Published online Aug 21, 2016. doi: 10.3748/wjg.v22.i31.7124
Peer-review started: March 18, 2016
First decision: March 31, 2016
Revised: April 28, 2016
Accepted: May 21, 2016
Article in press: May 23, 2016
Published online: August 21, 2016
Processing time: 152 Days and 18 Hours
AIM: To further improve the endoscopic detection of intestinal mucosa alterations due to celiac disease (CD).
METHODS: We assessed a hybrid approach based on the integration of expert knowledge into the computer-based classification pipeline. A total of 2835 endoscopic images from the duodenum were recorded in 290 children using the modified immersion technique (MIT). These children underwent routine upper endoscopy for suspected CD or non-celiac upper abdominal symptoms between August 2008 and December 2014. Blinded to the clinical data and biopsy results, three medical experts visually classified each image as normal mucosa (Marsh-0) or villous atrophy (Marsh-3). The experts’ decisions were further integrated into state-of-the-art texture recognition systems. Using the biopsy results as the reference standard, the classification accuracies of this hybrid approach were compared to the experts’ diagnoses in 27 different settings.
RESULTS: Compared to the experts’ diagnoses, in 24 of 27 classification settings (consisting of three imaging modalities, three endoscopists and three classification approaches), the best overall classification accuracies were obtained with the new hybrid approach. In 17 of 24 classification settings, the improvements achieved with the hybrid approach were statistically significant (P < 0.05). Using the hybrid approach classification accuracies between 94% and 100% were obtained. Whereas the improvements are only moderate in the case of the most experienced expert, the results of the less experienced expert could be improved significantly in 17 out of 18 classification settings. Furthermore, the lowest classification accuracy, based on the combination of one database and one specific expert, could be improved from 80% to 95% (P < 0.001).
CONCLUSION: The overall classification performance of medical experts, especially less experienced experts, can be boosted significantly by integrating expert knowledge into computer-aided diagnosis systems.
Core tip: A hybrid system for the detection of villous atrophy integrating human texture recognition into computer-aided diagnosis systems outperforms human judgement alone in the diagnosis of pediatric celiac disease. In the classification of 2835 endoscopic images from the duodenum into one of two categories (“normal mucosa or villous atrophy”) using 27 different classification settings the hybrid system was superior to human experts in 24 settings. This superiority was significant in 17 of these 24 settings. Less experienced endoscopists in particular can benefit from this new method because their diagnostic accuracy can be improved the most.
- Citation: Gadermayr M, Kogler H, Karla M, Merhof D, Uhl A, Vécsei A. Computer-aided texture analysis combined with experts' knowledge: Improving endoscopic celiac disease diagnosis. World J Gastroenterol 2016; 22(31): 7124-7134
- URL: https://www.wjgnet.com/1007-9327/full/v22/i31/7124.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i31.7124
Recently, significant research has been performed to evaluate computer-aided endoscopic diagnosis[1], e.g., in celiac disease (CD)[2], colon polyp classification[3], the classification of dysplasia in Barrett’s esophagus[4], and the classification of Crohn’s disease lesions[5].
CD[6,7] is a common autoimmune disorder triggered by dietary gluten primarily affecting the small bowel. CD is characterized by inflammation affecting the mucosa of the small intestine, which finally loses its absorptive villi, while enteric crypts become hyperplastic. Endoscopy combined with intestinal biopsies is currently considered the gold standard for the diagnosis of CD. The severity of the microscopic changes found in CD biopsies is staged by Marsh and Oberhuber[6,7]. However, the histological staging is subject to significant intra- and inter-observer variability[8,9]. This variability gives strong incentive to seek a second opinion based on the objective assessment of image data that are captured during endoscopy. Furthermore, the number of biopsies could be reduced in the future if such a reliable second opinion was easily available. A further limitation of the current diagnostic gold standard arises from the possibly patchy distribution of CD-affected mucosal areas[10]. If, unfortunately, biopsies are taken only from areas with healthy mucosa within the duodenum, a proper diagnosis of CD could be missed.
Visual classification during endoscopy can be realized either by the endoscopist[11,12] or by computer-based methods[2,13-18]. Experienced endoscopists are able to classify with a high accuracy of up to 95%[19] considering two classes case [classifying between normal mucosa (Marsh-0) and villous atrophy (Marsh-3)]. However, the accuracy can drop to approximately 80% depending on the image data when less experienced endoscopists perform the classification[19]. Instead, computer-based techniques provide largely objective and user-independent classification performances. However, recent work showed that the accuracy of highly experienced endoscopists currently cannot be reached by such automated systems. The accuracies of the state-of-the-art approaches[16,18,20] range from only 85% to 90%, thus hampering their clinical use. Thus far, it is unclear whether and how a computer-aided CD diagnosis can be implemented in clinical practice in the foreseeable future.
To overcome these shortcomings we developed a hybrid classification system combining medical experts’ knowledge with state-of-the-art computer-based texture analysis methods. The aim of this study was to determine whether such a hybrid system can improve the classification accuracies of experts. Additionally, the impact of transferring a model trained with one expert’s knowledge to another expert was investigated.
Children who underwent upper endoscopy with biopsies of the duodenal mucosa for suspected CD or non-celiac upper abdominal symptoms were enrolled in this comparative diagnostic study. Among 668 consecutive children referred to the endoscopy unit of the St. Anna Children’s Hospital between August 2008 and December 2014, a total of 290 were willing to participate. Children on a gluten-free diet were excluded from this study, as were those undergoing follow-up biopsies for the surveillance of previously diagnosed CD.
The study protocol was approved by the Institutional Review Board of the St. Anna Children’s Hospital. Written informed consent was obtained from the parents or legal guardians, and assent was obtained from the children when appropriate.
During endoscopy, images of the regions of interest were recorded with Olympus endoscopes (GIF-Q165, GIF-H180 and GIF-N180) immediately before biopsy specimens (Jumbo forceps, Olympus FB-25 K) from the same mucosal area were taken as a part of routine care.
Images were recorded by applying the modified immersion technique (MIT), which is based on the instillation of water into the duodenal lumen for better visualization of the villi[21]. For MIT, an accuracy rate between 93% and 100% was found in detecting villous atrophy. Previous work[22] also found that the MIT is more suitable for automated classification purposes than the classical image capturing technique.
One part of the image data were captured using narrow-band imaging[11], which has been reported to improve the diagnostic accuracy in various fields of endoscopy[12,23]. This technology utilizes specific blue (440 to 460 nm) and green (540 to 560 nm) wavelengths for illumination to enhance the contrast of vascular patterns on the mucosal surface. It is employed to specifically delineate the outline of the residual villous structures (if present) due to a better visualization of the villous height and shape than traditional white-light endoscopy.
For each image, the ground truth was determined by histopathologic evaluation of the corresponding biopsy specimens, which were taken from the center of the preceding endoscopic image. The biopsies were classified according to a modified Marsh classification (Oberhuber) by pathologists, who were blinded to the endoscopic findings and clinical information of the children. In children with biopsy results consistent with CD the diagnosis was confirmed by positive CD serology and HLA-DQ2 and/or HLA-DQ8 positivity. CD was ruled out by negative biopsy results.
Because a binary classification of the endoscopic images of normal mucosa (Marsh-0) or villous atrophy (Marsh-3A to Marsh-3C) was used, all images containing Marsh-1 or Marsh-2 lesions according to the histology report were excluded.
In total, 1155 endoscopic images of the duodenal bulb and the second part of the duodenum were available for further experimentation. These images were captured in 75 children with CD (479 images) and in 215 children without CD (676 images).
This image data were divided into three distinct image databases, DB-1, DB-2 and DB-3, as outlined in Table 1, collecting images acquired with a specific imaging protocol into a separate database. DB-3 contains images acquired with narrow-band imaging, whereas DB-1 and DB-2 contain images obtained with traditional white-light endoscopy. Images in DB-2 and DB-3 were obtained with newer endoscopic devices than those used for DB-1 (see Table 1). The separation was performed to avoid bias in the results due to variations within the image data sets[22].
| DB-1 | DB-2 | DB-3 | |
| Number patches Marsh-0 | 280 | 280 | 280 |
| Number patches Marsh-3 | 280 | 280 | 280 |
| Number images Marsh-0 | 246 | 210 | 220 |
| Number images Marsh-3 | 171 | 154 | 154 |
| Number patients Marsh-0 | 125 | 82 | 80 |
| Number patients Marsh-3 | 38 | 35 | 36 |
| Endoscope | GIF-Q165, N180 | GIF-H180 | GIF-H180 |
| Imaging technique | Traditional (white-light) imaging | Traditional (white-light) imaging | Narrow-band imaging[11] |
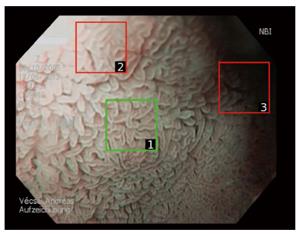
In this context, “image” or “original image” refers to the complete visible content if an image is captured with an endoscope (see Figure 1). The size of such an image is typically 768 × 576 pixels (GIF-H180). However, images partly suffer from degradation, such as noise, blur, under- or overexposure or reflections. As computer-aided diagnosis has been shown to be affected by these distortions[24], a manual selection of image sections was introduced[17,24,25] to obtain reliable, distortion-free image regions (square patches). This technique is exemplarily shown for an image in Figure 1.
A patch size of 128 × 128 pixels turned out to be optimal in previous work[24,25]. For this study, 280 patches per class (Marsh-0 and Marsh-3) and per database were extracted by a highly experienced consultant according to quality assessment criteria, such as sharpness, appropriate exposure, visibility of features and low degree of degradations. Because more than one patch can be extracted from one image, this patch extraction improved not only the quality but also the number of the data. In total, the database consisted of 2835 images. Of these 2835 images, 1155 were captured original (full-size) endoscopic images and 1680 were patches.
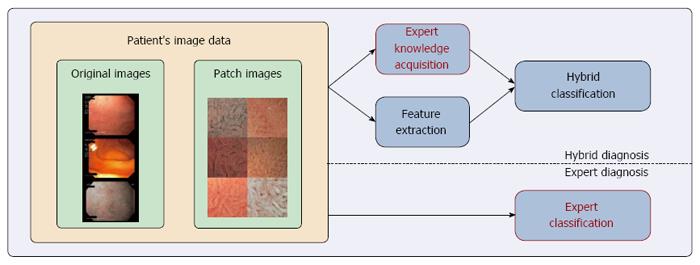
The general idea of hybrid CD diagnosis is to improve the classification performance by combining experts’ diagnoses with computer-based classification methods. As outlined in Figure 2, the proposed hybrid CD diagnosis approach consisted of three stages: expert knowledge acquisition, feature extraction and hybrid classification.
Expert knowledge acquisition and expert classification, as the baseline for experimental evaluation, was performed similarly by asking endoscopists to classify image data based on two classes, normal mucosa (Marsh-0) or villous atrophy (Marsh-3A to Marsh-3C). In this study, the three endoscopists involved were a highly experienced consultant (Expert-A), an experienced pediatric resident (Expert-B) and a less experienced intern (Expert-C). We decided on a binary classification (normal mucosa or villous atrophy) of the image data because this classification is the most relevant from the patient’s perspective. Each of the three medical experts classified the three available image data sets (Table 1) in a blinded fashion without any knowledge of the patient characteristics or histological results. Overall, we collected 5040 diagnoses corresponding to patch image data and 3465 diagnoses corresponding to original image data.
For experimentation, three high-performing yet conceptually divergent image representation methods were deployed: (1) Multi-resolution local binary patterns (LBP)[26] is an established, yet effective, texture feature extraction technique, that is highly efficient from a computational point of view. It extracts the joint-distribution of binary quantized local intensity differences. An eight-neighborhood was utilized with circular radii of two and three pixels; (2) The multi-fractal spectrum (MFS)[27] is obtained by computing the local fractal dimension per pixel based on three different measurements. The final image representation is obtained by concatenation of three different measurements and has already successfully been applied to endoscopic image data[3]; and (3) Improved Fisher vectors (IFV)[28] is a high-performing, state-of-the-art method that is based on standard low-level scale-invariant feature transform (SIFT) features and Gaussian-mixture modeling as a mid-level representation. SIFT features were extracted based on dense sampling (every fourth pixel), and the number of Gaussian components was fixed to 16.
We selected these three methods because LBP is an established, highly efficient standard method; MFS proved to be effective especially in endoscopic imaging[3,29]; and IFV is a general purpose state-of-the-art method for texture recognition[30,31].
For feature extraction, all images were converted to gray-scale data. This conversion was used because a recent study[32] showed that the utilization of color information does not lead to consistent improvements. In contrast, for the acquisition of the medical experts’ knowledge, such a conversion could easily cause a decrease in the accuracy of the classification by endoscopists, who rely on color information. Thus, no gray-scale data conversion was performed in the context of classification by human experts.
Hybrid classification first combined the feature vector (from feature extraction) with the binary expert diagnosis for each individual image by straight-forward vector concatenation. To obtain sensible weightings, first, the feature vector was L2 normalized and a multiplicative weight was evaluated (between 2-2 and 22). To avoid any bias, the evaluation was performed based on cross-validation. Finally, the obtained hybrid feature vector was classified by a linear support vector machine, which has been deployed in recent work on computer-aided CD diagnosis[17,33] and generally in recent work on texture recognition[30].
In a previous study[22], computer-based methods performed quite well based on manually extracted patches with a size of 128 × 128 pixels. These patches can either be selected by the physician during endoscopy or by automated systems[16,34]. Such an automated system can perform an analysis of the quality of the data and thus suggest a patch that is free from distortions to be assessed.
As the concept of hybrid classification is not considered fully automated per se, we considered only manual patch selection throughout this work because it performs slightly better than automated selection[16,34]. However, the classification performance of experts generally decreases in the case of the smaller, but ideal, patch image data[19].
To compensate for these performance difficulties specific to either computer-based or expert-based classification, we investigated three different strategies concerning the classification of processed image data.
In this scenario, only patch data were considered during classification by experts and computers. A realistic scenario in which to base human assessment on only patches would be an automated patch suggestion scheme, as described above. Additionally, a telemedical setting might benefit from a patch-based approach because smaller patches of high quality may be transferred for review by external human experts as opposed to transmitting few entire frames of highly varying quality.
In any case, to reveal whether there is a positive effect of using the hybrid classification method, we investigated this scenario, which is based on a large quantity of data because the number of patches is higher than the number of original images. Additionally, beforehand, it was not clear which type of data is the best (patch, image or patient-based) for hybrid classification.
The second scenario relied on the original image data obtained during endoscopy. As experts are more familiar with these data, we expected slightly better overall classification rates for the original images. Feature extraction again considered only the smaller patch data, as patches turned out to be more appropriate for the methods used[16].
One reason to stick with this approach instead of choosing video data is to conserve human resources because assessing several frames is more time-consuming, and furthermore, images are captured in only relevant mucosal areas. In addition, a tele-endoscopical setting (incorporating a remote expert’s knowledge) benefits from the reduced transmission bandwidth required for this approach.
It is common practice during endoscopy to capture several images from the duodenum. Consequently, the image data sets used for experimentation contained more than one image per patient (Table 1). On average, per database 3.0 images (between one and eight) per patient were available. To obtain one final diagnosis per patient, all image data from one specific patient were utilized. Because it exploited all available image data, this scenario was assumed to be the most relevant and realistic one.
To obtain patient-based decisions, hybrid diagnosis was performed as described for image-based classification. The classification stage was followed by a soft decision level fusion. Here, soft decision level fusion means that the majority vote was applied based on binary decisions [normal mucosa (Marsh-0) or villous atrophy (Marsh-3)] for each image and, in case of a tie, the signed distances to the linear decision boundary of the support vector machine were averaged and thresholded (with a threshold of zero) to determine the final overall decision.
Expert diagnosis was performed similarly by majority voting on the basis of image-based classification. As only hard decisions were available, ties had to be resolved by means of random choice.
Thus far, we considered a scenario where the classification model (in our case, the support vector machine) is trained and evaluated with decisions from the same expert. Hence, the obtained classification accuracies correspond to a scenario where the classifier needs to be trained individually for each expert. However, this individual training is expensive, and it is questionable whether it is required. To find out whether the individual training was necessary, we also investigated the impact of changing experts between the training and evaluation phases.
All overall accuracies presented were based on the mean accuracy of 50 random splits. Each distinct split divided the data set into approximately balanced training (80%) and evaluation sets (20%), restricting the images of one patient to the same set to avoid any bias (due to similarities within the data from one patient).
To determine whether the performance of two techniques was significantly different, we applied the Mann-Whitney-Wilcoxon rank-sum test[35]. Specifically, based on various settings, we investigated whether hybrid diagnosis significantly outperformed expert diagnosis. As is commonly accepted, the significance level was set to 0.05.
Considering the three endoscopists, Expert-A achieved the best average patient-based classification accuracy of 98.4%. The less experienced Expert-B and Expert-C achieved average patient-based classification rates of 88.4% and 89.6%, respectively. Among all three endoscopists, the accuracies decreased when classifying in an image-based manner (compared to patient-based), and the lowest accuracies were achieved in the patch-based classifications (Table 2). The patient-based average accuracies obtained in distinct image data sets were between 88.1% (DB-1) and 95.9% (DB-2). All other individual and averaged classification rates are given in Tables 2 and 3 (columns: expert diagnosis).
| Feature extraction | Data set | Expert | Patch-based | Image-based | Patient-based | |||||||||
| Expert diagnosis | Hybrid diagnosis | Expert diagnosis | Hybrid diagnosis | Expert diagnosis | Hybrid diagnosis | |||||||||
| mean | ± | mean | ± | mean | ± | mean | ± | mean | ± | mean | ± | |||
| LBP | DB-1 | A | 0.870 | 0.057 | 0.910 | 0.042 | 0.970 | 0.027 | 0.964 | 0.033 | 0.970 | 0.058 | 0.972 | 0.033 |
| LBP | DB-2 | A | 0.857 | 0.042 | 0.910 | 0.040 | 0.957 | 0.026 | 0.955 | 0.038 | 0.988 | 0.047 | 0.999 | 0.005 |
| LBP | DB-3 | A | 0.773 | 0.051 | 0.902 | 0.059 | 0.963 | 0.020 | 0.965 | 0.036 | 0.995 | 0.055 | 0.994 | 0.016 |
| LBP | DB-1 | B | 0.834 | 0.063 | 0.909 | 0.051 | 0.879 | 0.053 | 0.908 | 0.046 | 0.873 | 0.066 | 0.947 | 0.062 |
| LBP | DB-2 | B | 0.827 | 0.051 | 0.903 | 0.048 | 0.896 | 0.040 | 0.905 | 0.048 | 0.946 | 0.046 | 0.998 | 0.009 |
| LBP | DB-3 | B | 0.627 | 0.058 | 0.883 | 0.057 | 0.818 | 0.040 | 0.913 | 0.047 | 0.832 | 0.070 | 0.960 | 0.064 |
| LBP | DB-1 | C | 0.882 | 0.052 | 0.916 | 0.045 | 0.778 | 0.070 | 0.902 | 0.057 | 0.799 | 0.064 | 0.959 | 0.053 |
| LBP | DB-2 | C | 0.912 | 0.037 | 0.926 | 0.044 | 0.893 | 0.033 | 0.914 | 0.040 | 0.943 | 0.036 | 0.991 | 0.017 |
| LBP | DB-3 | C | 0.718 | 0.064 | 0.892 | 0.053 | 0.879 | 0.045 | 0.922 | 0.042 | 0.946 | 0.059 | 0.984 | 0.035 |
| MFS | DB-1 | A | 0.870 | 0.057 | 0.891 | 0.051 | 0.970 | 0.027 | 0.964 | 0.026 | 0.970 | 0.058 | 0.974 | 0.032 |
| MFS | DB-2 | A | 0.857 | 0.042 | 0.878 | 0.040 | 0.957 | 0.026 | 0.955 | 0.038 | 0.988 | 0.047 | 0.999 | 0.005 |
| MFS | DB-3 | A | 0.773 | 0.051 | 0.817 | 0.062 | 0.963 | 0.020 | 0.968 | 0.035 | 0.995 | 0.055 | 0.994 | 0.016 |
| MFS | DB-1 | B | 0.834 | 0.063 | 0.899 | 0.062 | 0.879 | 0.053 | 0.887 | 0.065 | 0.873 | 0.066 | 0.932 | 0.067 |
| MFS | DB-2 | B | 0.827 | 0.051 | 0.853 | 0.061 | 0.896 | 0.040 | 0.901 | 0.060 | 0.946 | 0.046 | 0.997 | 0.011 |
| MFS | DB-3 | B | 0.627 | 0.058 | 0.776 | 0.074 | 0.818 | 0.040 | 0.840 | 0.064 | 0.832 | 0.070 | 0.950 | 0.064 |
| MFS | DB-1 | C | 0.882 | 0.052 | 0.909 | 0.050 | 0.778 | 0.070 | 0.862 | 0.055 | 0.799 | 0.064 | 0.925 | 0.053 |
| MFS | DB-2 | C | 0.912 | 0.037 | 0.929 | 0.049 | 0.893 | 0.033 | 0.888 | 0.053 | 0.943 | 0.036 | 0.993 | 0.017 |
| MFS | DB-3 | C | 0.718 | 0.064 | 0.811 | 0.071 | 0.879 | 0.045 | 0.888 | 0.067 | 0.946 | 0.059 | 0.944 | 0.090 |
| IFV | DB-1 | A | 0.870 | 0.057 | 0.903 | 0.046 | 0.970 | 0.027 | 0.968 | 0.032 | 0.970 | 0.058 | 0.976 | 0.033 |
| IFV | DB-2 | A | 0.857 | 0.042 | 0.889 | 0.044 | 0.957 | 0.026 | 0.957 | 0.038 | 0.988 | 0.047 | 0.999 | 0.005 |
| IFV | DB-3 | A | 0.773 | 0.051 | 0.880 | 0.061 | 0.963 | 0.020 | 0.968 | 0.035 | 0.995 | 0.055 | 0.996 | 0.010 |
| IFV | DB-1 | B | 0.834 | 0.063 | 0.903 | 0.048 | 0.879 | 0.053 | 0.910 | 0.049 | 0.873 | 0.066 | 0.961 | 0.049 |
| IFV | DB-2 | B | 0.827 | 0.051 | 0.878 | 0.055 | 0.896 | 0.040 | 0.908 | 0.059 | 0.946 | 0.046 | 0.996 | 0.010 |
| IFV | DB-3 | B | 0.627 | 0.058 | 0.883 | 0.054 | 0.818 | 0.040 | 0.903 | 0.053 | 0.832 | 0.070 | 0.997 | 0.037 |
| IFV | DB-1 | C | 0.882 | 0.052 | 0.908 | 0.047 | 0.778 | 0.070 | 0.892 | 0.058 | 0.799 | 0.064 | 0.954 | 0.061 |
| IFV | DB-2 | C | 0.912 | 0.037 | 0.923 | 0.052 | 0.893 | 0.033 | 0.906 | 0.048 | 0.943 | 0.036 | 0.993 | 0.012 |
| IFV | DB-3 | C | 0.718 | 0.064 | 0.887 | 0.055 | 0.879 | 0.045 | 0.925 | 0.044 | 0.946 | 0.059 | 0.981 | 0.024 |
| Feature extraction | Data set | Expert | Patch-based | Image-based | Patient-based | |||||||||
| Expert diagnosis | Hybrid diagnosis | Expert diagnosis | Hybrid diagnosis | Expert diagnosis | Hybrid diagnosis | |||||||||
| mean | ± | mean | ± | mean | ± | mean | ± | mean | ± | mean | ± | |||
| LBP | mean | mean | 0.811 | 0.091 | 0.906 | 0.013 | 0.893 | 0.065 | 0.928 | 0.026 | 0.921 | 0.070 | 0.978 | 0.019 |
| MFS | mean | mean | 0.811 | 0.091 | 0.863 | 0.052 | 0.893 | 0.065 | 0.906 | 0.046 | 0.921 | 0.070 | 0.968 | 0.030 |
| IFV | mean | mean | 0.811 | 0.091 | 0.895 | 0.065 | 0.893 | 0.065 | 0.926 | 0.030 | 0.921 | 0.070 | 0.981 | 0.017 |
| mean | DB-1 | mean | 0.862 | 0.022 | 0.905 | 0.007 | 0.876 | 0.083 | 0.917 | 0.039 | 0.881 | 0.074 | 0.956 | 0.018 |
| mean | DB-2 | mean | 0.865 | 0.037 | 0.899 | 0.031 | 0.915 | 0.031 | 0.921 | 0.027 | 0.959 | 0.022 | 0.996 | 0.003 |
| mean | DB-3 | mean | 0.706 | 0.064 | 0.859 | 0.045 | 0.887 | 0.063 | 0.921 | 0.042 | 0.924 | 0.072 | 0.975 | 0.020 |
| mean | mean | A | 0.833 | 0.046 | 0.887 | 0.006 | 0.963 | 0.006 | 0.963 | 0.006 | 0.984 | 0.011 | 0.989 | 0.012 |
| mean | mean | B | 0.763 | 0.102 | 0.876 | 0.041 | 0.864 | 0.036 | 0.897 | 0.023 | 0.884 | 0.050 | 0.968 | 0.024 |
| mean | mean | C | 0.837 | 0.090 | 0.900 | 0.036 | 0.850 | 0.054 | 0.900 | 0.020 | 0.896 | 0.073 | 0.969 | 0.025 |
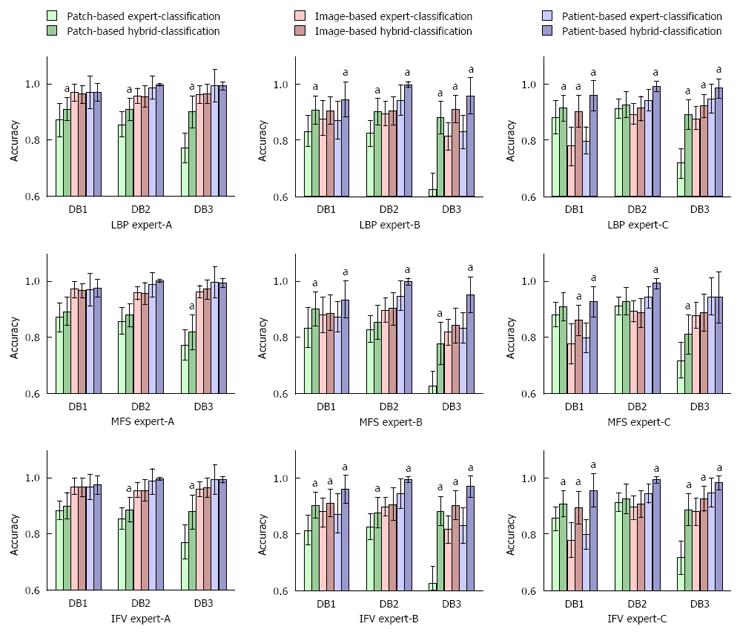
The hybrid classification method outperformed the average experts’ classification accuracies regardless of whether the classification was based on patches, images or using the patient-based approach (see Figure 3 for an overview and Tables 2 and 3 for exhaustive results). These performance improvements were mostly statistically significant in the case of patch-based and patient-based classifications in combination with Expert-B and Expert-C. Although the performance improvements of Expert-A did not reach the significance level, applying this hybrid classification method reduced error by 31% (from 1.6% to 1.1%).
Similar to expert-only classification, in the hybrid setting, the highest accuracies were obtained in the patient-based approach, with average accuracies ranging from 96.8% to 98.9%. The image- and patch-based hybrid classification achieved accuracies of 89.7% to 96.3% and 87.6% to 90%, respectively. Of the different feature-extracting techniques used, LBP obtained the highest accuracies in the cases of patch- and image-based hybrid classification (90.6% and 92.8% accuracy, respectively), while IFV was superior in the case of patient-based hybrid classification (98.1% accuracy). However, these performance differences did not reach the significance level. MFS was outperformed by LBP and IFV in all 3 classification approaches.
With hybrid classification, the rates of the three experts converged. In the case of DB-2, the final rates (with a decision level fusion on a per patient basis) were always above 98%. Based on the other data sets, the rates were never below 94%.
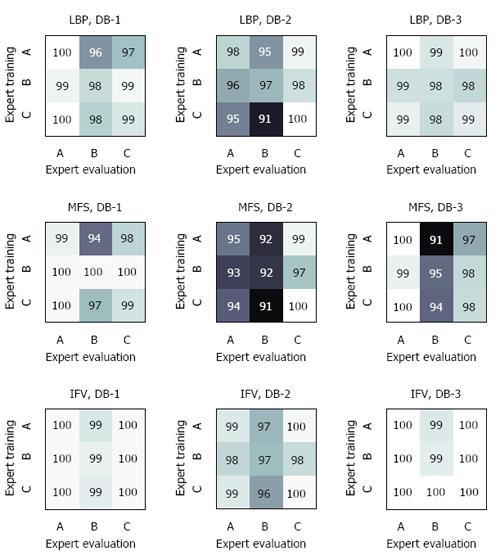
In a second experiment, we investigated whether a model trained with a certain expert can be simply transferred to another expert (Figure 4). We observed that the accuracies remained quite stable if the evaluating expert was more accurate than the expert for training (see values at the diagonal of Figure 4). When using the combination of DB-1 and the evaluating of Expert-A, the training set had virtually no impact (first column of Figure 4). More distinct decreases in accuracy were observed if the evaluating expert was less accurate. According to the results of LBP and DB-2, Expert-B obtained an accuracy of 97% if his training data were used (center cell of Figure 4), whereas the use of the trained model of the more accurate Expert-C led to a distinct loss of performance (91% accuracy). The extent of this effect varied between the different data sets. Considering the different image description methods, IFV was more stable than LBP and MFS.
Here, we report that a hybrid classification approach combining medical expert knowledge with state-of-the-art computer-based texture analysis methods actually improves the diagnostic accuracy of medical experts. Performance improvements are most distinct for (but not limited to) less experienced physicians.
From the literature[19], we know that state-of-the-art computer-aided diagnosis is currently unable to compete with experienced endoscopists considering the overall classification rates, which are roughly between 85% and 90%. With our new approach, however, even the diagnostic accuracy of the most experienced expert, Expert-A, could be outperformed. Without the hybrid system Expert-A achieved on average 98.4% accurate diagnoses with fusion (i.e., patient-based diagnosis), which is probably the most relevant variable for clinical practice. This classification rate reflects the reported accuracy of MIT[36,37].
In contrast, when applying the proposed hybrid system the accuracy increased slightly, to 98.9%. This increase seems to be negligible; however, the classification error was reduced by 31%. A less experienced expert (Expert-C) achieved on average 89.6% accuracy without the hybrid approach and 96.9% with the new technique, which was a statistically significant improvement. This relatively high classification rate obtained by Expert-C is in accordance with the easy applicability of the MIT even with regard to beginners[38].
The outcome of Expert-B was also significantly improved. Using patch-based classification, the accuracies were on average lower than using image-based classification. However, this loss of accuracy was not completely consistent. The general effect can potentially be attributed to the less accurate expert’s diagnosis, which influenced the hybrid system results. Obviously, the fusion of images from different regions of the duodenum led to distinctly enhanced accuracy.
Comparing hybrid diagnosis to expert diagnosis, the most distinct improvements were noticed in the case of patch-based classification but also in the case of patient-based classification, which is highly relevant from a practical point of view. For this patient-based approach, a panel of images for each patient is necessary to assist the diagnostic procedure. In this study, on average 3.0 images per patient were available. Due to the frequently observed patchy pattern of tissue damage in CD, using a higher number of endoscopic images might improve the detection of villous atrophy.
Furthermore, we noticed more distinct enhancements in combination with less experienced experts (e.g., see rates of Expert-3). In general, this effect holds true for all classification scenarios, all data sets and all feature extraction methods.
Considering the different image data sets, we found that different experts have individual strengths and weaknesses. Hybrid classification is especially able to compensate for the experts’ weaknesses, as shown by the combination of Expert-B and DB-3. This expert supposedly was less experienced with the narrow-band image data, thus reaching a maximum accuracy of only approximately 85%. In contrast, using hybrid diagnosis, an accuracy of 95% was obtained.
Looking at the outcome, the impact of the feature extraction method was quite small. Although the best average outcome was obtained with the most sophisticated method IFV, the other two methods were only slightly (insignificantly) inferior.
Additional experiments showed that a model can generally be transferred from one expert to another if their performances are similar. We found that a model can be trained with a slightly less accurate expert without expecting a severe loss of performance. Consequently, it should not be trained with a more accurate one, as this can lead to distinct drops in accuracy.
The clinical relevance of the reported hybrid classification approach would be primarily to support endoscopists in identifying whether and where biopsies from the duodenum are to be taken. Especially in the case of a patchy distribution of villous atrophy in the midst of normal mucosa, the hybrid system could indicate areas with villous atrophy, thus targeting the biopsy. Subsequently, such a diagnostic approach including selective and targeted tissue sampling might improve the accuracy of CD diagnosis, especially for less experienced endoscopists. In the foreseeable future, it would be conceivable that with this reported hybrid approach, biopsies could be avoided or reduced in some carefully selected scenarios, such as endoscopic evidence of villous atrophy in patients with positive celiac antibodies[36] or monitoring the histologic recovery of CD patients on a gluten-free diet[39] Hence, the hybrid approach could finally result in cost savings by reducing the number of biopsy specimens. However, one limitation is that with the hybrid approach, it is not possible to detect Marsh-1 or Marsh-2 lesions. Therefore, the hybrid approach is not suitable to completely substitute for diagnostic biopsy. In cases where villous atrophy is not detected, biopsies and subsequent histopathologic evaluation will still be indispensable. Biopsies should always be performed in the case of macroscopic wall abnormalities, which indicate CD-associated intestinal lymphomas.
A potential limitation of our study is the limitation of the study population to children and the relatively small number of experts involved in the evaluation of the endoscopic imagery. However, based on the vast amount of image data evaluated and the different levels of experience of the study endoscopists we strongly assume that the diagnostic accuracy obtained by our new approach is generalizable to other settings and holds true for routine patient care.
One strength of computer-aided endoscopic diagnosis of CD is its observer independence. However, the significant intra- and inter-observer variability in the histological staging of CD described in the literature refers to only the use of the Marsh classification. This classification variability might be significantly less if pathologists also used a binary histological staging (normal mucosa vs villous atrophy) instead of the Marsh classification.
In conclusion, our results indicate that a hybrid classification approach combining medical expert knowledge with state-of-the-art computer-based texture analysis methods can improve the diagnostic accuracy of endoscopists in detecting duodenal areas with villous atrophy. It is possible that in the near future, an automated CD diagnosis tool will be on hand to support endoscopists in identifying whether and where biopsies from the duodenum should be taken. However, beyond these possibilities, several further potentials of this new technique lie in its application in capsule endoscopy, in the cost reduction of reliable biopsy-avoiding approaches in CD diagnosis and in the complete prevention of biopsy-associated complications.
Celiac disease (CD) is one of the most common autoimmune diseases, and it can occur at any point in life. However, because 50% of cases are diagnosed in childhood, care for these children with celiac disease is a prominent task within the field of Pediatric Gastroenterology. One of the issues in celiac disease care is obtaining a reliable diagnosis before embarking on a life-long strict gluten-free diet, which is a highly effective treatment modality. In contrast, if celiac disease goes undiagnosed, or in the case of lacking dietary adherence, severe complications can arise. Endoscopy combined with intestinal biopsies is currently considered the gold standard for the diagnosis of CD.
Visual assessment of mucosal celiac disease markers during upper endoscopy was shown to be of modest reliability. In particular, less experienced endoscopists are prone to diagnostic errors. Furthermore, histopathological examination of biopsies is subject to a significant intra- and inter-observer variability. For the diagnostic exploitation of visual disease markers, especially in the case of less experienced physicians, a reliable automated and observer-independent decision support system is missing. Computer-based methods not incorporating expert knowledge proved not to be as reliable as experienced endoscopists, a finding that prevents such systems from being deployed in the endoscopic routine.
The proposed hybrid approach constitutes a technique improving the diagnostic performance of both less experienced and experienced physicians. The main finding that the computer-aided hybrid system mostly outperforms, but never underperforms, human diagnostic accuracy will ease the establishment of this new system as a diagnostic support tool.
The proposed method can be utilized for the diagnosis of celiac disease based on visual markers. Using this method, biopsies can be targeted, the histopathological assessment of biopsies can be supported to increase diagnostic accuracy, and biopsies can even be omitted in the case of a clear-cut detection of villous atrophy.
CD is an autoimmune disorder triggered by dietary gluten. It is marked by intestinal inflammation, finally leading to villous atrophy.
This study investigates a computer-aided decision support system based on image data captured during endoscopies. By fusing computer-based image features with endoscopists’ decisions, the diagnostic accuracy of medical experts can be outperformed on average. The performance increase is most significant in the case of less experienced physicians.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Bonaz B, Ciaccio EJ, Gassler N, Ludvigsson JF S- Editor: Gong ZM L- Editor: A E- Editor: Wang CH
| 1. | Liedlgruber M, Uhl A. Computer-aided decision support systems for endoscopy in the gastrointestinal tract: a review. IEEE Rev Biomed Eng. 2011;4:73-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 2. | Ciaccio EJ, Tennyson CA, Bhagat G, Lewis SK, Green PH. Use of basis images for detection and classification of celiac disease. Biomed Mater Eng. 2014;24:1913-1923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Häfner M, Tamaki T, Tanaka S, Uhl A, Wimmer G, Yoshida S. Local fractal dimension based approaches for colonic polyp classification. Med Image Anal. 2015;26:92-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | Qi X, Pan Y, Sivak MV, Willis JE, Isenberg G, Rollins AM. Image analysis for classification of dysplasia in Barrett’s esophagus using endoscopic optical coherence tomography. Biomed Opt Express. 2010;1:825-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Jebarani W, Daisy V. Assessment of Crohn’s disease lesions in Wireless Capsule Endos-copy images using SVM based classification. : Proc of ICSIPR 2013; 303-307. [DOI] [Full Text] |
| 6. | Marsh MN. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’). Gastroenterology. 1992;102:330-354. [PubMed] |
| 7. | Oberhuber G, Granditsch G, Vogelsang H. The histopathology of coeliac disease: time for a standardized report scheme for pathologists. Eur J Gastroenterol Hepatol. 1999;11:1185-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1142] [Cited by in RCA: 1205] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 8. | Mubarak A, Nikkels P, Houwen R, Ten Kate F. Reproducibility of the histological diagnosis of celiac disease. Scand J Gastroenterol. 2011;46:1065-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 9. | Arguelles-Grande C, Tennyson CA, Lewis SK, Green PH, Bhagat G. Variability in small bowel histopathology reporting between different pathology practice settings: impact on the diagnosis of coeliac disease. J Clin Pathol. 2012;65:242-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Hopper AD, Cross SS, Sanders DS. Patchy villous atrophy in adult patients with suspected gluten-sensitive enteropathy: is a multiple duodenal biopsy strategy appropriate? Endoscopy. 2008;40:219-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 123] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Emura F, Saito Y, Ikematsu H. Narrow-band imaging optical chromocolonoscopy: advantages and limitations. World J Gastroenterol. 2008;14:4867-4872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Valitutti F, Oliva S, Iorfida D, Aloi M, Gatti S, Trovato CM, Montuori M, Tiberti A, Cucchiara S, Di Nardo G. Narrow band imaging combined with water immersion technique in the diagnosis of celiac disease. Dig Liver Dis. 2014;46:1099-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Hämmerle-Uhl J, Höller Y, Uhl A, Vécsei A. Endoscope distortion correction does not (easily) improve mucosa-based classification of celiac disease. Med Image Comput Comput Assist Interv. 2012;15:574-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | Ciaccio EJ, Tennyson CA, Bhagat G, Lewis SK, Green PH. Implementation of a polling protocol for predicting celiac disease in videocapsule analysis. World J Gastrointest Endosc. 2013;5:313-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 15. | Hegenbart S, Uhl A, Vécsei A, Wimmer G. Scale invariant texture descriptors for classifying celiac disease. Med Image Anal. 2013;17:458-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Gadermayr M, Uhl A, Vécsei A. Getting One Step Closer to Fully Automatized Celiac Disease Diagnosis. : Proc of IPTA 2014; 13-17. [DOI] [Full Text] |
| 17. | Kwitt R, Hegenbart S, Rasiwasia N, Vécsei A, Uhl A. Do we need annotation experts? A case study in celiac disease classification. Med Image Comput Comput Assist Interv. 2014;17:454-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Hegenbart S, Uhl A, Vécsei A. Survey on computer aided decision support for diagnosis of celiac disease. Comput Biol Med. 2015;65:348-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Gadermayr M, Uhl A, Vécsei A. The Effect of Endoscopic Lens Distortion Correction on Physicians’ Diagnosis Performance. : Proc of BVM 2014; 174-179. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Gadermayr M, Liedlgruber M, Uhl A, Vécsei A. Shape Curvature Histogram: A Shape Feature for Celiac Disease Diagnosis. : Proc of MICCAI-MCV 2014; 175-184. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Gasbarrini A, Ojetti V, Cuoco L, Cammarota G, Migneco A, Armuzzi A, Pola P, Gasbarrini G. Lack of endoscopic visualization of intestinal villi with the “immersion technique” in overt atrophic celiac disease. Gastrointest Endosc. 2003;57:348-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Hegenbart S, Kwitt R, Liedlgruber M, Uhl A, Vécsei A. Impact of Duodenal Image Capturing Techniques and Duodenal Regions on the Performance of Automated Diagnosis of Celiac Disease. : Proc of ISPA 2009; 718-723. [DOI] [Full Text] |
| 23. | Sinha SK, Basha JA, Vaiphei K, Appasani S, Singh K, Kochhar R. Can Narrow-Band Imaging Predict Duodenal Histology in Celiac Disease? A Prospective Double Blind Pilot Study. Gastroenterology. 2013;144:S889-S890. |
| 24. | Hegenbart S, Uhl A, Vécsei A. Impact of Endoscopic Image Degradations on LBP based Features using One-Class SVM for Classification of Celiac Disease. : Proc of ISPA 2011; 715-720. |
| 25. | Vécsei A, Amann G, Hegenbart S, Liedlgruber M, Uhl A. Automated Marsh-like classification of celiac disease in children using local texture operators. Comput Biol Med. 2011;41:313-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Ojala T, Pietikäinen M, Mäenpää T. Multiresolution Gray-Scale and Rotation Invariant Texture Classification with Local Binary Patterns. IEEE Trans Pattern Anal Mach Intell (TPAMI). 2002;24:971-987. [DOI] [Full Text] |
| 27. | Xu Y, Ji H, Fermüller C. Viewpoint invariant texture description using fractal analysis. Int J Comput Vision. 2009;83:85-100. [RCA] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Sánchez J, Perronnin F, Mensink T, Verbeek JJ. Image Classification with the Fisher Vector: Theory and Practice. Int J Comput Vision. 2013;105:222-245. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1059] [Cited by in RCA: 299] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 29. | Häfner M, Uhl A, Wimmer G. (2014). Shape and Size Adapted Local Fractal Dimension for the Classification of Polyps in HD colonoscopy. : Proc of ICIP 2014; 2299-2303. [DOI] [Full Text] |
| 30. | Cimpoi M, Maji S, Kokkinos I, Mohamed S, Vedaldi A. Describing Textures in the Wild. : Proc of IEEE CVPR 2014; 3606-3613. [DOI] [Full Text] |
| 31. | Perronnin F, Larlus D. Fisher Vectors Meet Neural Networks: A Hybrid Classification Architecture. : Proc of CVPR 2015; 3743-3752. [DOI] [Full Text] |
| 32. | Vécsei A, Fuhrmann T, Liedlgruber M, Brunauer L, Payer H, Uhl A. Automated classification of duodenal imagery in celiac disease using evolved Fourier feature vectors. Comput Methods Programs Biomed. 2009;95:S68-S78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Hegenbart S, Uhl A. A Scale- and Orientation-Adaptive Extension of Local Binary Pat-terns for Texture Classification. Pattern Recognit. 2015;48:2633-2644. [RCA] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 34. | Gadermayr M, Uhl A, Vécsei A. Quality Based Information Fusion in Fully Automatized Celiac Disease Diagnosis. : Proc of GCPR 2014; 1-12. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 35. | Mann HB, Whitney DR. On a test of whether one of two random variables is stochasti-cally larger than the other. Ann Math Stat. 1947;18:50-60. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7063] [Cited by in RCA: 7154] [Article Influence: 91.7] [Reference Citation Analysis (0)] |
| 36. | Cammarota G, Cesaro P, Martino A, Zuccalà G, Cianci R, Nista E, Larocca LM, Vecchio FM, Gasbarrini A, Gasbarrini G. High accuracy and cost-effectiveness of a biopsy-avoiding endoscopic approach in diagnosing coeliac disease. Aliment Pharmacol Ther. 2006;23:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Cammarota G, Cazzato A, Genovese O, Pantanella A, Ianiro G, Giorgio V, Montalto M, Vecchio FM, Larocca LM, Gasbarrini G. Water-immersion technique during standard upper endoscopy may be useful to drive the biopsy sampling of duodenal mucosa in children with celiac disease. J Pediatr Gastroenterol Nutr. 2009;49:411-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Cammarota G, Cesaro P, Cazzato A, Cianci R, Fedeli P, Ojetti V, Certo M, Sparano L, Giovannini S, Larocca LM. The water immersion technique is easy to learn for routine use during EGD for duodenal villous evaluation: a single-center 2-year experience. J Clin Gastroenterol. 2009;43:244-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 39. | Cammarota G, Cuoco L, Cesaro P, Santoro L, Cazzato A, Montalto M, La Mura R, Larocca LM, Vecchio FM, Gasbarrini A. A highly accurate method for monitoring histological recovery in patients with celiac disease on a gluten-free diet using an endoscopic approach that avoids the need for biopsy: a double-center study. Endoscopy. 2007;39:46-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |