Published online Aug 21, 2016. doi: 10.3748/wjg.v22.i31.7099
Peer-review started: March 28, 2016
First decision: May 12, 2016
Revised: May 21, 2016
Accepted: June 15, 2016
Article in press: June 15, 2016
Published online: August 21, 2016
Processing time: 140 Days and 21.9 Hours
In mammals, the intestinal epithelium is a tissue that contains two distinct pools of stem cells: active intestinal stem cells and reserve intestinal stem cells. The former are located in the crypt basement membrane and are responsible for maintaining epithelial homeostasis under intact conditions, whereas the latter exhibit the capacity to facilitate epithelial regeneration after injury. These two pools of cells can convert into each other, maintaining their quantitative balance. In terms of the active intestinal stem cells, their development into functional epithelium is precisely controlled by the following signaling pathways: Wnt/β-catenin, Ras/Raf/Mek/Erk/MAPK, Notch and BMP/Smad. However, mutations in some of the key regulator genes associated with these signaling pathways, such as APC, Kras and Smad4, are also highly associated with gut malformations. At this point, clarifying the biological characteristics of intestinal stem cells will increase the feasibility of preventing or treating some intestinal diseases, such as colorectal cancer. Moreover, as preclinical data demonstrate the therapeutic effects of colon stem cells on murine models of experimental colitis, the prospects of stem cell-based regenerative treatments for ulcerous lesions in the gastrointestinal tract will be improved all the same.
Core tip: Although the specific roles of intestinal stem cells in epithelial homeostasis and regeneration have been explored, the specific markers for identifying intestinal stem cells (ISCs) remain unclear. The reserve pool of intestinal stem cells is located at the 4+ position of crypts, and their biological characteristics are distinct from the intestinal stem cells at the crypt basement membrane. Intestinal stem cells are important cellular sources for initiating colorectal cancers. Managing murine models of ulcerous colitis using colon organoids indicates the therapeutic effects of ISCs.
- Citation: Cui S, Chang PY. Current understanding concerning intestinal stem cells. World J Gastroenterol 2016; 22(31): 7099-7110
- URL: https://www.wjgnet.com/1007-9327/full/v22/i31/7099.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i31.7099
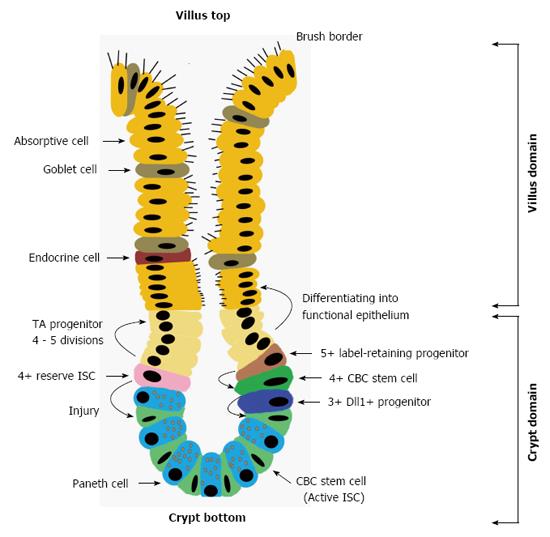
The intestinal epithelium in mammals turns over every 4-5 d[1]. The fast-cycling cells located the at crypt basement membrane, termed intestinal stem cells (ISCs), are responsible for maintaining epithelial homeostasis under intact circumstances[1]. In the homeostatic process, ISCs produce daughter cells, the transit-amplifying (TA) progenitors, who undergo 4-5 divisions and migrate along the villus-crypt axis to differentiate into functional cells within the epithelium. In the small intestine for example, TA progenitors are committed into absorptive cells, endocrine cells, goblet cells and Paneth cells[2]. Herein, the former three cell types constitute the villus-domain of the intestinal epithelium, and the Paneth cells move towards the crypt bottom because of active Wnt signals within this domain regulating their maturation[3] (Figure 1). However, ISCs within the crypts are not uniform because a reserve pool of ISCs at the 4+ position of the crypts exhibit special roles in epithelial regeneration[4]. Relative to the reserve ISCs, ISCs located at the crypt basement membrane, which are involved in epithelial homeostasis, are classified into a more active pool. In recent years, great effort has been made to compare the differences between these two pools of ISCs in their biological characteristics and in investigating their relative origins. Moreover, as the growth signals for active ISCs in vivo are clarified, emerging protocols for culturing these ISCs have been established and are constantly being optimized. In addition, colon-derived ISCs have exhibited therapeutic potential in experimental models of colitis[5,6]. Based on these advancements, this review will first introduce the mechanisms by which niche-signals regulate the development of active ISCs into functional epithelial cells under intact conditions. Then, issues concerning the locations of ISCs and their diverse populations will be presented. Subsequently, advancements involved in identifying and expanding ISCs will be summarized in this review. For ISC-related malformations of the gut, sequential mutations of the APC, p53, Smad4 and Kras genes are exclusively associated with the transformation of ISCs into colorectal cancer stem cells (CSCs), which are regarded as the primary sources for initiating colorectal cancers (CRCs)[1]. Additionally, the most important event for mediating cancer progression, namely, cross-talk between colorectal CSCs and their niche cells, will be summarized in this review in relation to recently published findings. In reviewing the topics above, the prospects for the clinical use of ISCs for managing some epithelial injuries will be analyzed along with presenting our insights on the transplantation of ISCs.
Within crypt domains, robust self-renewing active ISCs enable constitutive epithelial turnover, and the development of active ISCs into functional epithelial cells is generally mediated by the following signaling pathways: Wnt/β-catenin, Ras/Raf/Mek/Erk/MAPK, Notch and BMP/Smad[1,4,7]. In this process, Paneth cells are capable of secreting niche signals for ISCs, including Wnt3 (an agonist of Wnt/β-catenin), epithermal growth factor (EGF), and Delta-like ligand1/4 (Dll1/4, ligands of Notch receptors)[8]. Another population of niche cells include the myofibroblasts located around the crypts[9,10]. These cells can produce some bioactive proteins for ISCs, such as R-spondin1 (an amplifier of Wnt3-activated signals) and Noggin (an antagonist of BMP/Smad)[10,11]. All these proteins are essential for maintaining the proliferative status in ISCs (Table 1).
| Niche cell | Bioactive protein | Receptor | Target pathway | ISC proliferation | ISC differentiation |
| Paneth cell | Wnt 3 | LRP5/6 and Frizzled | Wnt/β-catenin | ↑ | / |
| Paneth cell | Dll1/4 | Notch 1/2 | Notch | ↑ | ↓ |
| Paneth cell | EGF | EGFR | Ras/Raf/Mek/Erk/MAPK | ↑ | / |
| Myofibroblast | R-spondin1 | Lgr4/5 | Wnt/β-catenin | ↑ | / |
| Myofibroblast | Noggin | BMP4 | BMP/Smad | / | ↓ |
Active Wnt signaling is believed to be the main driving force leading to ISC proliferation[1]. In this process, acting via a co-receptor binding approach, Wnt3 couples with LRP5/6 and Frizzled receptors, leading to the cytoplasmic accumulation of β-catenin, which up-regulates c-Myc expression through β-catenin/TCF4-mediated transcriptional activation[7]. R-spondin1 is capable of protecting LRP6 against Dkk1/Kremen-mediated internalization by binding to its receptors (Lgr4/5), resulting in an increase in LRP6 on the cell surface[12-14]. As a result of the actions of R-spondin1, ISCs become more sensitive to Wnt3. Moreover, the inactivation of Lgr4 gene function results in a significant reduction of Paneth cells in the crypts[15]. Likewise, a loss of TCF4 gene function hampers the maturation of Paneth cells[3]. All these results suggest that Wnt signals are not only essential for driving the proliferation of ISCs but also for their commitment into mature Paneth cells.
The other driving force for ISC proliferation relies on the EGF-mediated activation of the Ras/Raf/Mek/Erk/MAPK signaling pathway. Previous data suggest that more than 50% of mitosis in ISCs and TA progenitors relies on high levels of EGF within the crypt-domains[16]. In addition, Dll1/4-mediated activation of the Notch pathway also contributes to the proliferative potential of ISCs[17]. This is supported by evidence showing that the proliferative potential of ISCs from Dll1/4 knock-out mice are decreased, but this depletion of Dll1/4 expression in vivo increases the potential for ISCs to differentiate into secretory cell lineages, including goblet cells, endocrine cells and Paneth cells. In contrast, ISCs from Dll1/4 over-expressing mice show accelerated proliferation, leading to a decreased number of secretory cells within the epithelium[17]. Therefore, Dll1/4 appears to maintain the proliferative status of ISCs within the crypts, preventing ISCs from differentiating into secretory cell lineages. Similar effects have also been observed in relation to Noggin expression. Noggin binds to and inactivates the BMP4 protein, resulting in a blockade of the BMP/Smad signaling pathway, which helps ISCs maintain their proliferative status[18].
Thus, the fast turnover of the intestinal epithelium not only relies on ISC proliferation but also on the differentiation of ISCs into functional cells. As described above, TA progenitors become mature when they migrate along the villus-crypt axis. In this process, the levels of Wnt3, EGF and R-spondin1 progressively decrease from the crypt basement membrane to the upper regions of crypt, whereas BMP4 levels are obviously increased[7]. Combined with the expressions of some lineage-devoted genes, such as Math1 (secretory lineage)[19] and Hes1 (absorptive lineage)[20], these TA progenitors stop dividing and ultimately differentiate into functional epithelial cells. In all, the spatial differences in the expressions of niche cell-derived signals along the villus-crypt axis maintain the proliferative status of ISCs and prevent these cells from differentiating within the crypt-domains.
Within the crypt-domains of the small intestine, three types of cells are arranged, including ISCs, TA progenitor cells and Paneth cells[2]. ISCs are exclusively located within the lower domain of the crypts because the development of active ISCs is dependent on essential signals from Paneth cells[3]. In 1974, Cheng and Leblond pointed out that the column cells in the crypt basement membrane, located next to the Paneth cells, seemed to be the progenitors of intestinal epithelial cells because they observed that these column cells were rapidly cycling[21]. However, due to a lack of available markers for identifying these epithelial progenitors at that time, the “stemness” of these cells was difficult to define. As recent clarification that the Lgr5 gene is a target of the Wnt/β-catenin signaling pathway, the transgenic (Tg) mouse (Strain name: Lgr5-eGFP-IRES-CreERT2) was established by Clevers, H.’s group[22]. Based on this model, this team first demonstrated, both in vitro and in vivo, that these crypt basement column cells (CBC), which highly express Lgr5, were ISCs (also called CBC stem cells)[22,23]. Since then, various promising data concerning the characteristics of these Lgr5+ ISCs have been published, such as their numbers in the crypts (14-16 per crypt)[22], their resistance to foreign stimuli (less radiosensitive than mature epithelial cells)[24], their cell-cycle duration (about 21.5 h), their mitotic process (symmetrical division) and their DNA segregation pattern (random distribution to offspring)[25] (Table 2). Apart from these points, several genes have been reported to be highly expressed by Lgr5+ ISCs, such as Musashi-1[26], Sox9[27], Ascl2[28], Smoc2[25], Rnf43[29], Znrf3[30], Olfm4[31], Cd24I[32], Cd44 variant 4-10 (Cd44v4-10)[33], Cd133[34] and Cd166[35]. Hence, these genes are referred to as ISC-related genes.
| Location | Representative research by | Number per crypt | Sensitivity to IR | Cell-cycle | Cell-division | DNA segregation |
| CBC | Clevers H’s team | 14-16 | Less than mature cells | 21.5 h | Symmetry | Random |
| 4+ position | Potten CS’s team | 4-6 | Apoptosis upon receiving 1 cGy | Approximately 24 h | Asymmetry | Hierarchy |
However, Potten[36] believed that the cells located at the 4+ position of the crypts were active ISCs. It has been shown that ISCs at this position expand their numbers every 24 h and that they are very sensitive to ionizing irradiation, with only 1 cGy of ionizing irradiation being enough to initiate apoptosis among these ISCs[36]. Thereafter, through the double labeling of DNA using 3HTdR and BrdU, Potten et al[37] found that when these ISCs divided, parental DNA strands were allocated to their larger daughter cells, while the newly synthesized strands were allocated to the smaller progeny[37]. Because of this action, the larger cells keep their stem-cell identities and genomic stability, and the smaller cells become TA progenitors for replacing dead cells within the epithelium[37]. Although evidence for the hierarchical distribution of parental DNA strands is well supported, there has been an inconclusive debate concerning this issue. Data from Clevers H’s team recently demonstrated that the dividing Lgr5+ ISCs randomly segregate the parental DNA stands into equal-sized offspring[25]. Thereafter, the daughters adhering to Paneth cells remain at the crypt basement membrane and continue to function as ISCs, whereas the cells not receiving resources from Paneth cells develop into TA progenitors[38,39]. In spite of such discrepancies in the characteristics of ISCs at different locations, recent evidence indeed supports the idea that the ISCs at the 4+ position are more sensitive to foreign stimuli-induced cell-death than are ISCs at the crypt basement membrane[40]. A study on this was carried out by Zhu et al[40], and they found that tamoxifen-preconditioning for Cre-expression in Lgr5-eGFP-IRES-CreERT2 Tg mice induced apoptosis in a small portion of ISCs. Herein, the dead ISCs were fast-cycling cells, positive for Lgr5, and mainly located at the 4+ position of crypts. Additionally, ionizing irradiation (1 cGy) also caused the death of ISCs at the 4+ position[40]. Lgr5+ ISCs at the crypt basement membrane replenished such cell loss[21]. Accordingly, nearly 10% of Lgr5+ ISCs were confirmed to be located around the same sites in crypts[21]. To a certain extent, these findings indicate the heterogenicity among Lgr5+ ISCs.
The ISCs mentioned above are referred to as an active pool for maintaining epithelial homeostasis in intact circumstances. Thus, the niches of the ISCs manipulate their fates for tissue regeneration[4]. Recently, a distinct population of ISCs located at the 4+ position of crypts, has been extensively investigated, and the relevant factors that identify these cells are as follows: (1) Marker genes: Bmi1, HopX, mTERT and Lrig1[41-44]; (2) Status under intact conditions: No more than 2% of these ISCs are proliferative (termed label-retaining cells, LRC)[41]; (3) Response to foreign stimuli: They activate their cell-cycles to replenish dead cells through direct differentiation into functional epithelial cells or by converting themselves into CBC stem cells[41,45-47]; and (4) Forces driving proliferation: Independent of the Wnt/β-catenin signaling pathway[41] (Table 3). Because of such characteristics, these ISCs are classified as a reserve population. In mammals, some tissues or organs also share the same pattern of maintaining their homeostasis using a system of diverse stem cells, such as hematopoietic stem cells in bone marrow and hair follicle stem cells in skin[4].
| ISC type | Main location | Cell-cycle status under intact conditions | Differentiating capability | Foreign stimuli-induced response | Leading pathway | Typical marker genes |
| CBC stem cell | Crypt basement | Active | Yes | Apoptosis | Wnt/β-catenin | Lgr5 |
| Reserve ISC | 4+ position | Slow | Yes | Proliferation | Unknown but independent of Wnt/β-catenin | Bmi1, HopX, Lrig1, mTERT |
In terms of the inter-conversion between reserve ISCs and CBC stem cells, Tian et al[45] firstly reported their findings through the use of a Tg mouse strain whose Lgr5 gene contained the sequence encoding the human diphtherotoxin receptor (DTR). When treated with diphtherotoxin, the Lgr5+ cells were depleted in the crypts, while numbers of Bmi1+ cells increased. However, Lgr5+cells emerged within the crypts 48 hours later. Thereafter, the number of Lgr5+ compartments increased and they migrated progressively from the 4+ position to the crypt basement membrane, indicating a revival of CBC stem cells. Based on these findings, the offspring of Tg mice (Strain: Bmi-CreER) crossed with an Lgr5-DTR/+ mouse strain were then crossed with different Tg mice (Strain: Rosa26-LacZ), and these offspring was used to trace the lineage of the restored CBC stem cells. The results of this analysis demonstrate that the CBC stem cells were derived from Bmi1+ ISCs. Similar results were also reported in studies using murine models of radiation-induced ISC apoptosis[46,47] Moreover, when affected by foreign stimuli, some TA progenitors are also capable of converting themselves into ISCs, such as the Dll1+ secretory progenitors at the 3+ position[48] and the label-retaining progenitors at the 5+ position[49]. Apart from the issues mentioned above, the Lgr5+ ISCs were shown to be able to convert into HopX+ ISCs both in vitro and in vivo[42]. All these findings suggest niche conversion between distinct stem cell populations, but the relevant mechanisms remained unclear until now. Thus, it can be confirmed that the expressions of the above mentioned marker genes of reserve ISCs overlap in active ISCs but at relatively low levels, meaning that ISCs still lack specific definable markers[21]. Hence, it is only feasible to discriminate the distinct populations of ISCs based on their locations and the genes that are highly expressed[50].
Precisely discriminating Lgr5 expressing levels using FACS-sorting techniques facilitates the isolation of ISCs from Tg mice (Strain: Lgr5-eGFP-IRES-CreERT2)[23]. For the wild-type hosts, recent works have also reported that several cell-surface markers may serve as potential candidates for sorting ISCs, including CD24[51], CD44[52], EphB2[5,53], CD133[34] and CD166[35]. However, the main obstacle to this lies in the fact that not all sorted cells are ISCs because all these markers can be found not only among ISCs but also among TA progenitors and mature cells, which results in a low purity for the target cells. To solve this problem, several novel strategies that use combinations of several surface makers, such as CD24/Sox9[54], CD24/CD44[55], CD44/CD133[56] and CD24/CD44/CD166/GRP78/c-Kit[57], enable the more precise identification of ISCs than is possible using one single marker for FACS analyses.
For culturing ISCs, extensive studies have been performed during the last two decades. At the beginning of this period, due to a profound lack of knowledge about the biological characteristics of ISCs, human colon cancer cell lines or an epithelial cell line (IEC6 from the small intestine of rats) were used as substitutes for ISCs[58]. Thereafter, as advances were made in understanding the biological characteristics of ISCs, a novel culturing system for ISCs was established by Clevers, H. and colleagues[23]. These ISCs are wrapped by Matrigel containing laminin-α1/α2 and supplemented with Wnt3, EGF, Noggin, R-spondin1 and Jagged-1 in a serum-free medium to enable their growth, together with Y-27632 dihydrochloride to protect ISCs against ROCK pathway-induced anoikis. In this three-dimensional (3D) system, one single isolated small intestine-derived ISC is capable of expanding in number, while also differentiating into functional epithelial cells for up to 1 year[23]. The ISC-derived structures are termed intestinal organoids. Moreover, through the use of similar models, Lgr5+ stem cells from various organs or tissues, such as the liver[59], pancreas[60], stomach[61] and colon[5], can also be cultured and induced to form organoids. Another successful system was established by Ootani et al[62], which was based on a fetal calf serum-containing system for the long-term expansion of ISC-derived organoids. A remarkable difference from the system established by Clevers H’s team lies in the fact that pieces of the small intestine were directly used in vitro. Details are shown in Table 4. Moreover, intestinal myofibroblasts were obviously expanded in this system. As we mentioned above, the myofibroblasts around the crypts form the niches for ISCs. To some extent, we propose that increased myofibroblast numbers will facilitate the development of ISCs into organoids based on previous findings that intestinal myofibroblasts are capable of increasing the organoid-forming potential of crypt fragments in vitro[10]. Although these 3D-culture systems allow for ISC expansion, the commitment of ISCs into functional cells is hard to be prevented. Moreover, the colony-forming efficacy (CFE) of the ISCs is only about 10%[23]. To solve this problem, Yin et al[63] used two small molecules, CHIR99021 and valproic acid (VPA), which were found to synergistically improve the expansion of ISCs in vitro, showing a CFE that was about 100-fold greater than that of ISCs without these two molecules. In this case, CHIR99021 mainly targets the activation of the Wnt/β-catenin signaling pathway, inhibiting absorptive cell differentiation, and VPA is capable of preventing ISCs from differentiating into secretory-lineage cells[63].
| Established by | Published year | System type | Substrate | Supplemental factor | Medium | Cultured content | Expanding duration of cultured content |
| Sato et al[23] | 2009 | 3D | Matrigel containing laminin α1 and α2 | N2, B27, Wnt3, EGF, Noggin, R-spondin1, Jagged-1 and Y-27632 dihydrochloride | Serum-free | ISCs | Up to 1 yr |
| Ootani et al[62] | 2009 | 3D | Collagen gel containing collagen I and III | / | Containing 20% of FCS in medium | Small pieces of intestinal tissue | Up to 1 yr |
CRC is the third leading cause of cancer-related death in developed countries, and several factors, such as a high fat diet, chronic inflammation and genomic instability, are highly associated with its occurrence[64,65]. For example, the constitutive activation of NF-κB in mature epithelial cells will initiate the formation of CRCs[66]. Likewise, chronic inflammation also induces the generation of CRCs from APC-mutant DCLK1+ Tuft cells[67]. Apart from these mature cells, ISCs are very important cellular sources for initiating CRCs[68]. Recent evidence suggested that factors including aging[69] and the total number of stem cell divisions[70], increase the frequency of gene mutations in ISCs in addition to the inherent mutations from the germline. Accordingly, CRCs can be identified and typed based on their genetic or epigenetic profiles[28,65,71].
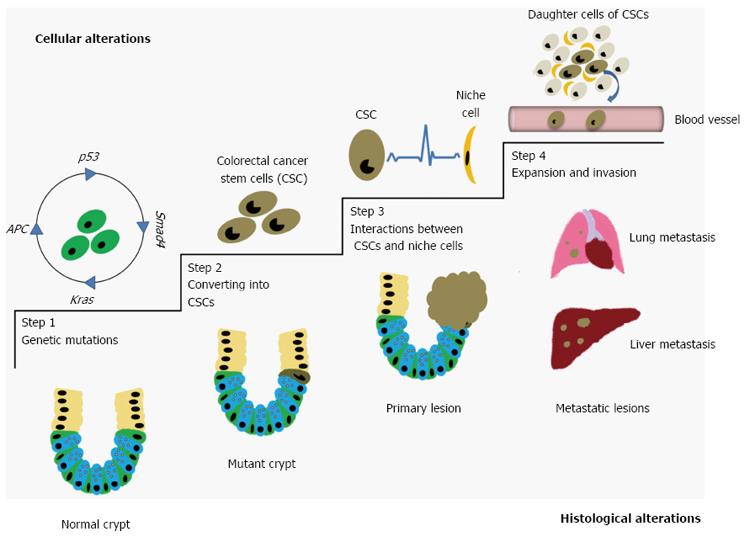
As mentioned above, four main pathways precisely control the proper development of one single ISC. However, aberrant Wnt signals overwhelmingly drive the pathogenesis of CRCs in most cases[65]. For example, mutant APC-induced robust activation of the Wnt/β-catenin signaling pathway is regarded as the first step in ISC-initiated gut malformation[72]. Along with the loss of function in p53, aberrant ISCs will wildly expand their numbers and show poor differentiating capabilities[73]. In addition, when mutations in Smad4 and Kras occur, invasive characteristics are conferred to these cells[74]. As a result of these actions, normal ISCs are converted into CSCs (Figure 2), and several ISC-related genes are expressed by CSCs, such as Lgr5[75], Wip1[76], Yap[77,78],EphB2[79], Cd24[80], Cd44 variant 6[33,81], Cd133[82,83] and Cd166[35,84], which indicate a poor prognosis for CRC patients when highly expressed due to the fact that they encode products that play roles in CSC expansion and invasion. However, not all CSCs share the same genetic profile. Recent data suggest that some subpopulations of colorectal CSCs have their own markers, such as CD26[85], CD110 and CUB-domain-containing protein-1(CDCP1)[86], which confer metastatic potential to CSCs. In addition, DCLK1 can be a specific marker of colorectal CSCs, which makes a distinction between colorectal CSCs and ISCs possible[87]. These results suggest that colorectal CSCs possess diverse genetic profiles, a paradigm reflecting the heterogenicity of CRCs[64,65,71].
As in ISCs, the development of colorectal CSCs also relies on their niches, which are composed of several types of cells, including endothelial cells, immune cells, myofibroblasts, cancer-associated fibroblasts (CAFs) and a small portion of mesenchymal cells[65]. Under such circumstances, colorectal CSCs progressively expand their numbers and produce daughter cells by using the resources from the niche-cells. Moreover, CSCs communicate with their niche-cells, obtaining invasive phenotypes to promote tumor progression, although the relevant signals are quite complicated. For example, within the tumor microenvironment, high levels of prostaglandin E2 can decrease the activity of GSK-3β through activating a cAMP/PKA cascade, leading to the cytoplasmic accumulation of β-catenin and facilitating the expansion and metastasis of colorectal CSCs[88,89]. In addition, prostaglandin E2 has an immunosuppressive capacity, which acts on several types of immune cells, including NK cells (decreased proliferation and cytolysis), dendritic cells (increased IL-10 production) and regulatory T cells (increased proliferation)[90]. Additionally, some CAF-derived cytokines, including HGF, osteopontin (OPN), SDF-1[81], TGF-β[91], IL-6[92] and IL-17A[93], also exhibit effects that increase the frequencies of colorectal CSC-induced malformations. For example, the TGF-β-induced epithelial-mesenchymal transition (EMT) in colonic CSCs increases their potential for liver metastasis[91,94],and HGF, OPN and SDF-1 are capable of up-regulating the expression of Cd44v6 in colorectal CSCs by activating the Wnt/β-catenin signaling pathway. In addition, colorectal CSCs show improved survival via the activation of the PI3K/Akt cascade, which is regulated by the interaction between HGF and the CD44 receptor[81]. Similarly, the survival and invasive capacities of CSCs can be increased through the interaction between the CD44 receptor and hyaluronan, a molecule mediating cell adhesion[95]. Additionally, SDF-1 is an important attractant for some bone marrow progenitors, such as CD133+ hematopoietic progenitor cells and endothelial progenitor cells, to the tumor microenvironment, which facilitates tumor growth and metastasis[96,97]. For the immune mediators mentioned above, it has been reported that preconditioning using IL-6 could increase the proportion of ALDH+ cancer stem-like cells among cultured colonic cancer cells and increase the expression of Lgr5 by these cells. Moreover, another tumor-facilitating action of IL-6 lies in its capacity to increase the number of Th17 cells and increase their production of IL-17A, a cytokine favoring tumor growth through its support of the expansion of colorectal CSCs[92,93]. CAFs have been reported to be capable of increasing IL-17A secretion after chemotherapy, and the increase in this immune mediator enables colorectal CSCs to acquire tolerance to anti-cancer drugs[93].
In addition to the cytokines mentioned above, at the subcellular level, exosomes also participate in establishing the network mediating intercellular communications. Exosomes are bioactive nanoparticles originating from multiple cell types (including cancer cells), and can be endocytotically taken up by adjacent or distant cells. Exosomes regulate the biological responses of target cells using their cargoes of proteins, lipids, miRNAs and mRNAs[98]. Of the specific roles of exosomes in regulating the generation of CRCs, fibroblast-derived exosomes have been demonstrated to be capable of enhancing the expansion of CRCs and conferring a resistance to 5-fluorouracil[99]. In providing feedback to niche-cells, exosomes from SW480 CRC cells have been shown to enhance the proliferation of endothelial cells via their cell cycle-facilitating mRNAs[100]. Moreover, tumor exosomes have antagonistic effects on immune cells, decreasing the cytotoxicity of NK cells, impairing the anti-tumor response of CTL cells and increasing the numbers of regulatory T/B cells, which establishes an immunosuppressive microenvironment, favoring tumor growth in vivo[101].
However, not all exosomes from CRC cells carry the same cargo profiles. For example, Cha et al[102] found that miRNA-100 was the main type of exosomal miRNA in Kras-mutant CRC cells, whereas miRNA-10b is predominant in wild-type exosomes. Even when CRCs relapse, the main type of serum miRNA in patients with recurrent CRC are different from those in patients without recurrent CRC[103]. It is possible that seven exosomal miRNAs, let-7a, miRNA-1299, miRNA-1246, miRNA-150, miRNA-21, miRNA-223 and miRNA-23a, may be useful candidates for assessing the progression of CRCs due to the high correlation between their serum levels and tumor burdens in vivo[104]. In addition, exosomal miRNAs differ in type among different CRC cells, reflecting the epigenetic heterogenicity of CRCs.
Based on the findings presented above, we conclude that genomic mutations are prerequisites for the transformation from normal cells into CSCs, which will ultimately form solid tumors through interactions with cancer niche-cells.
Clearly, the generation of Tg mice (Strain: Lgr5-eGFP-IRES-CreERT2) and establishment of 3D-culture systems for ISCs initiated the era of treating intestinal diseases using ISCs. Recently, data from two separate studies demonstrate that murine Lgr5+ colonic stem cells have the potential to healing epithelial injuries in immunodeficient mice with dextran sodium sulfate (DSS)-induced colitis[5,6]. Moreover, these studies also suggest that engrafted clusters could survive within lesioned sites for more than 6 mo, undergoing crypt-fission and commitment into functional cells, indicating the effectiveness and feasibility of homogenous transplantation[6]. Moreover, the protocols for expanding human ISCs from stomach, small intestine and colon tissues have been established, which holds great promise for treating some gastrointestinal diseases, such as inflammatory bowel disease (IBD), gastric ulcers (GCs) or microvillus inclusion disease[5,53,58]. To this end, Watanabe[105] has described a strategy for the autologous transplantation of colonic stem cells into patients with IBD. In brief, normal colonic tissues can be isolated through enteroscopy-guided biopsy. After expanding their number in vitro, the colonic stem cells can be transplanted into the lesioned sites[105]. Based on this, we recommend that GCs can also be managed using a similar approach. Of note, we believe that the influence of the microenvironment within the site of the injury on the viability of the graft must be taken into consideration. For IBD, extended inflammation within intestinal lesions limits epithelial healing[106]. To some extent, it would be beneficial to decrease apoptosis in engrafted stem cells in vivo through the use of immunosuppressive therapy. To date, it remains unclear whether stomach stem cells are acid-resistant. If so, this would help increase the viability of such grafts through the use of certain anti-acid drugs. Another issue concerning the application of autologous stem cells for treating IBD or GCs lies in the lack of evidence indicating whether the genomes of stem cells derived from such patients are stable. As mentioned above, chronic inflammation within the colon will initiate CRC formation. In addition, it is hard to determine whether freshly isolated samples contain mutant cells, especially when such cells are cultured with supplemental growth factors. Recent data suggest that ISCs from diabetic mice are more inclined to differentiate into absorptive cells and Paneth cells than are ISCs from healthy mice[107]. To avoid the above issues, we recommend that the genomic stability be determined before clinical transplantation. If instable genomes are detected, allogenic transplantations could be used as substitutes. Li et al[58] noted that a standardized strategy including HLA-matching between donors and recipients and the use of immunosuppressive drugs could be an option for dealing with graft rejection. However, the shortcomings of allogeneic transplantation lie in the long-term use of immunosuppressants and the limited availability of donor tissues.
For ISCs with gene mutations, it is hoped that they can be reprogrammed into normal cells using CRISPR/Cas9 technology, which is a powerful tool for editing multiple genes synchronously within an individual cell[108]. Using this technology, sequential mutations of four key genes, APC, p53, Smad4 and Kras, in human ISCs were first carried out in vitro to investigate the specific roles of these mutant ISCs in initiating CRCs in vivo[74]. Apart from this, it is also possible that ISCs may be massively cultured within biocompatible frameworks to form artificial mucosa, which could be realized by using 3D-bioprinting technology[109]. Therefore, any advancement concerning new theories in stem cell biology, new technologies for culturing stem cells or new applications of stem cells in treating diseases will push regenerative medicine forward.
Overall, the mechanisms by which ISCs maintain epithelial homeostasis are highly specific, and any stimuli altering the biological characteristics of ISCs will interrupt the homeostatic process, leading to lesions involving mucosal atrophy, hypertrophy or even malformation. Any information concerning biological alterations in ISCs will improve the public’s awareness on how to prevent some intestinal diseases. Moreover, ISCs are valuable tools, and their potentials for healing ulcerous lesions in experimental models have been demonstrated. In spite of this, a detailed protocol, including quality control for ISC culturing, criteria for evaluating the effectiveness of grafts, and criteria for evaluating transplantation-related adverse effects, needs to be established before carrying out relevant clinical trials.
We thank Bo Chen for his kind English language editing.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Manguso F S- Editor: Ma YJ L- Editor: O’Neill M E- Editor: Wang CH
| 1. | van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1156] [Cited by in RCA: 1327] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 2. | Umar S. Intestinal stem cells. Curr Gastroenterol Rep. 2010;12:340-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 3. | van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 515] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 4. | Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 954] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 5. | Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 546] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 6. | Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med. 2012;18:618-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 620] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 7. | Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 803] [Cited by in RCA: 878] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 8. | Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2119] [Cited by in RCA: 1933] [Article Influence: 138.1] [Reference Citation Analysis (0)] |
| 9. | Hirokawa Y, Yip KH, Tan CW, Burgess AW. Colonic myofibroblast cell line stimulates colonoid formation. Am J Physiol Gastrointest Liver Physiol. 2014;306:G547-G556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Lei NY, Jabaji Z, Wang J, Joshi VS, Brinkley GJ, Khalil H, Wang F, Jaroszewicz A, Pellegrini M, Li L. Intestinal subepithelial myofibroblasts support the growth of intestinal epithelial stem cells. PLoS One. 2014;9:e84651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Peiris D, Pacheco I, Spencer C, MacLeod RJ. The extracellular calcium-sensing receptor reciprocally regulates the secretion of BMP-2 and the BMP antagonist Noggin in colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2007;292:G753-G766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Roth S, Fodde R. The nature of intestinal stem cells’ nurture. EMBO Rep. 2011;12:483-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 13. | Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 462] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 14. | Binnerts ME, Kim KA, Bright JM, Patel SM, Tran K, Zhou M, Leung JM, Liu Y, Lomas WE, Dixon M. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc Natl Acad Sci USA. 2007;104:14700-14705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 218] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 15. | Mustata RC, Van Loy T, Lefort A, Libert F, Strollo S, Vassart G, Garcia MI. Lgr4 is required for Paneth cell differentiation and maintenance of intestinal stem cells ex vivo. EMBO Rep. 2011;12:558-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 114] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 16. | Holmberg J, Genander M, Halford MM, Annerén C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisén J. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 240] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 17. | Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, Kaestner KH, Kopan R, Lewis J, Radtke F. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230-1240.e1-7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 344] [Cited by in RCA: 325] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 18. | He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117-1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 814] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 19. | Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155-2158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 699] [Cited by in RCA: 718] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 20. | Suzuki K, Fukui H, Kayahara T, Sawada M, Seno H, Hiai H, Kageyama R, Okano H, Chiba T. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochem Biophys Res Commun. 2005;328:348-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Barker N, van Oudenaarden A, Clevers H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell. 2012;11:452-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 243] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 22. | Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3854] [Cited by in RCA: 4335] [Article Influence: 240.8] [Reference Citation Analysis (0)] |
| 23. | Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4394] [Cited by in RCA: 5172] [Article Influence: 323.3] [Reference Citation Analysis (0)] |
| 24. | Hua G, Thin TH, Feldman R, Haimovitz-Friedman A, Clevers H, Fuks Z, Kolesnick R. Crypt base columnar stem cells in small intestines of mice are radioresistant. Gastroenterology. 2012;143:1266-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 175] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 25. | Schepers AG, Vries R, van den Born M, van de Wetering M, Clevers H. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011;30:1104-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 172] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 26. | Maria Cambuli F, Rezza A, Nadjar J, Plateroti M. Brief report: musashi1-eGFP mice, a new tool for differential isolation of the intestinal stem cell populations. Stem Cells. 2013;31:2273-2278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1108-G1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 188] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 28. | van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 567] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 29. | Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 735] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 30. | Koo BK, van Es JH, van den Born M, Clevers H. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43; Znrf3-mutant neoplasia. Proc Natl Acad Sci USA. 2015;112:7548-7550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 31. | VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud Å, Grosse AS, Gumucio DL, Ernst SA, Tsai YH, Dempsey PJ, Samuelson LC. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 442] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 32. | Van Limbergen J, Geddes K, Henderson P, Russell RK, Drummond HE, Satsangi J, Griffiths AM, Philpott DJ, Wilson DC. Paneth cell marker CD24 in NOD2 knockout organoids and in inflammatory bowel disease (IBD). Gut. 2015;64:353-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Zeilstra J, Joosten SP, van Andel H, Tolg C, Berns A, Snoek M, van de Wetering M, Spaargaren M, Clevers H, Pals ST. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene. 2014;33:665-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 34. | Snippert HJ, van Es JH, van den Born M, Begthel H, Stange DE, Barker N, Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136:2187-2194.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, Swain JR, Wong MH. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139:2072-2082.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 36. | Potten CS. Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos Trans R Soc Lond B Biol Sci. 1998;353:821-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 381] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 37. | Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381-2388. [PubMed] |
| 38. | Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1401] [Cited by in RCA: 1501] [Article Influence: 100.1] [Reference Citation Analysis (0)] |
| 39. | Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 553] [Cited by in RCA: 490] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 40. | Zhu Y, Huang YF, Kek C, Bulavin DV. Apoptosis differently affects lineage tracing of Lgr5 and Bmi1 intestinal stem cell populations. Cell Stem Cell. 2013;12:298-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 41. | Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915-920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1017] [Cited by in RCA: 966] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 42. | Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420-1424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 606] [Cited by in RCA: 604] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 43. | Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 568] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 44. | Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci USA. 2011;108:179-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 432] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 45. | Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 836] [Cited by in RCA: 935] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 46. | Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 656] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 47. | Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 450] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 48. | van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 610] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 49. | Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 614] [Cited by in RCA: 606] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 50. | Clevers H. Stem Cells: A unifying theory for the crypt. Nature. 2013;495:53-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 51. | von Furstenberg RJ, Gulati AS, Baxi A, Doherty JM, Stappenbeck TS, Gracz AD, Magness ST, Henning SJ. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2011;300:G409-G417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 52. | Chang PY, Jin X, Jiang YY, Wang LX, Liu YJ, Wang J. Mensenchymal stem cells can delay radiation-induced crypt death: impact on intestinal CD44(+) fragments. Cell Tissue Res. 2016;364:331-344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 53. | Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762-1772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2253] [Cited by in RCA: 2723] [Article Influence: 194.5] [Reference Citation Analysis (0)] |
| 54. | Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010;298:G590-G600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 125] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 55. | Gracz AD, Fuller MK, Wang F, Li L, Stelzner M, Dunn JC, Martin MG, Magness ST. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31:2024-2030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 56. | Hou NY, Yang K, Chen T, Chen XZ, Zhang B, Mo XM, Hu JK. CD133+ CD44+ subgroups may be human small intestinal stem cells. Mol Biol Rep. 2011;38:997-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 57. | Wang F, Scoville D, He XC, Mahe MM, Box A, Perry JM, Smith NR, Lei NY, Davies PS, Fuller MK. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145:383-95.e1-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 166] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 58. | Li VS, Clevers H. In vitro expansion and transplantation of intestinal crypt stem cells. Gastroenterology. 2012;143:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 59. | Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 966] [Cited by in RCA: 1163] [Article Influence: 96.9] [Reference Citation Analysis (0)] |
| 60. | Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708-2721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 460] [Cited by in RCA: 538] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 61. | Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1056] [Cited by in RCA: 1197] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 62. | Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701-706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 754] [Cited by in RCA: 708] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 63. | Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 446] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 64. | Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118:671-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 373] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 65. | Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 287] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 66. | Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 832] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 67. | Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, Brandtner A, Setlik W, Remotti H, Muley A. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest. 2014;124:1283-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 308] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 68. | Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1797] [Cited by in RCA: 1668] [Article Influence: 104.3] [Reference Citation Analysis (0)] |
| 69. | Adams PD, Jasper H, Rudolph KL. Aging-Induced Stem Cell Mutations as Drivers for Disease and Cancer. Cell Stem Cell. 2015;16:601-612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 70. | Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1440] [Cited by in RCA: 1274] [Article Influence: 127.4] [Reference Citation Analysis (0)] |
| 71. | Biswas S, Davis H, Irshad S, Sandberg T, Worthley D, Leedham S. Microenvironmental control of stem cell fate in intestinal homeostasis and disease. J Pathol. 2015;237:135-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 72. | Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 403] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 73. | Davidson LA, Callaway ES, Kim E, Weeks BR, Fan YY, Allred CD, Chapkin RS. Targeted Deletion of p53 in Lgr5-Expressing Intestinal Stem Cells Promotes Colon Tumorigenesis in a Preclinical Model of Colitis-Associated Cancer. Cancer Res. 2015;75:5392-5397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 74. | Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 796] [Article Influence: 79.6] [Reference Citation Analysis (0)] |
| 75. | Liu Z, Dai W, Jiang L, Cheng Y. Over-expression of LGR5 correlates with poor survival of colon cancer in mice as well as in patients. Neoplasma. 2014;61:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 76. | Demidov ON, Timofeev O, Lwin HN, Kek C, Appella E, Bulavin DV. Wip1 phosphatase regulates p53-dependent apoptosis of stem cells and tumorigenesis in the mouse intestine. Cell Stem Cell. 2007;1:180-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 94] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 77. | Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 386] [Cited by in RCA: 455] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 78. | Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 476] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 79. | Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Muñoz P. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 742] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 80. | Sagiv E, Starr A, Rozovski U, Khosravi R, Altevogt P, Wang T, Arber N. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res. 2008;68:2803-2812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 128] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 81. | Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 578] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 82. | Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2952] [Cited by in RCA: 3035] [Article Influence: 159.7] [Reference Citation Analysis (0)] |
| 83. | Zhang SS, Han ZP, Jing YY, Tao SF, Li TJ, Wang H, Wang Y, Li R, Yang Y, Zhao X. CD133(+)CXCR4(+) colon cancer cells exhibit metastatic potential and predict poor prognosis of patients. BMC Med. 2012;10:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 84. | Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158-10163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1541] [Cited by in RCA: 1663] [Article Influence: 92.4] [Reference Citation Analysis (0)] |
| 85. | Pang R, Law WL, Chu AC, Poon JT, Lam CS, Chow AK, Ng L, Cheung LW, Lan XR, Lan HY. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 2010;6:603-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 428] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 86. | Gao W, Chen L, Ma Z, Du Z, Zhao Z, Hu Z, Li Q. Isolation and phenotypic characterization of colorectal cancer stem cells with organ-specific metastatic potential. Gastroenterology. 2013;145:636-646.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 87. | Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 330] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 88. | Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136-1147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 604] [Cited by in RCA: 580] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 89. | Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 Promotes Colorectal Cancer Stem Cell Expansion and Metastasis in Mice. Gastroenterology. 2015;149:1884-1895.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 228] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 90. | Chang PY, Qu YQ, Wang J, Dong LH. The potential of mesenchymal stem cells in the management of radiation enteropathy. Cell Death Dis. 2015;6:e1840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 91. | Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, Sevillano M, Palomo-Ponce S, Tauriello DV, Byrom D. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 745] [Cited by in RCA: 826] [Article Influence: 82.6] [Reference Citation Analysis (0)] |
| 92. | Huynh PT, Beswick EJ, Coronado YA, Johnson P, O’Connell MR, Watts T, Singh P, Qiu S, Morris K, Powell DW. CD90(+) stromal cells are the major source of IL-6, which supports cancer stem-like cells and inflammation in colorectal cancer. Int J Cancer. 2016;138:1971-1981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 93. | Lotti F, Jarrar AM, Pai RK, Hitomi M, Lathia J, Mace A, Gantt GA, Sukhdeo K, DeVecchio J, Vasanji A. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. J Exp Med. 2013;210:2851-2872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 323] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 94. | Zubeldia IG, Bleau AM, Redrado M, Serrano D, Agliano A, Gil-Puig C, Vidal-Vanaclocha F, Lecanda J, Calvo A. Epithelial to mesenchymal transition and cancer stem cell phenotypes leading to liver metastasis are abrogated by the novel TGFβ1-targeting peptides P17 and P144. Exp Cell Res. 2013;319:12-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 95. | Jung T, Gross W, Zöller M. CD44v6 coordinates tumor matrix-triggered motility and apoptosis resistance. J Biol Chem. 2011;286:15862-15874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 96. | Zhang C, Zhou C, Wu XJ, Yang M, Yang ZH, Xiong HZ, Zhou CP, Lu YX, Li Y, Li XN. Human CD133-positive hematopoietic progenitor cells initiate growth and metastasis of colorectal cancer cells. Carcinogenesis. 2014;35:2771-2777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 97. | Varol U, Yildiz I, Salman T, Karabulut B, Uslu R. Markers to predict the efficacy of bevacizumab in the treatment of metastatic colorectal cancer. Tumori. 2014;100:370-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 98. | Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3378] [Cited by in RCA: 3537] [Article Influence: 321.5] [Reference Citation Analysis (0)] |
| 99. | Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, Wu Y, Qin J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS One. 2015;10:e0125625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 100. | Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, Kim JH, Choi DS, Kim YK, Hwang D. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 330] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 101. | The exosomes in tumor immunity. Oncoimmunology. 2015;4:e1027472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 182] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 102. | Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 289] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 103. | Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113:275-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 332] [Cited by in RCA: 408] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 104. | Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 647] [Article Influence: 58.8] [Reference Citation Analysis (1)] |
| 105. | Watanabe M. Adult tissue stem cell therapy for gastrointestinal diseases. J Gastroenterol Hepatol. 2014; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 106. | Quetglas EG, Mujagic Z, Wigge S, Keszthelyi D, Wachten S, Masclee A, Reinisch W. Update on pathogenesis and predictors of response of therapeutic strategies used in inflammatory bowel disease. World J Gastroenterol. 2015;21:12519-12543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 107. | Zhong XY, Yu T, Zhong W, Li JY, Xia ZS, Yuan YH, Yu Z, Chen QK. Lgr5 positive stem cells sorted from small intestines of diabetic mice differentiate into higher proportion of absorptive cells and Paneth cells in vitro. Dev Growth Differ. 2015;57:453-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 108. | Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10336] [Cited by in RCA: 11171] [Article Influence: 930.9] [Reference Citation Analysis (0)] |
| 109. | Bhattacharjee T, Zehnder SM, Rowe KG, Jain S, Nixon RM, Sawyer WG, Angelini TE. Writing in the granular gel medium. Sci Adv. 2015;1:e1500655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 349] [Article Influence: 34.9] [Reference Citation Analysis (0)] |