 5 and 6 are responsible for the remaining < 5%. While genotypes 1 and 3 are common worldwide, the largest proportion of genotypes 4 and 5 is in lower-income countries. Although HCV genotypes 1 and 3 infections are the most prevalent globally (67.0% if considered together), other genotypes are found more commonly in lower-income countries where still account for a significant proportion of HCV cases.
A more precise knowledge of HCV genotype distribution will be helpful to best inform national healthcare models to improve access to new treatments.
Core tip: Hepatitis C virus (HCV) infection is a global public health burden, causing an increasing level of liver-related morbidity and mortality due to the disease progression. Unfortunately, in many countries, there is a lack of robust epidemiological data, especially HCV genotypes distribution, upon which to base country-specific prevention, diagnosis and treatment strategies in order to reduce the disease burden represented by HCV. Stratification by viral genotypes at national and regional level, and a better understanding of viral diversity within target populations, might also critically inform the rational design and testing of future HCV vaccines.
- Citation: Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol 2016; 22(34): 7824-7840
- URL: https://www.wjgnet.com/1007-9327/full/v22/i34/7824.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i34.7824
Hepatitis C virus (HCV) is one of the major globally cause of death and morbidity[1] and recent estimates showed an increase in its seroprevalence over the last decade to 2.8%, corresponding to > 185 million infections worldwide[2].
Chronic HCV infection is often associated with the development of liver cirrhosis, hepatocellular cancer, liver failure, and death[3] especially in HIV-positive patients during active antiretroviral therapy[4]. It has been estimated that while the incidence of HCV infection seems to decrease in the developed world, mortality secondary related to HCV infection will continue to increase over the next 20 years[5]. So, although many data suggest that HCV infection could be eliminated in the next 15-20 years with focused therapeutic strategies[6,7], a good understanding of HCV infections should be required to develop strategies to prevent new infections.
Previous and more recent studies have reported regional prevalence estimates, but always considering a limited number of countries[6,8-13]. A more recent analysis, instead, estimates a global HCV prevalence, but provides only regional estimates[2]. In all the cases, studies focused only on the presence of HCV antibodies generally overestimate the disease burden because they include also patients healed spontaneously or through treatments. So, although antibodies to HCV (anti-HCV) are at present the most commonly available marker of HCV infection, used both to estimate its prevalence and to compare HCV infection levels globally, the most important indicator of HCV diffusion seems to be its classification into different genetic variants. At present, in fact, the length of the therapy and the opportunity to associate interferon and/or ribavirin with the new direct-acting antiviral (DAA) therapies still remain partially dependent on HCV genotype. A detailed understanding of the regional HCV genotype distribution might led to the development of specific national treatment strategies.
Up to now, HCV is classified into seven recognized genotypes[1-5,14-16] on the basis of sequence of the viral genome[17], each differing at 30%-35% of nucleotide sites and into 67 confirmed and 20 provisional subtypes, differing at < 15% of nucleotide sites[18].
There are several methods used to determine HCV genotypes, all using the direct sequencing of specific polymerase chain reaction (PCR)-amplified portions of the virus (NS5, core, E1 and 5’ UTR regions)[18-21], often in combination with the phylogenetic analysis[17]. Apart from the restriction fragment length polymorphisms, in which restriction enzymes are able to recognize genotype-specific cleavage sites in a PCR-amplified DNA fragments[22] but whose sensitivity and specificity seems to need further investigations[23] and Kinetic amplification[24,25], whose reproducibility will need surely more investigations in the future, the most common method for its simple interpretation is surely line probe assays (LiPA)[14-16], in which PCR amplified fragments are able to hybridize to genotype-specific probes immobilized on nitrocellulose strips.
Genotype-specific antibodies able to recognize the NS4 region of HCV have also been exploited[20], even if this type of serological genotyping still lacks specificity and sensitivity with huge limits of its clinical applications.
The geographic distribution of HCV genotypes is rather complex. The so called “epidemic subtypes” - specifically 1a, 1b, 2a, and 3a - are widely distributed worldwide and account for a great proportion of the totality of HCV cases, especially in high income countries. They were probably spread in the 70’s and 80’s, before HCV sequencing, through transfusion, blood products and drug abusers[26-29].
The so called “endemic” strains, instead, are comparatively more rare and have been restricted for long time in specific regions, as West Africa, Southern Asia, Central Africa and South Eastern Asia[26,30,31]. At present, only one genotype 7 infection has been reported from a Central African immigrant in Canada[32].
The present global distribution of HCV genotypes has undoubtedly been influenced by historical events (for example the trans-Atlantic slave trade) or by the contemporary human migration trends[33].
Since the duration and the cost of clinical treatments useful to fight HCV infection are still mostly impacted by the different clinical evolution that each HCV subtype seems to have, especially until pan-genotypic therapies will not able to reach the global market, a correct knowledge of the HCV distribution is surely crucial to contain this global burden disease. At the present, however, more than half of the countries in the world do not have robust studies of the HCV infected population.
The purpose of this study was to conduct a comprehensive review of recently published literature to estimate anti-HCV prevalence, the viraemic rate (HCV-RNA positive) and genotype distribution to generate a global estimate of HCV disease burden.
A comprehensive review of the literature from 2000 to 2015 was used to gather country-specific data on prevalence, number of diagnosed individuals and genotype distribution. References were identified through two sources: indexed journals and non-indexed sources. Indexed articles were found by searching PubMed and regional databases using the following terms: ‘‘[Country Name] and [hepatitis c or HCV] and [prevalence]’’ or “[genotypes] or [viraemia]’’. Furthermore, references cited within the articles were used.
Regions included in the analysis were those defined by the Global Burden of Diseases, Injuries, and Risk Factors 2010 (GBD) study[34,35]. This study defined 21 regions that were ‘‘epidemiologically homogenous as possible so that information from detailed studies in one country can plausibly be extrapolated to other countries in the region to create burden estimates that are useful to individual countries in planning for health sector activities”[36].
The average HCV prevalence for each continent was calculated by dividing the sum of data reported from each region to the total number of countries within the region. The first- and second generation immunoassay tests which usually provide false-positive results overestimating the total infected population[37,38] were not used to estimate the country’s HCV prevalence, selecting only studies in which it was used a third generation test. Similarly, the genotyping and the viraemic rate was obtained only by considering studies in which LiPA test and a well described PCR RT system (range of sensitivity and linearity) was used.
Article titles and abstracts were reviewed for relevance and the following data were extracted from full articles or abstracts: anti-HCV prevalence, viraemic prevalence, viraemic rate and genotypes distribution.
The infected general population was composed of high-risk groups [e.g., people who inject drugs (PWID’s), dialysis patients, haemophiliacs, minority ethnic groups, etc.] as well as non-high-risk groups that contracted the disease through contact with infected blood (e.g. nosocomial infections, dental procedures, etc.). Studies with a sample size of less than 1000 and studies published prior to 2000 or not in English were excluded from the analysis.
Five hundred and fifty-seven articles were selected based on relevance. In addition, non-indexed sources were identified through searches of individual country’s Ministry of Health’s websites and international health agency reports. If articles contained the same patient cohort then this cohort was only counted once.
Because the first- and second generation immunoassay tests may provide false-positive results, which can overestimate the total infected population, care was taken to use only studies that used the latest generation tests to estimate the country’s prevalence.
In the majority of studies HCV cases were classified by LiPA Method at the genotype level, but not at the subtype level, so we decided to use only genotype classification using as general method that proposed by Simmonds et al[17]. In case of one or more genotypes identified in the same patient, we classified it as “mixed”. We did not include genotype 7 in the analysis.
Countries for which we were unable to obtain HCV genotype prevalence data were excluded from calculations of regional proportions, although their populations were included in the total population size of each region when generating regional genotype prevalence estimates.
The GBD subdivides the African continent into 4 macro areas: Central, East, West and Southern, whereas the Saharan area (North Africa) is generally associated with Middle East countries.
The estimated prevalence of anti-HCV in the whole Sub- Saharan Africa is 2.9% with an estimated 26.9 million of cases (Table 1), ranging from 6.0% in the Central area and 0.9% in the Southern countries (Table 2).
| Continent | Anti- HCV prevalence (%) | Viraemic rate (%) | 2013 population (millions) | Anti- HCV infected (millions) | Viraemic HCV infected (millions) |
| Africa | 2.9 | 70.5 | 927.0 | 26.9 | 19.0 |
| North Africa/Middle East | 2.7 | 68.8 | 469.0 | 12.7 | 8.7 |
| America | 1.3 | 74.0 | 953.7 | 12.4 | 9.2 |
| Asia | 2.8 | 64.4 | 3985.0 | 111.6 | 71.9 |
| Australasia | 1.8 | 74.8 | 28.0 | 0.5 | 0.4 |
| Europe | 1.8 | 72.4 | 742.5 | 13.4 | 9.7 |
| Total | 2.5 | 67.0 | 7105.2 | 177.5 | 118.9 |
| Regions | Anti-HCV prevalence (%) | Viraemic rate (%) |
| Central Sub-Saharan Africa | 6.0 | 68.5 |
| EastSub-Saharan Africa | 2.4 | 65.0 |
| Southern Sub-Saharan Africa | 0.9 | 69.0 |
| WestSub-Saharan Africa | 2.4 | 79.6 |
| North Africa and Middle East | 2.7 | 68.8 |
| North America, High Income | 1.2 | 75.7 |
| Caribbean | 1.5 | 70.0 |
| Andean Latin America | 1.2 | 70.0 |
| Central Latin America | 1.4 | 75.8 |
| Southern Latin America | 1.5 | 79.5 |
| Tropical Latin America | 1.6 | 80.2 |
| Central Asia | 5.8 | 48.7 |
| East Asia | 2.8 | 63.6 |
| Pacific Asia, High-income | 1.1 | 70.5 |
| South Asia | 2.5 | 78.5 |
| Southeast Asia | 1.6 | 60.5 |
| Australasia | 1.8 | 74.8 |
| Europe, Central | 1.3 | 76.6 |
| Europe, Eastern | 3.1 | 69.6 |
| Europe, Western | 0.9 | 71.0 |
The viraemic rate is estimated at 70.5%, with a peak of 79.6% in the Western Africa (Table 2), accounting almost 20.0 millions of active HCV replication cases (Table 1).
The most representative genotype is the genotype 4 (G4) (28.1%), followed by genotypes 1 (G1) (26.3%), genotype 2 (G2) (23.7%), genotype 5 (G5) (12.2%), and genotype 3 (G3) (6.3%). No cases of genotype 6 (G6) and only a small percentage of mixed genotypes were reported (Table 3). The genotype distribution shows high variability among the four macro-areas studied, ranging between 82.9% (Central Africa) and 0.6% (West Africa) for G4, 35.7% (Southern Africa) and 0% (West and Central Africa) for G5, 62.9% (West Africa) and 1.2% (Southern Africa) for G2 and 7.4 (East Africa) and 0.8% (Central Africa) for G3. No high variability was observed for G1 (36.2%, 31.4% and 25.5% in East, Southern and West Africa respectively), except for Central Africa (12.3%) (Table 4).
| Continents | G1 (%) | G2 (%) | G3 (%) | G4 (%) | G5 (%) | G6 (%) | Mixed |
| Africa | 26.3 | 23.7 | 6.3 | 28.1 | 12.2 | - | 3.4 |
| North Africa/Middle East | 27.3 | 0.8 | 6.3 | 65.3 | 0.3 | - | - |
| America | 74.5 | 10.2 | 10.6 | 1.7 | 0.1 | 0.3 | 2.6 |
| Asia | 46.6 | 18.6 | 22.4 | 1.0 | 0.1 | 7.0 | 4.3 |
| Australasia | 55.0 | 6.5 | 36.0 | 1.2 | - | 1.3 | - |
| Europe | 64.4 | 5.5 | 25.5 | 3.7 | 0.1 | 0.1 | 0.7 |
| Total (excludes Oceania) | 49.1 | 11.0 | 17.9 | 16.8 | 2.0 | 1.4 | 1.8 |
| Regions | G1 (%) | G2 (%) | G3 (%) | G4 (%) | G5 (%) | G6 (%) | Mixed |
| Central Sub-Saharan Africa | 12.3 | 4.0 | 0.8 | 82.9 | - | - | - |
| East Sub-Saharan Africa | 36.2 | 26.8 | 7.4 | 16.6 | 13.0 | - | - |
| Southern Sub-Saharan Africa | 31.4 | 1.2 | 12.6 | 12.4 | 35.7 | - | 6.7 |
| West Sub-Saharan Africa | 25.5 | 62.9 | 4.4 | 0.6 | - | - | 6.6 |
| North Africa and Middle East | 27.3 | 0.8 | 6.3 | 65.3 | 0.3 | - | - |
| North America | 66.3 | 13.1 | 15.7 | 4.3 | - | 0.6 | - |
| Caribbean | 83.0 | 7.2 | 2.1 | 0.6 | - | 0.1 | 7.0 |
| Andean Latin America | 86.0 | 2.0 | 10.0 | - | - | - | 2.0 |
| Central Latin America | 74.6 | 21.6 | 3.3 | 0.1 | 0.1 | - | 0.3 |
| Southern Latin America | 72.0 | 13.3 | 13.5 | 0.9 | 0.1 | 0.1 | 0.1 |
| Tropical Latin America | 64.8 | 4.6 | 30.2 | 0.2 | 0.1 | - | 0.1 |
| Central Asia | 70.4 | 8.6 | 19.6 | - | - | - | 1.4 |
| East Asia | 53.5 | 31.7 | 5.4 | 0.1 | - | 3.3 | 6.0 |
| Pacific Asia, High-Income | 58.7 | 39.7 | 0.4 | 0.1 | - | 0.5 | 0.6 |
| South Asia | 15.5 | 1.9 | 66.7 | 3.7 | 0.1 | 0.5 | 11.6 |
| Southeast Asia | 35.2 | 11.1 | 19.9 | 0.9 | 0.4 | 30.8 | 1.7 |
| Australasia | 55.0 | 6.5 | 36.0 | 1.2 | - | 1.3 | - |
| Central Europe | 70.0 | 3.2 | 21.0 | 4.9 | - | 0.1 | 0.8 |
| Eastern Europe | 68.1 | 4.3 | 26.6 | 0.5 | - | - | 0.5 |
| Western Europe | 55.1 | 8.9 | 29.0 | 5.8 | 0.2 | 0.1 | 0.8 |
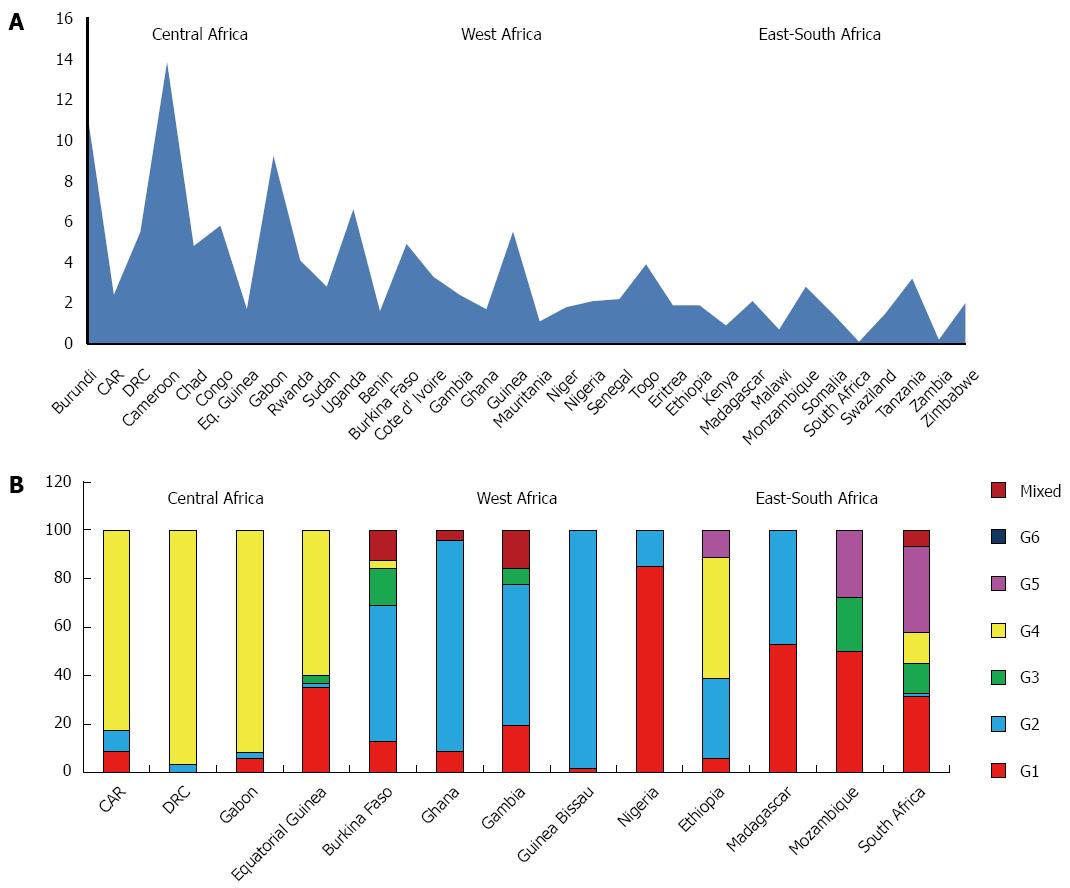
Central Sub-Saharan Africa: The countries studied in this area are Burundi, Cameroon, Central African Republic (CAR), Chad, Congo, Democratic Republic of Congo (DRC), Equatorial Guinea, Gabon, Rwanda, Sudan and Uganda.
The prevalence of HCV in the general population is 6.0%, ranging between 1.7% and 13.8%, depending on the country, with an average viraemic rate estimated at 68.5% (Table 2).The countries with the highest prevalence include Cameroon (13.8%), Burundi (11.3%) and Gabon (9.2%). The countries with the lowest prevalence Equatorial Guinea (1.7%), CAR (2.4%) and Sudan (2.8%) (Figure 1A).
The predominant genotype is G4 (82.9%), followed by G1 (12.3%), G2 (4.0%) and G3 (0.8%). No cases of G5 and G6 were reported (Table 4). In some countries, like DRC and Gabon, G4 is quite the only genotype observed (96.8% and 91.9%, respectively), whereas in others, as Equatorial Guinea, it has been detected an increasing percentage of G1 (35.0%) (Figure 1B). No genotypes distribution data are available from Burundi, Cameroon, Chad, Congo, Rwanda, Sudan and Uganda
East Sub-Saharan Africa: Analysis of this area includes studies from Eritrea, Ethiopia, Kenya, Madagascar, Mozambique, Somalia and Tanzania and shows an average prevalence of HCV infection of 2.4%, ranging between 0.9% and 3.2%, depending on the country, and the lowest viraemic rate of the whole African continent (65.0%) (Table 2). The countries with the highest prevalence are Tanzania (3.2%) and Mozambique (2.8%), those with the lowest Kenya (0.9%) and Somalia (1.5%) (Figure 1A).
The predominant genotype in this area is G1 (36.2%), followed by G2 (26.8%), G4 (16.6%), G5 (13.0%) and G3 (7.4%). No cases of G6 are reported (Table 4). Except for Ethiopia, where G4 represents about the half of all the genotypes described and Madagascar, where the only two genotypes found are G1 (52.9%) and G4 (47.1%), in other countries all the genotypes are equally present (Figure 1B). No genotypes distribution data are available from Eritrea, Kenya, Somalia and Tanzania.
Southern Sub-Saharan Africa: Data concerning this macro area are reported from Malawi, South Africa, Swaziland, Zambia and Zimbabwe and show an average HCV prevalence of 0.9% and a viraemic rate estimated at 69.0% (Table 2). The countries with the highest prevalence are Zimbabwe (2.0%) and Swaziland (1.5%), whereas South Africa and Zambia show the lowest prevalence (0.1% and 0.2%, respectively) (Figure 1A).
The more common genotype is the G5 (35.7%), as reported from the only country where genotypes prevalence has been studied (South Africa), followed by G1 (31.4%), G3 (12.6%) and G4 (12.4%). No cases of G6 were reported and only a small percentage of G2 is described (Figure 1B). No genotypes distribution data are available from Malawi, Swaziland, Zambia and Zimbabwe.
West Sub-Saharan Africa: The countries studied in this area are Benin, Burkina Faso, Cote d’Ivoire, Gambia, Ghana, Guinea Bissau, Mauritania, Niger, Nigeria, Senegal and Togo. The prevalence of HCV is 2.4%, ranging between 1.1% and 5.5%, depending on the country, with the higher viraemic rate of all the African continent (79.6%) (Table 2). The countries with the highest prevalence include Guinea Bissau (5.5%) and Burkina Faso (4.9%), those with the lowest Mauritania (1.1%) and Benin (1.6%) (Figure 1A).
The predominant genotype is G2 (62.9%), followed by G1 (25.5%) and G3 (4.4%) (Table 4). No cases of G5 and G6 were reported and only a small percentage of G4 was described from Burkina Faso. The genotype distribution shows a great heterogeneity from country to country. If G2 accounts nearly the totality of genotyped cases from Guinea Bissau (98.2%) and Ghana (87.0%), Nigeria shows a prevalent circulation of G1 (85.0%) (Figure 1B). No genotypes distribution data are available from Benin, Cote d’Ivoire, Mauritania, Niger, Senegal and Togo.
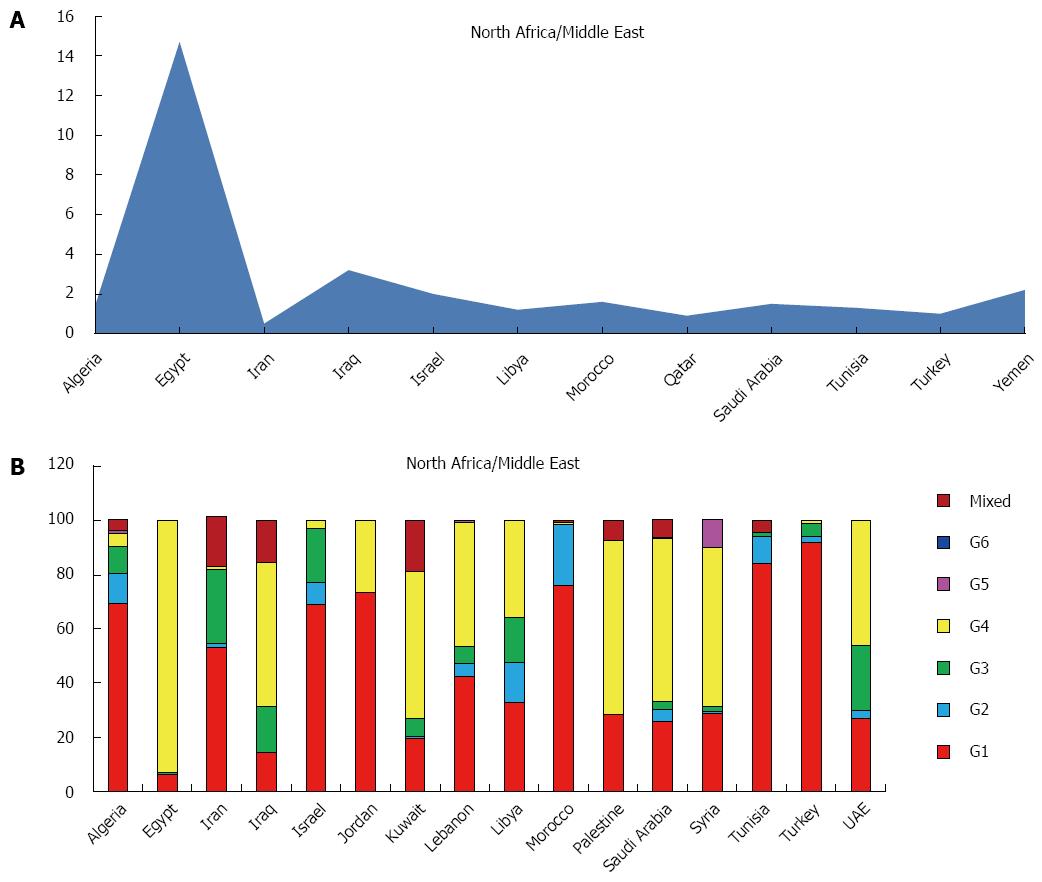
This region includes Algeria, Egypt, Iran, Iraq, Israel, Jordan, Kuwait, Lebanon, Libya, Morocco, Oman, Palestine, Qatar, Saudi Arabia, Syria, Tunisia, Turkey, United Arab Emirates and Yemen with an estimated prevalence of HCV in the general population of 2.7%, corresponding to an estimated 12.7 million of cases. The HCV viraemic rate is estimated at 68.8%, accounting over 8 millions of active HCV replication cases (Table 1). No adult HCV prevalence and/or viraemic data are available from Jordan, Kuwait, Lebanon, Oman, Palestine, Syria and United Arab Emirates. The countries with the highest prevalence include, over Egypt (14.7%), Iraq (3.2%) and Yemen (2.2%). The countries with the lowest Qatar (0.9%) and Turkey (1.0%) (Figure 2A).
The predominant genotype is the genotype 4 (65.3%), followed by 1 (27.3%), 3 (6.3%). Only small percentages of genotype 2 and 5 and no genotype 6 were reported (Table 3).
In this area genotypes distribution is highly heterogenic. If genotype 4 accounts almost the totality of genotyped cases from Egypt (93.1%) and over the half of the total cases in Iraq, Kuwait, Palestine, Saudi Arabia and Syria (52.9%, 54.2%, 64.1%, 60.0% and 59.0%), Turkey, Tunisia, Jordan and Morocco show a prevalent circulation of genotype 1 (91.8%, 84.0%, 77.3% and 75.9%, respectively). The majority of cases of genotype 3 are described in Iran and United Arab Emirates (27.5% and 23.8%), whereas in Libya all the genotypes are equally distributed. No data of genotypes distribution are available from Israel, Oman, Qatar, and Yemen (Figure 2B).
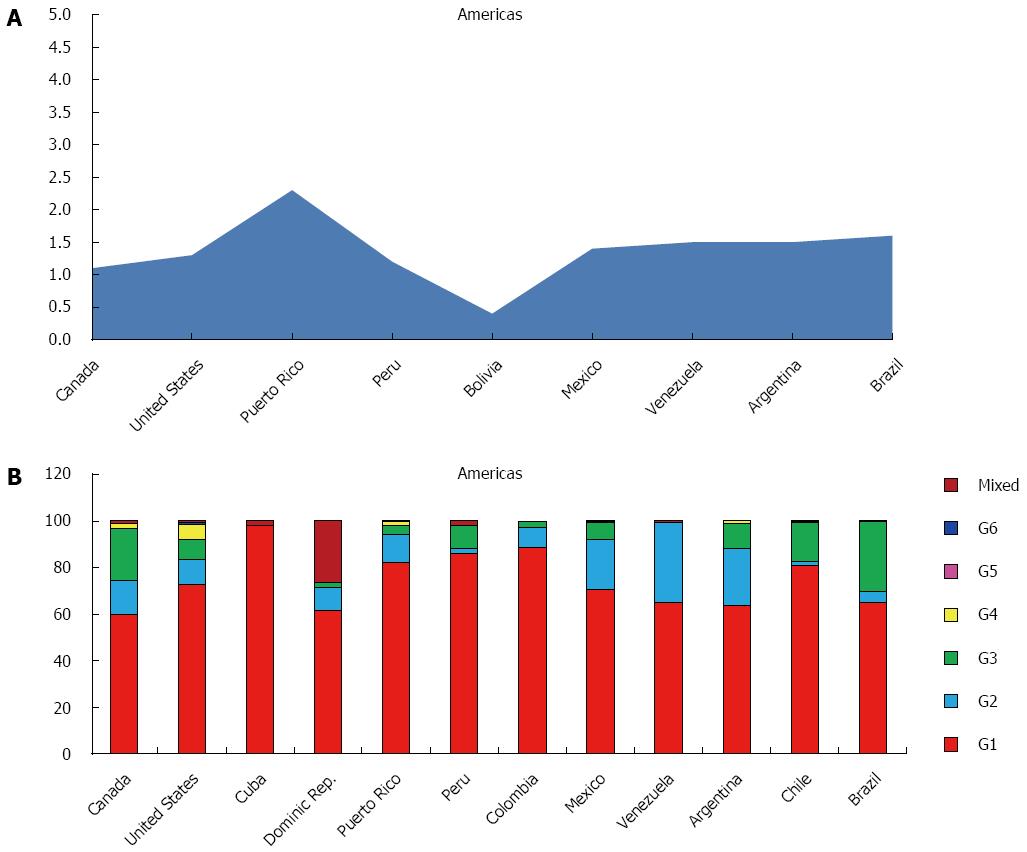
The GBD subdivides the American continent in three macro areas: North America, Caribbean and Latin America.
The estimated prevalence of HCV in the whole continent is 1.3%, with more than 12 million of estimated cases, while the viraemic rate is 74.0%, accounting over 9 million of cases with active HCV replication (Table 1).
The most representative genotype is G1 (74.5%), followed by G3 (10.6%), G2 (10.2%) and G4 (1.7%). Only small percentages of G5, G6 and not well classified are reported (Table 3). Genotype distribution shows high variability among the three macro-areas studied, ranging between 83.0% of the Caribbean area and 66.3% in Northern America for G1, 21.6% (Central Latin America) and 2.0% (Andean Latin America) for G2, 15.7% (Northern America) and 2.1% (Caribbean) for G3 (Table 4).
North America, High Income: The countries studied in this area are Canada and United States, with an average prevalence of HCV in the general population of 1.2%, ranging between 1.1% in Canada and 1.3% in the United States, and a viraemic rate estimated at 75.7% (Figure 3A).
The predominant genotype is G1 (66.3%), followed by G3 (15.7%), G2 (13.1%) and G4 (4.3%). No cases of G5 and only a small percentage of G6 has been reported (Table 4). In both the two studied countries, G1 represents over the half of all genotypes observed (60.0% and 72.5%, respectively), whereas G3 is more frequent in Canada (22.3%), if compared to the United States (8.9%) (Figure 3B).
Caribbean: In this area, studies responding to the selected parameters are available only from Cuba, Dominican Republic and Puerto Rico.
The prevalence of HCV is 1.5%, ranging between 0.8% in Cuba and 2.3% in Puerto Rico (Figure 3A), with a viraemic rate estimated at 70.0% (Table 2). No adult HCV prevalence and/or viraemic data are available from Cuba and Dominican Republic.
The predominant genotype in this area is G1 (83.0%), followed by G2 (7.2%), G3 (2.1%). No cases of G5 and only a small percentage of G4 and G6 are reported (Table 4). In all the studied countries (Cuba, Dominican Republic and Puerto Rico), G1 is almost the only observed (98.0%, 62.6% and 82.1%, respectively) (Figure 3B).
Latin America: GBD sub divides Latin America in four different areas: Andean, which includes Bolivia and Peru, Central, formed by Colombia, Mexico and Venezuela, Southern, represented by Argentina, Chile and Uruguay and Tropical that includes only Brazil.
The HCV prevalence is 1.4%, ranging between 1.2% in the Andean area and 1.6% in the Tropical zone, with a viraemic rate of 76.4% (Table 2).
All the countries in the 4 macro areas show almost the same HCV prevalence: Brazil (1.6%), Argentina and Venezuela (1.5%), Mexico (1.4%) and Peru (1.2%) (Figure 3A). No adult HCV prevalence and/or viraemic data are available from Colombia and Chile.
The predominant genotype is G1 (74.3%), followed by G3 (14.2%), G2 (10.4%). Only a small percentage of G4, G5, G6 and mixed genotypes are reported (Table 4). In some of the countries studied (Peru, Colombia and Chile) G1 is almost the only observed (86.0%, 88.5% and 80.6%, respectively), whereas in others (Venezuela, Mexico and Argentina) G2 shows a significantly percentage (34.4%, 21.8% and 24.7%). Brazil is the only one country where G3 has a high percentage (30.2%). No genotypes distribution data are available from Bolivia and Uruguay (Figure 3B).
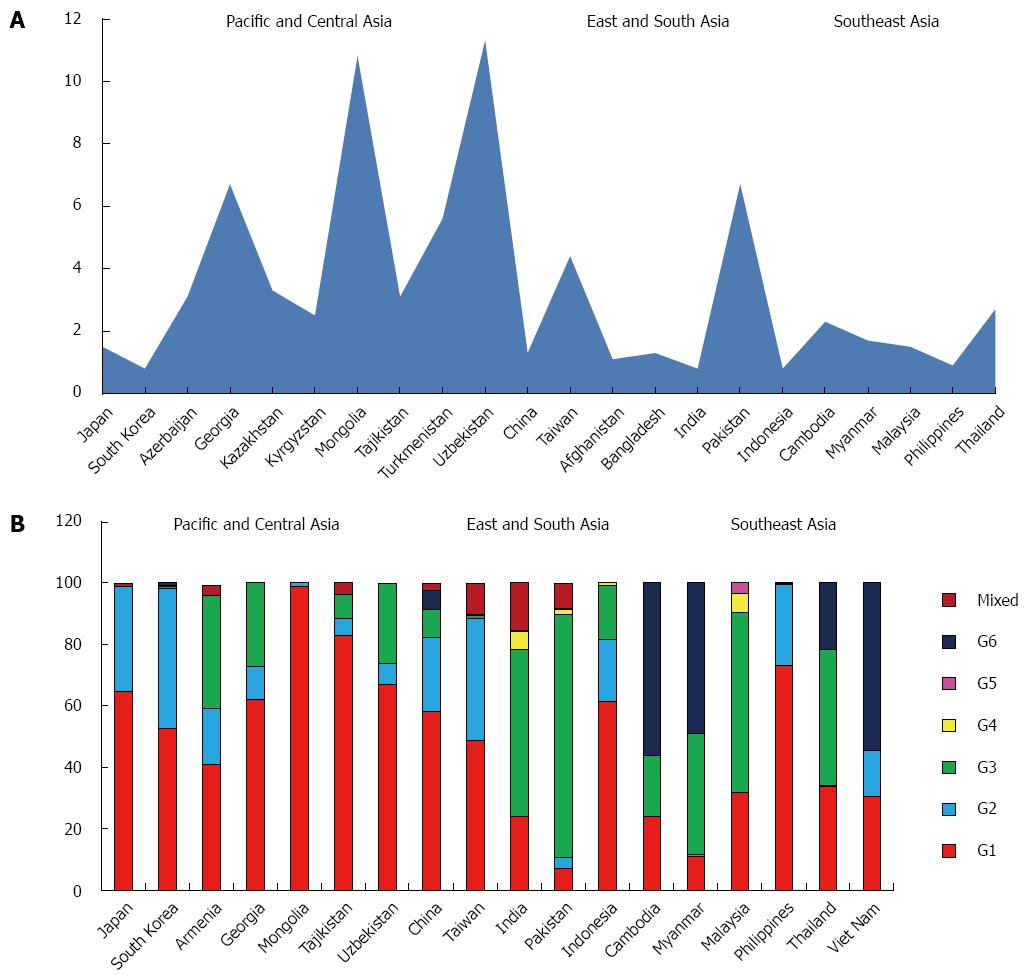
GBD subdivides Asia in five macro areas: Pacific, Central, East, South, and Southeast.
The estimated prevalence of HCV in the whole Asian continent is 2.8%, accounting over 60% of the estimated cases worldwide. The HCV viraemic rate is 64.4%, with 71.9 million of active HCV replication cases (Table 1).
The predominant genotype is the 1 (46.6%), followed by G3 (22.4%), G2 (18.6%) and G6 (7.0%). Only small percentages of G4, G5 and mixed or not further classified genotypes are reported (Table 3).
Genotype distribution shows high variability among the five macro-areas studied, ranging between 70.4% (Central Asia) and 15.5% (Southeast Asia) for G1, 39.7% (Pacific area) and 1.9% (Southeast Asia) for G2, 66.7% (Southeast Asia) and 0.4% (Pacific Asia) for G3, 30.8% (Southeast Asia) and 0.5% (Pacific and South Asia) for G6 and 3.7% (Southeast Asia) and 0.1% (Pacific and East Asia) for G4. Only few cases of G5 are reported and all from South and Southeast areas (Table 2).
Asia Pacific, High Income: Data coming from this zone are available only from Japan and South Korea and show a prevalence of HCV of 1.1%, ranging between 1.5% in Japan and 0.8% in the South Korea, with a viraemic rate estimated at 70.5% (Figure 4A).
The most common genotype is G1 (58.7%), followed by G2 (39.7%). Only small percentages of G3, G4 and G6 and no cases of G5 are reported (Table 4). In the only two countries studied G1 and G2 represent over the 90% of the totality of the genotypes observed (Figure 4B).
Central Asia: The countries studied in this area are Armenia, Azerbaijan, Georgia, Kazakhstan, Kyrgyzstan, Mongolia, Tajikistan, Turkmenistan, Uzbekistan.
The prevalence of HCV in the general population is 5.8% (Table 2), ranging between 11.3% in Uzbekistan and 2.5% in Kyrgyzstan (Figure 4A), with a viraemic rate estimated at 48.7%. No adult HCV prevalence and/or viraemic data are available from Armenia.
The predominant genotype in this area is clearly G1 (70.4%), followed by G3 (19.6%) and G2 (8.6%). Only a small percentage of mixed genotypes were found, whereas no G4, G5 and G6 cases are reported (Table 4).
In Mongolia and Tajikistan, G1 is almost the only genotype found (98.8% and 82.7%, respectively), whereas in two of the five countries in which genotype distribution was studied (Georgia and Uzbekistan) is over the half of the totality of the genotypes identified (62.0% and 67.1%). A considerable percentage of G3 was described in Armenia (37.0%), Georgia (27.0%) and Uzbekistan (26.0%). No genotypes distribution data are available from Azerbaijan, Kazakhstan, Kyrgyzstan and Turkmenistan (Figure 4B).
East Asia: The HCV prevalence in this macroarea, including only China and Taiwan, is 2.8%, ranging between 4.4% in Taiwan and 1.3% in China (Figure 4A), with a viraemic rate estimated at 63.6% (Table 2).
The predominant genotypes in this area are G1 (53.5%) and G2 (31.7%). Only a small percentage of G3 and G6 (5.4% and 3.3%, respectively) have been found, whereas no G5 cases are reported (Table 4).
In the only two countries studied, G1 and G2 represent over the 80% of the totality of the genotypes observed, although a significant portion of G3 is described in China (9.1%) (Figure 4B).
South Asia: The prevalence of anti-HCV in the general population of this large area, including Afghanistan, Bangladesh, India and Pakistan, is 2.5%, ranging between 6.7% in Pakistan and 0.8% in India, with a viraemic rate estimated at 78.5% (Figure 4A).
The most common genotype is G3 (66.7%), followed by G1 (15.5%) and G4 (3.7%). Low percentage of G2, G5 and G6 are reported (Table 4). In both the studied countries (India and Pakistan) G3 counts over the half of all the genotypes described (54.4% and 79.0%). No genotypes distribution data are available from Afghanistan and Bangladesh (Figure 4B).
Southeast Asia: This area includes Cambodia, Indonesia, Laos, Myanmar, Malaysia, Philippines, Sri Lanka, Thailand and Viet Nam, where the prevalence of anti-HCV in the general population is 1.6%, ranging between 2.7% in Thailand and 0.8% in Indonesia (Figure 4A), with a viraemic rate estimated at 60.5% No adult HCV prevalence and/or viraemic data are available from Laos, Sri Lanka and Viet Nam.
G1 and G6 account over the 60% of all the genotypes identified (35.2% and 30.8%, respectively), followed by G3 (19.9%) and G2 (11.1%). Only a small percentage of G4 and G5 has been found (Table 4). Except for Laos where G6 represents about the totality of genotypes observed (95.6%) and Indonesia and Philippines in which G1 accounts over the 60% of the totality of cases reported, a high heterogeneity is observed in all the other countries. In some countries (Cambodia, Myanmar and Viet Nam) G6 is predominant (56.0%, 49.0% and 54.4%, respectively), whereas in others (Sri Lanka, Philippines and Indonesia) G2 shows a significantly percentage (37.5%, 26.4% and 20.2%). Malaysia and Thailand are the countries where G3 shows the highest percentages (58.6% and 44.2%) (Figure 4B).
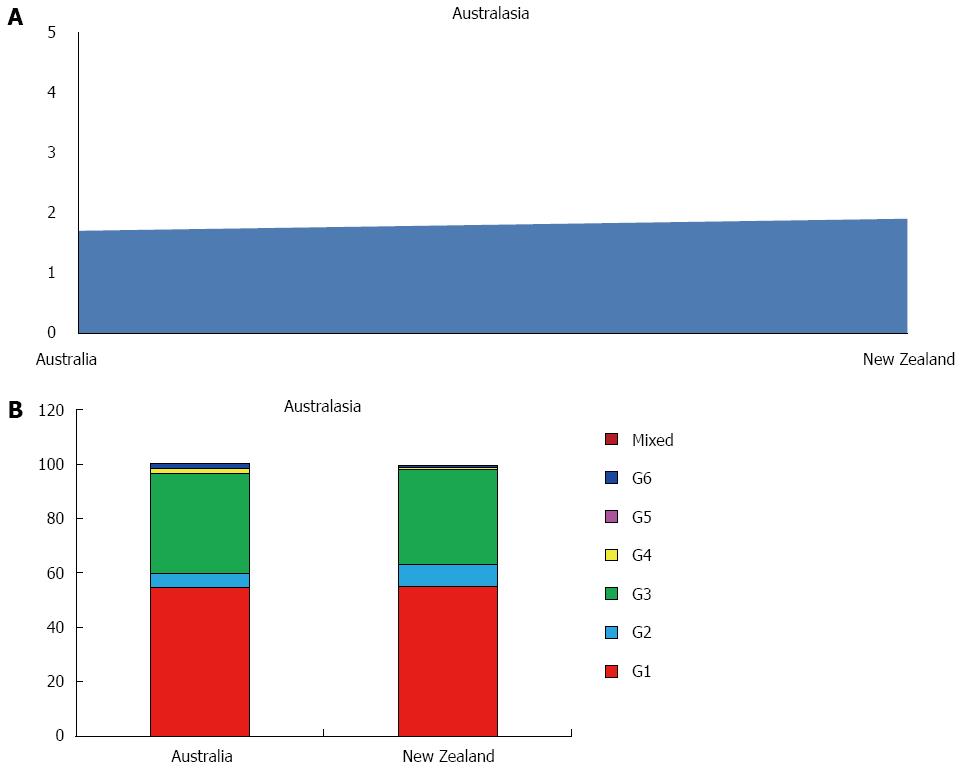
Studies concerning this area has been found only from Australia and New Zealand, with a prevalence of HCV of 1.8%, ranging between 1.9% in New Zealand and 1.7% in Australia, accounting 0.5 millions of estimated cases (Figure 5A). The viraemic rate is 74.8%, with 0.4 million of active HCV replication cases (Table 1).
The more common genotype is G1 (55.0%) followed by G3 (36.0%) and G2 (6.6%). Only a small percentage of G4 and G5 has been found. No G5 cases are described (Table 3, Figure 5B).
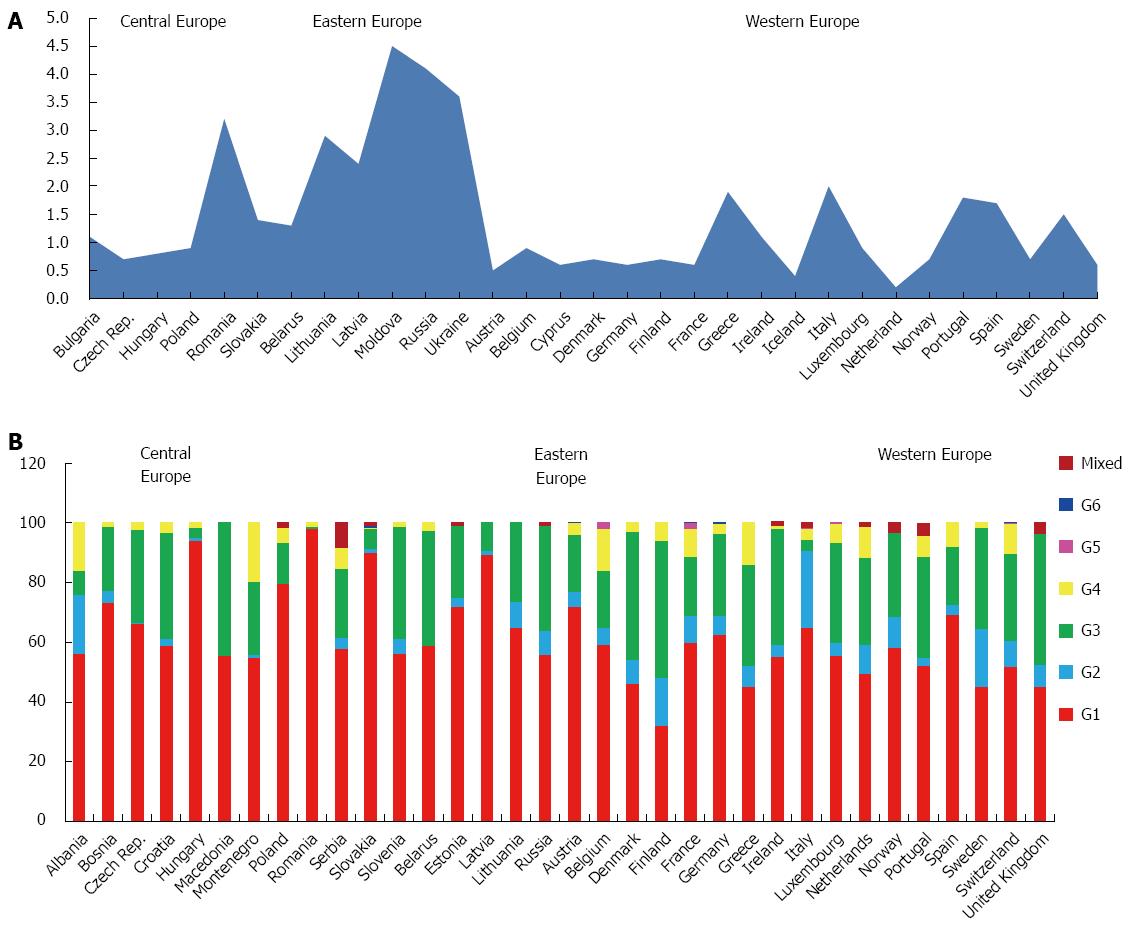
The GBD subdivides European countries into 3 main areas: Central, Eastern and Western.
The estimated prevalence of HCV of the whole continent is 1.8%, accounting over 13 million of estimated cases. The average HCV viraemic rate is 72.4%, with a population of almost 10 million of HCV RNA positive patients (Table 1).
The predominant genotype is G1 (64.4%), followed by G3 (25.5%), G2 (5.5%) and G4 (3.7%). Only small percentages of G5, G6 and mixed or not further classified genotypes are reported (Table 3).
Genotype distribution does not show high variability among the three macro-areas studied, ranging between 70.0% (Central Europe), 68.1% (Eastern Europe) and 55.1% (Western Europe) for genotype 1, 29.0% (Western Europe), 26.6% (Eastern Europe) and 21.0% (Central Europe) for genotype 3.
G2 seems to have a major prevalence in the Western Europe (8.9%), if compared to Eastern (4.3%) or Central (3.2%), whereas G4 is present especially in Central and Western area (4.9% and 5.8%, respectively).
Only few cases of G5 and G6 are reported and mainly from Western area (Table 4).
Central Europe: This large area includes countries like Albania, Bulgaria, Bosnia and Herzegovina, Czech Republic, Croatia, Hungary, Macedonia, Montenegro, Poland, Romania, Serbia, Slovakia and Slovenia, with a prevalence of HCV infection of 1.3%, varying between 1.4% in Slovakia and 0.7% in Czech Republic (Figure 6A) and a viraemic rate estimated at 76.6% (Table 2). We have not found representative data concerning the HCV prevalence and/or HCV viraemic rate from published studies in Albania, Bosnia and Herzegovina, Croatia, Macedonia, Montenegro, Serbia and Slovenia.
The predominant genotypes in this area is G1 (70.0%), followed by G3 (21.0%), G4 (4.9%) and G2 (3.2%). Only a small percentage of mixed genotypes and G6 has been found, whereas no G5 cases are reported (Table 4). In Romania, Hungary and Slovakia, G1 is almost the only genotype found (98.0%, 94.1% and 89.9%, respectively). A considerable percentage of G3 was described in Macedonia (44.6%), Slovenia (37.8%) and Croatia (35.6%), while a significant prevalence of G2 was described only in Albania (20.0%) and of G4 in Montenegro (19.6%) and Albania (16.0%). No genotypes distribution data are available from Bulgaria (Figure 6B).
Eastern Europe: The prevalence of HCV infection in this zone, including Belarus, Estonia, Lithuania, Latvia, Moldova, Russia and Ukraine, is 3.1%, ranging between 4.5% in Moldova and 1.3% in Belarus (Figure 6A), with a viraemic rate estimated at 69.6% (Table 2). No adult HCV prevalence and/or viraemic data are available from Estonia.
The predominant genotypes in this area is G1 (68.1%), followed by G3 (26.6%) and G2 (4.3%). Only a small percentage of mixed genotypes and G4 (0.5%) are reported, whereas no G5 and G6 cases has been described (Table 4).
Only in Latvia G1 is the dominant genetic variant (89.2%). A considerable percentage of G3 was described in Belarus (38.5%) and Russia (35.1%). No genotypes distribution data are available from Moldova and Ukraine (Figure 6B).
Western Europe: The countries studied in this area are Austria, Belgium, Cyprus, Denmark, Finland, France, Germany, Greece, Iceland, Ireland, Italy, Luxembourg, Netherlands, Norway, Portugal, Spain, Sweden, Switzerland and United Kingdom
The prevalence of HCV in the general population of this area is 0.9%, ranging between 2.0% in Italy and 0.2% in Netherlands (Figure 6A), with a viraemic rate estimated at 71.0% (Table 2).
The predominant genotypes is G1 (55.1%), followed by G3 (29.0%), G2 (8.9%) and G4 (5.8%), whereas only small percentages of G5, G6 and mixed genotypes are reported (Table 4). In Austria, Spain, Germany and Italy G1 is over the sixty percent of all the genotypes found. A considerable percentage of G3 was described in some of the countries of the Northern Europe, as Finland (46.0%), United Kingdom (43.8%), Denmark (43.0%), whereas only Italy shows a significant percentage of G2 (26.0%) (Figure 6B). No genotypes distribution data are available from Cyprus and Iceland.
Hepatitis C virus (HCV) infection is one of the main global health burden[5,39-43] and its paradigm varies regionally depending on its historical and present risk factors. A country-specific policy of prevention, diagnosis and treatment could reduce this disease burden, but unfortunately, in many countries, there is a lack of robust epidemiological data upon which to base these strategies, being HCV infection rates[2,44-48] typically focused on quantifying only the anti-HCV prevalence with no attention to HCV genotypes distribution.
This study represents one of the most comprehensive effort to present in a systematic manner the actual situation of the epidemiology of HCV infection, using the best available published and unpublished data between 2000 and 2015, giving a special care not only on available data, but focusing the attention especially on more relevant data. For example, studies on HCV prevalence among blood donors, available in many countries and surely attractive for the large sample size, were excluded because this population, corresponding to healthy screened adults, is not representative of the total population. For the same reason, even if opposite, numerous high risk populations studies (e.g., PWIDs, haemodialysis patients, cancer patients, etc.) were not considered too. Finally, it is important to clear that all the studies published prior to 2000 were also excluded considering the global epidemiological changes that HCV infections has had in the latest twenty years[39,40,49-52].
Studying 138 countries worldwide (36 in Sub- Saharan Africa, 16 in Americas, 26 in Asia, 2 in Australasia, 19 in North Africa/Middle East area and 39 in Europe), total global HCV prevalence is estimated at 2.5% (177 million of HCV infected adults). Central Asia and Central Africa are estimated to have high prevalence (> 3.5%); East, South and Southeast Asia, West and East Africa, North Africa and Middle East, Southern and Tropical Latin America, Caribbean, Australasia, and Eastern Europe moderate prevalence (1.5%-3.5%); whereas Southern Africa, North America, Andean and Central Latin America, Pacific Asia and Western and Central Europe have low prevalence (< 1.5%) (Table 2).
No adult HCV prevalence studies were available from 19 countries (4 in Asia, 4 in Americas, 5 in North Africa/Middle East area and 6 in Europe). In order of their contribution, twenty five of the 138 countries studied account for almost the half of total viraemic infections worldwide and China, Egypt, India, Nigeria, Pakistan and Russia together for more than 70% of them.
Our analysis shows that globally the prevalence and number of HCV infected patients, if compared to a similar study concerning the period 1990-2005[2], has decreased from 2.8% [95% uncertainty interval (UI): 2.6%-3.1%] to 2.5% (95%UI: 2.3%-2.7%) and from 185 to 177millions. It is interesting to note that the most relevant decrease has been observed in the high income zones, especially in Western Europe (-1.5%), Southern Africa (-1.2%) and Australasia (-0.9%), whereas a massive increase it’s reported in some of the low income areas as Central Africa (+3.7%) and Central Asia (+2.0%)[53].
By estimating the total number of HCV RNA positive infections, our data show that the global average viraemic rate is at 67% (118.9 million of HCV RNA positive cases), varying from 48.7% in Central Asia to 80.2% in Tropical Latin America (Table 2). It is interesting notice that some countries, as Poland, where it has been reported a high anti-HCV prevalence and an increasing viraemic rate[54], using a confirmatory antibody test, the anti-HCV prevalence is significantly lower (< 1%)[55], suggesting that some historically high antibody prevalence estimates may be influenced by the use of low sensitive screening HCV tests.
Globally, G1 accounts for 49.1% of all anti-HCV infections among adults making it the most common, followed by G3 (17.9%), G4 (16.8%), G2 (11.0%), G5 (2.0%) and G6 (1.4%). Our data are quite different from those reported by other global studies[56], not only because we considered some countries before excluded, but mostly because our data were summed with other global or continental reports[43,57,58]. Undefined or mixed genotypes accounts for 1.8% of the total HCV infections (Table 1). However, significant regional, country and local variations exists. Infections in Caribbean, Latin America, North America and Europe are predominately caused by G1 (83.0%, 74.3%, 66.3% and 64.4%, respectively). North Africa and the Middle East has a large G4 population (65.3%), probably attributable to the high prevalence of G4 in Egypt (93.1%). If Egypt is excluded, in fact, G4 accounts only for 32.3% of all infections and the genotype distribution of this region is dominated by G1 (48.3%). In Asia the two most common genotypes are G1 (46.6%) and G3 (22.4%), the latter largely driven by its high prevalence in South Asia (66.7%), where in India and Pakistan G3 shows the highest percentages of the whole continent (54.4% and 79.0%, respectively). In Australasia, G1 dominated (55.0%), followed by G3 (25.5%). HCV genotypes by sub regions are shown in Table 4.
Aside from countries entirely lacking genotype information (Bulgaria, Cyprus, Moldova and Ukraine in Europe, Afghanistan, Azerbaijan, Bangladesh, Kazakhstan, Kyrgyzstan and Turkmenistan in Asia, Bolivia, Martinique, Suriname and Uruguay in America, Israel, Oman, Qatar and Yemen in North Africa/Middle East Area and several of the Western and Eastern Sub- Saharan African countries) our data suggest that data concerning the genotype prevalence are particularly weakest in the majority of Asia (accounting for 3.6% of the global population), followed by Africa (3.2% of the global population) and Latin America (1.4% of the global population), if considered in terms of proportion of the genotyped patients respect to overall populations.
It is interesting to note that G1, the most prevalent genotype in high income countries, well served by second generation DAA therapies with a viral eradication rates > 90%[59,60], is also the most prevalent globally. This may be related to the strict association between G1 and the transfusion risk very common prior to the discovery of HCV in 1989[22].
The high circulation of G3, instead, the second most common genotype, especially in Europe, which accounts for almost 20% of global infections, not susceptible to the first generation of DAA protease inhibitors and less susceptible to Sofosbuvir[61,62], is likely related to the association between subtype 3a and drug abuse[20,63,64] and also to migrations to Europe from countries where this subtype is dominant, such as India and Pakistan[65].
Although HCV G1 and G3 infections are the most prevalent globally (67.0% if considered together), other genotypes are particularly common in lower-income countries (Table 4), as G2 in West Africa (62.9%) and in some regions of South America, probably caused by population movements during the trans-Atlantic slave trade in the Eighteenth Century[26] and G4 and G6, highly present in Central and Northern Africa (82.9% and 65.3%) and in Southeast Asia (30.8%). The particularly high prevalence of subtype 4a in Egypt could be the result of unsafe injections during the anti-schistosomal public health campaigns in the past[66], whereas the diffusion of subtype 4d in Europe[67,68] and of subtype 6a in Hong Kong[69] have probably been amplified by the high number of PWIDs in these areas.
It has been estimated that G2, G4, and G6 combined account for nearly one third of all HCV cases globally. Higher proportions of G2 were found in Northern Europe and in some of the ex-soviet republics, probably in accordance to the Asian genotypes distribution, and in Italy, especially in the Southern regions[70,71].
Anyway, it is necessary to clarify that this analysis shows several limitations, especially related to the lack of information available from some extended regions (first of all, Africa and Asia) that has necessarily forced us to use regional estimates, sometimes coming from few high populated countries. For example, the Central Asia viraemic rate estimate, calculated on the basis of data published in Uzbekistan (viraemic rate of 39%) and in Mongolia (viraemic rate of 70%), is heavily influenced by Uzbekistan due to its much larger population. Data from additional countries would be helpful in the future in minimizing this bias.
Another limitation is the lack of robust epidemiology studies at the national level. Only 21% of the 138 countries included in this analysis shows a sample size > 10000, selected by multiple cities or regions, and a random sampling strategy, while the majority are generally conducted in a select population within one setting (e.g., hospital, clinic, city, etc.).
Anyway, in the absence of better data, this analysis suggests that HCV prevalence and the viraemic rate have decreased from 2005 to date, maybe for the impact of the new DAA therapies, even if a better knowledge of genotype distribution at a national level, especially in its subtype diversification, may be yet critic for the complete understanding of HCV disease and to design and testing of vaccines, especially in countries where genotype diversity is particularly high.
Hepatitis C virus (HCV) infection and distribution of its genetic variants throughout the world.
This manuscript provided the strategies to fight the diffusion of HCV infection worldwide.
The authors showed us the epidemiological up-date of HCV genotypes.
These findings can be used in clinical therapy and prevention strategies.
This is a pretty good manuscript. The analysis performed by the authors maximized the extraction of meaningful information from available literature.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Rodriguez-Frias F, Vaughan G S- Editor: Gong ZM L- Editor: A E- Editor: Ma S
| 1. | Cooke GS, Lemoine M, Thursz M, Gore C, Swan T, Kamarulzaman A, DuCros P, Ford N. Viral hepatitis and the Global Burden of Disease: a need to regroup. J Viral Hepat. 2013;20:600-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 2. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1770] [Cited by in RCA: 1847] [Article Influence: 153.9] [Reference Citation Analysis (3)] |
| 3. | Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2042] [Cited by in RCA: 2025] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 4. | Antiretroviral Therapy Cohort Collaboration. Causes of death in HIV-1-infected patients treated with antiretroviral therapy, 1996-2006: collaborative analysis of 13 HIV cohort studies. Clin Infect Dis. 2010;50:1387-1396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 482] [Cited by in RCA: 481] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 5. | Razavi H, Elkhoury AC, Elbasha E, Estes C, Pasini K, Poynard T, Kumar R. Chronic hepatitis C virus (HCV) disease burden and cost in the United States. Hepatology. 2013;57:2164-2170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 337] [Cited by in RCA: 360] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 6. | Wedemeyer H, Duberg AS, Buti M, Rosenberg WM, Frankova S, Esmat G, Örmeci N, Van Vlierberghe H, Gschwantler M, Akarca U. Strategies to manage hepatitis C virus (HCV) disease burden. J Viral Hepat. 2014;21 Suppl 1:60-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 146] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 7. | Martin NK, Hickman M, Hutchinson SJ, Goldberg DJ, Vickerman P. Combination interventions to prevent HCV transmission among people who inject drugs: modeling the impact of antiviral treatment, needle and syringe programs, and opiate substitution therapy. Clin Infect Dis. 2013;57 Suppl 2:S39-S45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 257] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 8. | Lavanchy D. The threat to public health of hepatitis C. Res Virol. 1997;148:143-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 9. | Lavanchy D. Hepatitis C: public health strategies. J Hepatol. 1999;31 Suppl 1:146-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 919] [Cited by in RCA: 945] [Article Influence: 67.5] [Reference Citation Analysis (2)] |
| 11. | Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 939] [Article Influence: 58.7] [Reference Citation Analysis (1)] |
| 12. | Global Burden Of Hepatitis C Working Group. Global burden of disease (GBD) for hepatitis C. J Clin Pharmacol. 2004;44:20-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 359] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 13. | Bruggmann P, Berg T, Øvrehus AL, Moreno C, Brandão Mello CE, Roudot-Thoraval F, Marinho RT, Sherman M, Ryder SD, Sperl J. Historical epidemiology of hepatitis C virus (HCV) in selected countries. J Viral Hepat. 2014;21 Suppl 1:5-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 183] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 14. | Timm J, Roggendorf M. Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J Gastroenterol. 2007;13:4808-4817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 70] [Cited by in RCA: 67] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Ford N, Kirby C, Singh K, Mills EJ, Cooke G, Kamarulzaman A, duCros P. Chronic hepatitis C treatment outcomes in low- and middle-income countries: a systematic review and meta-analysis. Bull World Health Organ. 2012;90:540-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 16. | Ford N, Singh K, Cooke GS, Mills EJ, von Schoen-Angerer T, Kamarulzaman A, du Cros P. Expanding access to treatment for hepatitis C in resource-limited settings: lessons from HIV/AIDS. Clin Infect Dis. 2012;54:1465-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Simmonds P, Alberti A, Alter HJ, Bonino F, Bradley DW, Brechot C, Brouwer JT, Chan SW, Chayama K, Chen DS. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994;19:1321-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 573] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 18. | Anderson JC, Simonetti J, Fisher DG, Williams J, Yamamura Y, Rodriguez N, Sullivan DG, Gretch DR, McMahon B, Williams KJ. Comparison of different HCV viral load and genotyping assays. J Clin Virol. 2003;28:27-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Bullock GC, Bruns DE, Haverstick DM. Hepatitis C genotype determination by melting curve analysis with a single set of fluorescence resonance energy transfer probes. Clin Chem. 2002;48:2147-2154. [PubMed] |
| 20. | Zein NN. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000;13:223-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 245] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 21. | Simmonds P, Mellor J, Sakuldamrongpanich T, Nuchaprayoon C, Tanprasert S, Holmes EC, Smith DB. Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J Gen Virol. 1996;77:3013-3024. [PubMed] |
| 22. | Furione M, Simoncini L, Gatti M, Baldanti F, Grazia Revello M, Gerna G. HCV genotyping by three methods: analysis of discordant results based on sequencing. J Clin Virol. 1999;13:121-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Widell A, Shev S, Månsson S, Zhang YY, Foberg U, Norkrans G, Frydén A, Weiland O, Kurkus J, Nordenfelt E. Genotyping of hepatitis C virus isolates by a modified polymerase chain reaction assay using type specific primers: epidemiological applications. J Med Virol. 1994;44:272-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 24. | Comanor L, Elkin C, Leung K, Krajden M, Kronquist K, Nicolas K, Horansky E, deMedina M, Kittichai P, Sablon E. Successful HCV genotyping of previously failed and low viral load specimens using an HCV RNA qualitative assay based on transcription-mediated amplification in conjunction with the line probe assay. J Clin Virol. 2003;28:14-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Zheng X, Pang M, Chan A, Roberto A, Warner D, Yen-Lieberman B. Direct comparison of hepatitis C virus genotypes tested by INNO-LiPA HCV II and TRUGENE HCV genotyping methods. J Clin Virol. 2003;28:214-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 26. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 981] [Article Influence: 89.2] [Reference Citation Analysis (1)] |
| 27. | Smith DB, Pathirana S, Davidson F, Lawlor E, Power J, Yap PL, Simmonds P. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78:321-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 194] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Pybus OG, Cochrane A, Holmes EC, Simmonds P. The hepatitis C virus epidemic among injecting drug users. Infect Genet Evol. 2005;5:131-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 29. | Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SY, Shapiro B, Pybus OG, Allain JP, Hatzakis A. The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med. 2009;6:e1000198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 155] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 30. | Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 166] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 31. | Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, Humphreys IS. Genetic history of hepatitis C virus in East Asia. J Virol. 2009;83:1071-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 167] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 32. | Murphy DG, Willems B, Deschênes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5’ untranslated region sequences. J Clin Microbiol. 2007;45:1102-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 215] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 33. | Markov PV, van de Laar TJ, Thomas XV, Aronson SJ, Weegink CJ, van den Berk GE, Prins M, Pybus OG, Schinkel J. Colonial history and contemporary transmission shape the genetic diversity of hepatitis C virus genotype 2 in Amsterdam. J Virol. 2012;86:7677-7687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Institute for Health Metrics and Evaluation. GBD 2010 common questions. Seattle, WA: Institute for Health Metrics and Evaluation 2013; . [RCA] [DOI] [Full Text] [Cited by in Crossref: 592] [Cited by in RCA: 794] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 35. | Institute for Health Metrics and Evaluation. GBD 2013 protocol. Seattle, Washington: Institute for health Metrics and Evaluation 2014; . [RCA] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | GBD 2005 Study. Global burden of diseases, injuries and risk factors study operations manual (final draft, January 2009). Accessed 2010-12-05. Available from: http: //www.globalburden.org. |
| 37. | Abdel-Hamid M, El-Daly M, El-Kafrawy S, Mikhail N, Strickland GT, Fix AD. Comparison of second- and third-generation enzyme immunoassays for detecting antibodies to hepatitis C virus. J Clin Microbiol. 2002;40:1656-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 38. | Kershenobich D, Razavi HA, Cooper CL, Alberti A, Dusheiko GM, Pol S, Zuckerman E, Koike K, Han KH, Wallace CM. Applying a system approach to forecast the total hepatitis C virus-infected population size: model validation using US data. Liver Int. 2011;31 Suppl 2:4-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 39. | Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-2128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9500] [Cited by in RCA: 9576] [Article Influence: 736.6] [Reference Citation Analysis (0)] |
| 40. | Deuffic-Burban S, Deltenre P, Buti M, Stroffolini T, Parkes J, Mühlberger N, Siebert U, Moreno C, Hatzakis A, Rosenberg W. Predicted effects of treatment for HCV infection vary among European countries. Gastroenterology. 2012;143:974-985.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 41. | Razavi H, Waked I, Sarrazin C, Myers RP, Idilman R, Calinas F, Vogel W, Mendes Correa MC, Hézode C, Lázaro P. The present and future disease burden of hepatitis C virus (HCV) infection with today’s treatment paradigm. J Viral Hepat. 2014;21 Suppl 1:34-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 294] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 42. | Gane E, Kershenobich D, Seguin-Devaux C, Kristian P, Aho I, Dalgard O, Shestakova I, Nymadawa P, Blach S, Acharya S. Strategies to manage hepatitis C virus (HCV) infection disease burden - volume 2. J Viral Hepat. 2015;22 Suppl 1:46-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 43. | Hatzakis A, Chulanov V, Gadano AC, Bergin C, Ben-Ari Z, Mossong J, Schréter I, Baatarkhuu O, Acharya S, Aho I. The present and future disease burden of hepatitis C virus (HCV) infections with today’s treatment paradigm - volume 2. J Viral Hepat. 2015;22 Suppl 1:26-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 44. | Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, Amarapurkar D, Chen CH, Dou X, El Khayat H. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011;31 Suppl 2:61-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 395] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 45. | Kershenobich D, Razavi HA, Sánchez-Avila JF, Bessone F, Coelho HS, Dagher L, Gonçales FL, Quiroz JF, Rodriguez-Perez F, Rosado B. Trends and projections of hepatitis C virus epidemiology in Latin America. Liver Int. 2011;31 Suppl 2:18-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 46. | Madaliński K, Zakrzewska K, Kołakowska A, Godzik P. Epidemiology of HCV infection in Central and Eastern Europe. Przegl Epidemiol. 2015;69:459-464, 581-584. [PubMed] |
| 47. | Cornberg M, Razavi HA, Alberti A, Bernasconi E, Buti M, Cooper C, Dalgard O, Dillion JF, Flisiak R, Forns X. A systematic review of hepatitis C virus epidemiology in Europe, Canada and Israel. Liver Int. 2011;31 Suppl 2:30-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 287] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 48. | Hope VD, Eramova I, Capurro D, Donoghoe MC. Prevalence and estimation of hepatitis B and C infections in the WHO European Region: a review of data focusing on the countries outside the European Union and the European Free Trade Association. Epidemiol Infect. 2014;142:270-286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 49. | Davis GL, Alter MJ, El-Serag H, Poynard T, Jennings LW. Aging of hepatitis C virus (HCV)-infected persons in the United States: a multiple cohort model of HCV prevalence and disease progression. Gastroenterology. 2010;138:513-521, 521.e1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 666] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 50. | Sypsa V, Touloumi G, Tassopoulos NC, Ketikoglou I, Vafiadis I, Hatzis G, Tsantoulas D, Akriviadis E, Delladetsima J, Demonakou M. Reconstructing and predicting the hepatitis C virus epidemic in Greece: increasing trends of cirrhosis and hepatocellular carcinoma despite the decline in incidence of HCV infection. J Viral Hepat. 2004;11:366-374. [PubMed] |
| 51. | Meffre C, Le Strat Y, Delarocque-Astagneau E, Dubois F, Antona D, Lemasson JM, Warszawski J, Steinmetz J, Coste D, Meyer JF. Prevalence of hepatitis B and hepatitis C virus infections in France in 2004: social factors are important predictors after adjusting for known risk factors. J Med Virol. 2010;82:546-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 159] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 52. | Denniston MM, Jiles RB, Drobeniuc J, Klevens RM, Ward JW, McQuillan GM, Holmberg SD. Chronic hepatitis C virus infection in the United States, National Health and Nutrition Examination Survey 2003 to 2010. Ann Intern Med. 2014;160:293-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 579] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 53. | Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61:S45-S57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1325] [Cited by in RCA: 1362] [Article Influence: 123.8] [Reference Citation Analysis (0)] |
| 54. | Flisiak R, Halota W, Horban A, Juszczyk J, Pawlowska M, Simon K. Prevalence and risk factors of HCV infection in Poland. Eur J Gastroenterol Hepatol. 2011;23:1213-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 55. | Godzik P, Kołakowska A, Madaliński K, Stepień M, Zieliński A, Góralewska A, Kazimierska M, Kunc-Kozioł R, Nadolska B, Pawłowska A. [Prevalence of anti-HCV antibodies among adults in Poland--results of cross-sectional study in general population]. Przegl Epidemiol. 2012;66:575-580. [PubMed] |
| 56. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1145] [Article Influence: 114.5] [Reference Citation Analysis (0)] |
| 57. | Karoney MJ, Siika AM. Hepatitis C virus (HCV) infection in Africa: a review. Pan Afr Med J. 2013;14:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 58. | Liakina V, Hamid S, Tanaka J, Olafsson S, Sharara AI, Alavian SM, Gheorghe L, El Hassan ES, Abaalkhail F, Abbas Z. Historical epidemiology of hepatitis C virus (HCV) in select countries - volume 3. J Viral Hepat. 2015;22 Suppl 4:4-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 59. | Lawitz E, Poordad FF, Pang PS, Hyland RH, Ding X, Mo H, Symonds WT, McHutchison JG, Membreno FE. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383:515-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 443] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 60. | Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med. 2014;370:211-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 911] [Article Influence: 82.8] [Reference Citation Analysis (0)] |
| 61. | Jacobson IM, Gordon SC, Kowdley KV, Yoshida EM, Rodriguez-Torres M, Sulkowski MS, Shiffman ML, Lawitz E, Everson G, Bennett M. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N Engl J Med. 2013;368:1867-1877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 846] [Cited by in RCA: 838] [Article Influence: 69.8] [Reference Citation Analysis (0)] |
| 62. | Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368:1878-1887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1322] [Cited by in RCA: 1325] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 63. | Roman F, Hawotte K, Struck D, Ternes AM, Servais JY, Arendt V, Hoffman P, Hemmer R, Staub T, Seguin-Devaux C. Hepatitis C virus genotypes distribution and transmission risk factors in Luxembourg from 1991 to 2006. World J Gastroenterol. 2008;14:1237-1243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 41] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 64. | Morice Y, Cantaloube JF, Beaucourt S, Barbotte L, De Gendt S, Goncales FL, Butterworth L, Cooksley G, Gish RG, Beaugrand M. Molecular epidemiology of hepatitis C virus subtype 3a in injecting drug users. J Med Virol. 2006;78:1296-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 65. | Uddin G, Shoeb D, Solaiman S, Marley R, Gore C, Ramsay M, Harris R, Ushiro-Lumb I, Moreea S, Alam S. Prevalence of chronic viral hepatitis in people of south Asian ethnicity living in England: the prevalence cannot necessarily be predicted from the prevalence in the country of origin. J Viral Hepat. 2010;17:327-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 66. | Frank C, Mohamed MK, Strickland GT, Lavanchy D, Arthur RR, Magder LS, El Khoby T, Abdel-Wahab Y, Aly Ohn ES, Anwar W. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet. 2000;355:887-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 683] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 67. | Esteban JI, Sauleda S, Quer J. The changing epidemiology of hepatitis C virus infection in Europe. J Hepatol. 2008;48:148-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 309] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 68. | de Bruijne J, Schinkel J, Prins M, Koekkoek SM, Aronson SJ, van Ballegooijen MW, Reesink HW, Molenkamp R, van de Laar TJ. Emergence of hepatitis C virus genotype 4: phylogenetic analysis reveals three distinct epidemiological profiles. J Clin Microbiol. 2009;47:3832-3838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 69. | Fu Y, Qin W, Cao H, Xu R, Tan Y, Lu T, Wang H, Tong W, Rong X, Li G. HCV 6a prevalence in Guangdong province had the origin from Vietnam and recent dissemination to other regions of China: phylogeographic analyses. PLoS One. 2012;7:e28006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 70. | Petruzziello A, Coppola N, Diodato AM, Iervolino V, Azzaro R, Di Costanzo G, Di Macchia CA, Di Meo T, Loquercio G, Pasquale G. Age and gender distribution of hepatitis C virus genotypes in the metropolitan area of Naples. Intervirology. 2013;56:206-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 71. | Petruzziello A, Coppola N, Loquercio G, Marigliano S, Giordano M, Azzaro R, Diodato AM, Iervolino V, Di Costanzo G, Di Macchia CA. Distribution pattern of hepatitis C virus genotypes and correlation with viral load and risk factors in chronic positive patients. Intervirology. 2014;57:311-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |