Published online May 28, 2016. doi: 10.3748/wjg.v22.i20.4824
Peer-review started: February 9, 2016
First decision: March 7, 2016
Revised: March 9, 2016
Accepted: March 30, 2016
Article in press: March 30, 2016
Published online: May 28, 2016
Processing time: 100 Days and 0 Hours
The present review describes the current status of multiplex quantitative real time polymerase chain reaction (qPCR) assays developed and used globally for detection and subtyping of hepatitis viruses in body fluids. Several studies have reported the use of multiplex qPCR for the detection of hepatitis viruses, including hepatitis A virus (HAV), hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus (HDV), and hepatitis E virus (HEV). In addition, multiplex qPCR has also been developed for genotyping HBV, HCV, and HEV subtypes. Although a single step multiplex qPCR assay for all six hepatitis viruses, i.e., A to G viruses, is not yet reported, it may be available in the near future as the technologies continue to advance. All studies use a conserved region of the viral genome as the basis of amplification and hydrolysis probes as the preferred chemistries for improved detection. Based on a standard plot prepared using varying concentrations of template and the observed threshold cycle value, it is possible to determine the linear dynamic range and to calculate an exact copy number of virus in the specimen. Advantages of multiplex qPCR assay over singleplex or other molecular techniques in samples from patients with co-infection include fast results, low cost, and a single step investigation process.
Core tip: The present review describes the worldwide application and the significance of multiplex quantitative real time polymerase chain reaction (qPCR) for simultaneous detection of hepatitis viruses and their subtypes in serum. The published literature has demonstrated that the multiplex qPCR assay is a fast, easy, cost-effective, and sensitive technique for the early diagnosis of hepatitis co-infections. Use of this technique, in comparison to other diagnostic procedures, is increasing in diagnostic laboratories.
- Citation: Irshad M, Gupta P, Mankotia DS, Ansari MA. Multiplex qPCR for serodetection and serotyping of hepatitis viruses: A brief review. World J Gastroenterol 2016; 22(20): 4824-4834
- URL: https://www.wjgnet.com/1007-9327/full/v22/i20/4824.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i20.4824
Viral hepatitis is a serious public health problem requiring early diagnosis and timely treatment. There are a number of hepatitis viruses that have already been characterized based on their molecular structure and named alphabetically as hepatitis viruses A, B, C, D, E, and G (HAV, HBV, HCV, HDV, HEV, and HGV), respectively. These are hepatotropic and non-cytopathic in nature and cause liver damage by immune mediated cell lysis[1]. There is an additional group of viruses that cause hepatitis but are not yet characterized. These viruses have been put under the category of non A-G hepatitis viruses. HAV infects mainly the pediatric age group, occurs both sporadically as well as in epidemics, and accounts for an estimated 1.4 million cases annually[2]. Two billion people are suspected to be infected with HBV globally, and approximately 350 million of them suffer from chronic hepatitis B infection[3]. About 25% of adults infected with HBV during childhood are reported to die from hepatocellular carcinoma (HCC) or liver cirrhosis[4]. In addition, 3-4 million people are infected with HCV each year, and a high proportion of them develop chronic HCV infection. A large population infected with HCV dies from serious liver diseases annually[5]. Similarly, reports are also available on HEV infection. In addition to individual viral infection, there are cases of co-infections reported from various parts of the world. Hepatitis A and E infections usually run a benign course of disease and resolve in due course of time without developing chronic diseases. In contrast, hepatitis B and C infections cause severe liver diseases, developing chronicity in a significant number of patients. Interestingly, hepatitis A and E infections in patients with pre-existing HBV or HCV infections lead to the development of serious diseases with a significant rise in morbidity and mortality[6].
The diagnosis of hepatitis viral infections is usually done with serological markers in blood. However, there are situations where serology loses its credibility. For example, serological markers can not differentiate between past and present infections. In addition, serological tests do not address the problem of antigenic variations in viruses, infections with different genotypes, presence of silent carriers, and absence of antibody in early phase of infection[7]. Moreover, the presence of maternal antibodies makes it impossible to detect infections in newborns[8]. In order to have an alternate system, the nucleic acid tests (NAT) based methods were developed for detecting the viral genome in serum for the diagnosis of viral hepatitis. NAT based methods have the benefit of direct examination of the infectious agent’s genome in serum[9,10].
The conventional polymerase chain reaction (PCR) is one such NAT based method that has been in practice in some laboratories for the diagnosis of viral hepatitis in the last few years[11]. However, conventional PCR is a lengthy procedure with several technical and operational problems, and so, it is of limited use. In addition, each marker needs to be investigated separately by PCR, and it takes a very long time to reach a final diagnosis. Because of these limitations of conventional PCR, the use of real time PCR was supposed to be a better option for early diagnosis of viral hepatitis in both sporadic and epidemic cases. Real time PCR is one of the latest techniques frequently used for the diagnosis of various infectious diseases, including viral hepatitis. It can detect causative pathogen-related nucleic acid in body fluids in a very short time period. It can also be used to determine different molecular forms and variant molecular species of pathogens, including bacteria, viruses, and several parasites[12,13]. Real time PCR is a specific and sensitive technique and uses specific probes and primers to detect target sequences in the genome. Moreover, this technique is performed on an automated machine without the need of post PCR procedures, thus minimizing cross contamination between samples, simultaneously accelerating the analysis[14].
The recent development of molecular technologies has relayed a strong message to medical researchers to explore ways to further improve the diagnostic procedures. Those researchers working in the area of medical virology have switched from traditional approaches of virus detection in clinical samples to multiplexing for simultaneous detection of multiple pathogens in a single assay[15]. Recently, several PCR based assays coupled with oligonucleotide microarray technology have been designed to allow for the simultaneous detection and genotyping of several viruses, including blood borne pathogens[16], respiratory viruses[17], and adenoviruses[18]. These assays show a significant increase in the sensitivity of detection, reaching 10-100 copies of target RNA/DNA in a sample[19]. Given the ease of performance, short reaction time, low cost, and the ability to monitor the results on a screen, these assays have proved attractive to all diagnostic laboratories furnished with minimal essential facilities. After surveying the literature on the use of PCR based multiplex assays for detecting and genotyping hepatitis viruses, we noticed several attempts to develop multiplex real time PCR assays for hepatitis in the last few years. Here, we provide an up to date review on the development, use, and significance of multiplex qPCR in the field of viral hepatitis.
In order to develop a multiplex qPCR assay for multiple pathogens, the first and foremost step is to explore and locate the target region on each pathogen’s genome for amplification purpose. Since variation in the genome is a dynamic process, it is necessary that a multiplex assay uses the most conserved region representing all the strains/variants for detection of the pathogen in body fluid. In the case of hepatitis viral infections, studies have reported a distinct conserved region that has been used as a target for amplification of each individual viral genome[4,20,21]. Table 1 shows the list of target regions used in various studies on multiplex qPCR assays developed for hepatitis viruses. The 5’ untranslated region (UTR) was reported to be the main target template in HAV, HCV, and HGV[20,22]. It was based on the availability of most conserved sequence in the 5’ UTR for amplification purpose. Similarly, S-gene or X-gene was used for HBV, ribozyme-1 gene for HDV, and open reading frame (ORF)-2 or ORF-3 region for HEV. Different studies have reported different sequences as templates in these selected conserved regions, though, there was very little information provided about the exact location of the sequences used.
| Virus | Conserved region | Ref. |
| HAV | 5’ UTR | [4,15,20,22] |
| HBV | S-gene | [4, 19-21] |
| X-gene | [15] | |
| HCV | 5’ UTR | [4,15,19-21] |
| HDV | Ribozyme-1 | [20] |
| HEV | ORF2 | [15,22] |
| ORF3 | [20] | |
| HGV | 5’ UTR | [20] |
After deciding which conserved region and location of the sequence were to be used as template, it is important to design the primers and probes for their use in the development of qPCR[23]. The selection of the primer is based on its specificity with the target template. At the same time, its length, melting temperature, GC content, 3’ end stability, sequence complexity, and location in the target sequence determine the length and melting temperature of the amplicon produced and the amplification efficiency of the assay[23,24]. Notably, the choice of chemistry and probe design are at the liberty of the user’s interest, with numerous options available to them[24]. During selection of chemistry and probe, one needs to determine whether to quantify DNA, profile mRNA, or perform allelic discrimination assays[25].
Real-time PCR and melting curve analysis (MCA) are good techniques for quantifying nucleic acids, detecting mutations, and conducting genotyping analysis. These methods often use TaqMan probes[26], Molecular beacons[27], Sunrise primers[28], Scorpion primers[29], and Light-up probes[30]. An alternative to probe-based methods is the use of DNA intercalating dyes that bind to double-stranded DNA. These dyes include ethidium bromide[31] and SYBR Green I[32,33]. However, certain drawbacks limit the use of SYBR Green I for resolving multiplex PCR based on MCA[34]. Other alternative dyes, such as BEBO[35], YO-PRO-1[36], LC Green[37], and SYTO-9[38,39] have also been tried for use in real time PCR. Table 2 provides a brief review of various chemistries/dyes offering several options for their use in qPCR assay developed for different purposes. Studies for detecting and genotyping hepatitis viruses with qPCR have reported different sets of dyes based on choice and their availability[40]. However, most of the studies conducted have reported a frequent use of hydrolysis probes despite many options available. This information is available in the data[41-71] compiled in Table 3.
| S. NO. | Class | Types | Structure | Mechanism of action | Advantages | Applications |
| 1 | DNA binding dyes | Ethidium Bromide, SYBR Green, SYBR Gold, YO-PRO-1, SYTO, BEBO, BOXTO, EvaGreen | Intercalating dyes | Bind to the minor groove of dsDNA during amplification | Inexpensive | Pathogen detection |
| Easily available | Gene expression | |||||
| SNP detection | ||||||
| Genotyping | ||||||
| 2 | Fluorophore labeled oligonucleotide | Primer probes | ||||
| Hairpins: Scorpions, Ampliflour, LUX | Loop based oligonucleotides | Bind to target during denaturation with emission of fluorescence | Inexpensive, Prevent formation of primer dimer, Less background signals | Pathogen detection | ||
| Genotyping | ||||||
| SNP allelic discrimination | ||||||
| Mutation detection | ||||||
| Cyclicons | Cyclic structure with reporter at 3’ end and quencher at 5’ end | Reporter and quencher in close proximity with energy transfer via FRET quenching. Their separation results in fluorescence emission during amplification | Inexpensive | Pathogen detection | ||
| Less contamination | Genotyping | |||||
| Less background signals | SNP allelic discrimination | |||||
| Mutation detection | ||||||
| Angler | Probe with DNA sequence bound to reverse primer through a HEG linker | During annealing step, DNA polymerase does extension of 3’ end reverse primer. Later on, SYBR Gold dye intercalates in dsDNA emitting fluorescence | Highly specific | Gene expression | ||
| Pathogen detection | ||||||
| SNP detection | ||||||
| Genotyping | ||||||
| Probes | ||||||
| Hydrolysis Probes: TaqMan probes, MGB-TaqMan, Snake assay | Oligonucleotide with reporter at 5’ and quencher at 3’ end | Probe is degraded by 5’ to 3’ exonuclease activity of DNA polymerase generating fluorescence during extension | Design and synthesis easy | Microarray validation | ||
| Pathogen detection | ||||||
| SNP allelic discrimination | ||||||
| Mutation detection | ||||||
| Hybridization probes: Hybprobes, Molecular Beacon, HyBeacon, MGB Probes | A pair of oligonucleotides having reporter dye on first and quencher on second oligonucleotide | Binding to target during hybridization and annealing brings fluorophore into proximity producing fluorescence by FRET | Design and synthesis quick and easy | Microarray validation | ||
| Pathogen detection | ||||||
| Viral/Bacterial genotyping | ||||||
| SNP allelic discrimination | ||||||
| Mutation detection | ||||||
| Nucleic acid analogues | ||||||
| PNAs, LNAs, ZNAs | Intercalating/inserting dyes | Identical to conventional oligonucleotides | Resistant to nuclease and proteases activity | Discriminate between DNA and cDNA in prokaryotes | ||
| Non-natural bases | ||||||
| No. | Assay systems | Instruments used | Group of pathogens detected | Types of chemistries/detection methods used | Ref. | |
| Hepatitis viruses | Other pathogens | |||||
| 1 | Multiplex real time PCR | Mx4000 (Stratagene) | HBV, HCV | HIV type-1, T. pallidum | TaqMan-LNA probe | [21] |
| 2 | Multiplex real time PCR | Light cycler 480 (Roche) | HEV genotypes | - | N.A. | [41] |
| 3 | Real time PCR assay | ABI 7500 (Applied Biosystems) | HAV, HBV, HCV, HDV, HEV | - | TaqMan Array card | [42] |
| 4 | Multiplex qPCR assay | Light cycler 480 (Roche) | HBV, HDV | - | TaqMan probe | [43] |
| 5 | Multiplex qPCR assay | ABI 7500 (Applied Biosystems) | HAV, HEV | - | Hydrolysis probe | [22] |
| 6 | Multiplex qRT-PCR | N.A. | HAV | Norovirus genotypes 1 and 2 | TaqMan probe | [44] |
| 7 | Multiplex ligation dependent probe real time PCR | Rotor-GeneQ (Qiagen) | HBV mutants | - | TaqMan probe | [45] |
| MLPA probe | ||||||
| 8 | Multiplex real time RT-PCR | N.A. | HEV genotypes | - | N.A. | [46] |
| 9 | Multiplex qPCR | N.A. | HBV genotypes | - | SYBR Green | [47] |
| 10 | Multiplex Real time PCR | N.A. | HAV | Norovirus, Rotavirus, Coxsackievirus | TaqMan probe | [48] |
| 11 | Multiplex Real time PCR | Light cycler 2.0 (Roche) | HAV, HBV, HCV and HEV | - | FRET probe | [15] |
| 12 | Multiplex RT-PCR | ABI 2720 (Applied Biosystems) | HCV | HIV type-1 | SYBR Green I | [8] |
| 13 | Multiplex qPCR | N.A. | HAV, HEV | Entero and Adeno-viruses | N.A. | [49] |
| 14 | Multiplex Real-Time PCR Assay | CFX96 (Bio-Rad) | HAV, HBV, HCV | - | READ technology based fluorophore | [4] |
| 15 | RT PCR assay | Smart cycler II (Cepheid) | HBV, HCV | - | TaqMan probe | [50] |
| 16 | Duplex real time PCR | ABI 7500 (Applied Biosystems) | HBV variants | - | Hydrolysis probe | [51] |
| 17 | Multiplex RT PCR | N.A. | HCV subtyping | - | Electrophoresis | [52] |
| 18 | Multiplex qPCR | N.A. | HBV genotypes | - | N.A. | [53] |
| 19 | Multiplex qPCR | N.A. | HCV | HIV type-1 | SYBR Green I | [54] |
| 20 | Duplex real-time RT-PCR | ABI Prism system (Applied Biosystems) | HCV variants | - | Hydrolysis probe | [55] |
| 21 | Multiplex real time PCR | N.A. | HAV | Norovirus genotypes 1 and 2 | N.A. | [56] |
| 22 | Duplex real-time qRT-PCR | ABI Prism 7000 (Applied Biosystems) | HAV | MS2 bacteriophage | MGB-TaqMan probe | [57] |
| 23 | Multiplex TaqMan RT-qPCR system | MX30005P (Stratagene) | HEV | FCV | TaqMan probe | [58] |
| 24 | Multiplex real time PCR | ABI 7300 (Applied Biosystems) | HBV genotypes | - | TaqMan probe | [59] |
| 25 | Real time PCR | N.A. | HBV genotypes | - | TaqMan probe | [60] |
| 26 | Multiplex real time PCR | Mx3005P (Stratagene) | HEV | FCV | TaqMan probe | [61] |
| 27 | Multiplex RT PCR assay | ABI Prism 7500 (Applied Biosystems) | HCV | PDV | MGB hybridization probe | [62] |
| 28 | Multiplex qPCR assay | N.A. | HBV | B19, HHV-8, EBV, CMV, VZV | N.A. | [63] |
| 29 | Multiplex qPCR | N.A. | HBV, HCV | HIV type-1 | SYBR Green I | [16] |
| 30 | Multiplex Real Time PCR | ABI 7500 (Applied Biosystems) | HBV mutants | - | LNA probes with SYBR Green I | [64] |
| 31 | Microarray multiplex assay | ABI Prism 7700 (Applied Biosystems) | HBV, HCV | HIV type-1 | Oligonucleotide array labeled with Cy5 and Cy3 | [65] |
| 32 | Real time multiplex PCR | N.A. | HAV | Entero and Adeno-viruses | Probes labeled with FAM, R6G, ROX, Cy5 | [66] |
| 33 | Multiplex real time RT-PCR | LightCycler (Roche) | HCV | HIV type-1 | SYBR Green | [67] |
| 34 | Real time multiplex PCR | icycler iQ (Bio-Rad) | HCV variants | - | TaqMan probes | [68] |
| 35 | Multiplex real-time RT PCR | ABI 7000 (Applied Biosystems) | HCV genotypes | - | Primer probes | [69] |
| 36 | Multiplex real-time qPCR | Mx4000 (Stratagene) | HBV, HCV | HIV type-1 | TaqMan probes | [70] |
| 37 | Automated multiplex PCR | ABI Prism 7700 (Applied Biosystems) | HBV, HCV | HIV type-1 | TaqMan probes | [71] |
The probe-based assays (e.g., TaqMan assays)[72] began to gain attention in mid-1990s with the development of quenched, fluorescent probes[73,74] and the commercialization of real-time thermal cyclers[26,75]. TaqMan (also known as Fluorogenic 5’ nuclease assay) probes contain two dyes, a reporter dye (e.g., 6-FAM) at the 5’ end and an acceptor dye at 3’ end, usually tetramethyl rhodamine (TAMRA). Recently, TAMRA fluorescent acceptor quencher dye was substituted with a non-fluorescent quencher, e.g., Black Hole Quencher[76]. The proximity of the quencher to the reporter in an intact probe quenches the fluorescence signal of the reporter dye through fluorescence resonance energy transfer. During amplification, the 5’ to 3’ nucleolytic activity of Taq polymerase cleaves the probe between the reporter and the quencher only if the probe hybridizes to the target. The probe fragments get displaced from the target, separating the reporter dye from the quencher dye, resulting in increased emission of fluorescence. Floating TaqMan probes are quenched due to random coiling in solution, where fluorophore- and quencher-labeled ends come together[77]. In contrast, Molecular Beacon probes are oligonucleotides designed in a way to induce hairpin formation and produce the quenched state[78]. TaqMan and Molecular Beacon probes have been shown to be less effective in discriminating closely related targets, as in single nucleotide polymorphisms, drug-resistant mutants, and somatic cancer mutations[79,80]. However, molecular beacons are useful in situations where it is not possible to isolate probe-target hybrids from an excess of the hybridization probes, for example in sealed tubes or within living cells[81]. An effective probe requires a careful balancing act based on melting temperature (Tm) and, therefore, repeated design and testing are needed to develop an effective probe[82]. Available evidence suggests that the use of TaqMan probes in qPCR assays for hepatitis viruses provide a good balancing act.
Today, several designing tools are available to guide the design of qPCR assays and analyze resulting quantitative data. Many of them are available online, and some are provided with qPCR instruments from different manufacturers[83]. Some important tools include Primer3, Primer-BLAST, PerlPrimer, FastPCR software, IDTSciTools, and UniPrime[84-89]. In addition, some of them have programming to analyze the secondary structure of primers. MP primer is used to design primers for multiplex PCR assays[90]. The Minimum Information for Publication of qPCR Experiments (MIQE) guidelines also provide clear instructions on the steps that are important for qPCR assay design[91]. Several research companies offer help for designing primers and probes with use of their designing tools. The studies reported in this article demonstrate a liberal use of tools without any specific need or choice affecting the results.
Various types of advanced technology-based equipment for multiplex qPCR assays with analysis of amplified products are available globally. A list of the instruments used with their brands in various studies conducted on qPCR for viral hepatitis is shown in Table 3. With increasing advances in technology, the number of filters and, accordingly, the resolution of the amplification curve during the PCR assay have also increased. Now it is possible to detect/discriminate more pathogens or allelic/mutational changes[92,93] in a single step multiplex assay. The choice of instrument is more a function of availability, without much difference in their analytical qualities. Multiplex qPCR assays developed for hepatitis viruses may use any brand, depending on a match between the number of component pathogens to be detected and the filters available for detection. Other features of equipment do not seem to affect the results.
For each pathogen used as a component in the multiplex assay, a carefully developed singleplex assay is needed. The design of primers and probes is dictated purely by the nature of the target template and clear guidelines for amplification. This exercise is followed in order to prepare a record of common amplification conditions noted in singleplex assays and for their application as such in multiplex assays. The multiplex protocol is reframed in a way to have minimum possible deviations from the working protocol of the singleplex assay. During the multiplex assay, the possibility of cross interaction/interference among different molecules is quite likely and may cause unsuccessful amplification. This interaction may or may not occur, but it has to be worked out cautiously in each multiplex assay.
There have been reports on singleplex as well as multiplex assays developed for detection of some hepatitis viruses and their genotypes[16] (Table 3). Such a study was conducted at our research center where a multiplex assay was developed for simultaneous detection of hepatitis virus A, B, C, and E[15]. These viruses are frequently prevalent in India, posing a serious problem, causing incidences of both sporadic and epidemic hepatitis from time to time[94,95]. The use of singleplex followed by the development of multiplex assay in these cases does not show many changes in the experimental protocol. This implies that the amplification protocol of individual viruses is not influenced during multiplex assays. We noted a clear amplification curve on the screen during multiplex assay that was the same exact pattern noted during singleplex assay[15].
Table 3 shows the global status of multiplex assays used for analyzing hepatitis viruses with or without other pathogens[41-71]. In all these assays, viral amplification by the simultaneous presence of other pathogenic genomes was indicated. An overall survey of the experimental designs reported in multiplex assays indicated that standard conditions of reverse transcription, denaturation, annealing, and extension temperature were followed without much deviation from the singleplex protocol.
The guidelines published by Bustin et al[91] in 2009 clearly defined the terms used and steps necessary to design the experiments for developing qPCR assay. Since 2009, many published reports in the area of viral hepatitis on multiplex qPCR were found to follow these guidelines and give interpretation of results referring to terminology and definitions outlined there. The guidelines state that multiplexing expands power of qPCR analysis but needs documentation for accurate quantification of multiple targets in a single tube assay.
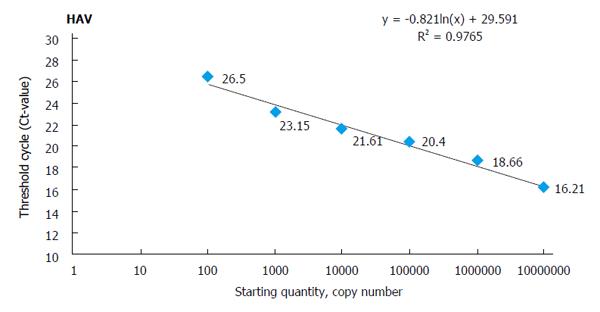
In order to generate a standard curve for each hepatitis virus, the standard control that includes the conserved region targeted for amplification/ detection is synthesized artificially and cloned into a suitable vector (e.g., pUC 57)[15] using cloning kits. These standards are used as a template for standardization of amplifications. The copy number of standard plasmids is calculated using their concentration and the size of linearized plasmids. Each standard template is added to PCR mix (Tris-HCl, KCl, MgCl2, four dNTPs, primers, and Taq DNA polymerase in a suitable concentration ratio), and PCR is performed under standardized conditions. For generation of the standard curve, a 10-fold serial dilution of each standard plasmid (101-108 copies/μL) is prepared and run in triplicate. At the end, data are analyzed by an automatic system that generates a standard curve[21]. The standard curves are used to quantify the amplification product and to assess the linear dynamic range using 10-fold dilution series of standard plasmid of each individual virus. One specimen standard plot is shown in Figure 1, which was prepared during development of quadruplex qPCR for hepatitis virus A, B, C, and E. Such plots are used to calculate copy number of individual template using correlation coefficient and Y-intercept value based on regression analysis.
Standard curve showing amplification plots of 10-fold serial dilution of HAV template using standard cloned plasmids. Such standard curves are generated from the amplification plots run in triplicate and show a linear dynamic range. The correlation coefficient and the slope of each standard plot are shown in the figure.
Using the standard curve prepared above, now it is possible to assess the sensitivity and determine the linear dynamic range of an individual virus. Moreover, observed Ct values may be used to calculate the exact copy number of virus in an unknown specimen[96,97]. Based on the data collected from various studies, including our study[15], it has been noticed that the linear dynamic range of each individual hepatitis virus usually falls in the range 101-108 copies/μL.
The specificity of qPCR assay is assessed by evaluating sera from healthy controls and patients with unrelated diseases negative for hepatitis markers by serology and all other NAT based techniques. Negative results from these sera and clear positive signals from serologically positive hepatitis sera demonstrate the high level of specificity of qPCR. To date, all studies on qPCR demonstrate the assay to be specific[15,22]. In reports on viral hepatitis, qPCR assays demonstrated high specificity with a very low chance of false positive results[19,71].
The multiplex qPCR assays were developed and used both for comparison as well as in combination with other molecular technologies to improve the sensitivity for detection of the viral genome[16,98]. Various other assay systems were also developed for simultaneous detection of HBV, HCV, and human immunodeficiency virus in addition to multiplex qPCR. The status of multiplex qPCR assay was assessed in comparison to other molecular techniques used for detection and genotyping of viruses, including hepatitis viruses. The other assay systems included flowcytometric microsphere based hybridization assay[99], transcription-mediated amplification (TMA)[100], and nucleic acid sequence based amplification (NASBA)[101]. Comparatively, TMA was reported to be an equally sensitive technique. However, when comparing qPCR with NASBA and TMA for the detection of hepatitis viruses, the level of sensitivity of TMA was found to be associated closely with qPCR[100]. Of course, qPCR assay was reported to be faster, more economic, and easier to perform compared to all other assays evaluated.
Multiplex qPCR assays are proving to be very good analytical and diagnostic procedures in medicine. Recently, these assays have been successfully used for both basic research and clinical applications[42,102]. Although the practice of doing separate assays for separate pathogens, including hepatitis viral markers, are still in place, the use of the multiplex assay is seen to be beneficial in terms of time and overall cost involved. Moreover, multiplex assays, when used for quantification of HCV- RNA, were found to resolve many problems with real time monitoring of the amplification process. In fact, in multiplex qPCR assays, real time PCR makes quantification of DNA and RNA of different organism more precisely and with better reproducibility because it depends on the threshold cycle value determined during the exponential phase of PCR rather than on end points[103]. In addition, these assays report a direct relationship between starting template copy number and the number of cycles required to get a positive signal. In this manner, real time qPCR appears to be a good option for laboratory diagnosis of viral hepatitis, both for screening as well as for the final diagnosis of suspected cases of viral hepatitis infections.
Based on the information compiled in the present review, there is an increasing trend/interest in the diagnostic area towards the development and use of multiplex qPCR assay for the simultaneous detection of hepatitis viruses or their subtypes in sera samples. Several studies have been conducted in last few years that clearly demonstrate the preferable use of qPCR over other techniques in the area of viral hepatitis. This technique has been used to detect hepatitis viruses in combination with various other viral and non-viral pathogens and reported to be a sensitive, fast, and cost-effective technique compared to other multi-step assay procedures. The use of multiplex qPCR in genotyping of hepatitis viral subtypes also provides great help in serotype detection. To date, multiplex qPCR has been successfully employed for the simultaneous detection of hepatitis virus A, B, C, D, and E and genotyping of their strains. It appears to be a good tool for screening blood donor samples in blood banks for hepatitis viruses. Moreover, a single step multiplex qPCR assay allows for an early diagnosis and timely treatment of patients with viral hepatitis. Several studies in this field are in progress, with more important information likely to be available until the next such update is necessary.
We appreciate the infrastructure provided by All India Institute of Medical Sciences, New Delhi, India, for conduct of this study.
P- Reviewer: da Silva NM, Pokorska-Spiewak M S- Editor: Qi Y L- Editor: Filipodia E- Editor: Ma S
| 1. | Oh IS, Park SH. Immune-mediated Liver Injury in Hepatitis B Virus Infection. Immune Netw. 2015;15:191-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 2. | Hadler S. Global impact of hepatitis A virus infection; changing patterns. Viral hepatitis and liver disease. Baltimore: Williams and Wilkins 1991; 4-20. |
| 3. | Custer B, Sullivan SD, Hazlet TK, Iloeje U, Veenstra DL, Kowdley KV. Global epidemiology of hepatitis B virus. J Clin Gastroenterol. 2004;38:S158-S168. [PubMed] |
| 4. | Park Y, Kim BS, Choi KH, Shin DH, Lee MJ, Cho Y, Kim HS. A novel multiplex real-time PCR assay for the concurrent detection of hepatitis A, B and C viruses in patients with acute hepatitis. PLoS One. 2012;7:e49106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-2128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9500] [Cited by in RCA: 9568] [Article Influence: 736.0] [Reference Citation Analysis (0)] |
| 6. | Tandon BN, Gupta H, Irshad M, Joshi YK, Chawla TC. Associated infection with non-A, non-B virus as possible cause of liver failure in Indian HBV carriers. Lancet. 1984;2:750-751. [PubMed] |
| 7. | Allain JP. Genomic screening for blood-borne viruses in transfusion settings. Clin Lab Haematol. 2000;22:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Paryan M, Forouzandeh MM, Kia V, Mohammadi-Yeganeh S, Abbasali RA, Mirab SS. Design and development of an in-house multiplex RT-PCR assay for simultaneous detection of HIV-1 and HCV in plasma samples. Iran J Microbiol. 2012;4:8-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Mine H, Emura H, Miyamoto M, Tomono T, Minegishi K, Murokawa H, Yamanaka R, Yoshikawa A, Nishioka K. High throughput screening of 16 million serologically negative blood donors for hepatitis B virus, hepatitis C virus and human immunodeficiency virus type-1 by nucleic acid amplification testing with specific and sensitive multiplex reagent in Japan. J Virol Methods. 2003;112:145-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Candotti D, Richetin A, Cant B, Temple J, Sims C, Reeves I, Barbara JA, Allain JP. Evaluation of a transcription-mediated amplification-based HCV and HIV-1 RNA duplex assay for screening individual blood donations: a comparison with a minipool testing system. Transfusion. 2003;43:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Heiat M, Ranjbar R, Alavian SM. Classical and modern approaches used for viral hepatitis diagnosis. Hepat Mon. 2014;14:e17632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Fukumoto H, Sato Y, Hasegawa H, Saeki H, Katano H. Development of a new real-time PCR system for simultaneous detection of bacteria and fungi in pathological samples. Int J Clin Exp Pathol. 2015;8:15479-15488. [PubMed] |
| 13. | Wong AA, Pabbaraju K, Wong S, Tellier R. Development of a multiplex real-time PCR for the simultaneous detection of herpes simplex and varicella zoster viruses in cerebrospinal fluid and lesion swab specimens. J Virol Methods. 2016;229:16-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27:95-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 850] [Cited by in RCA: 839] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 15. | Irshad M, Ansari MA, Irshad K, Lingaiah R. Novel single-step multiplex real-time polymerase chain reaction assay for simultaneous quantification of hepatitis virus A, B, C, and E in serum. J Gastroenterol Hepatol. 2013;28:1869-1876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Khodakov DA, Zakharova NV, Gryadunov DA, Filatov FP, Zasedatelev AS, Mikhailovich VM. An oligonucleotide microarray for multiplex real-time PCR identification of HIV-1, HBV, and HCV. Biotechniques. 2008;44:241-246, 248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, Schrag SJ, Taylor TH, Beall BW, Breiman RF. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol. 2011;49:2175-2182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 18. | Lin B, Vora GJ, Thach D, Walter E, Metzgar D, Tibbetts C, Stenger DA. Use of oligonucleotide microarrays for rapid detection and serotyping of acute respiratory disease-associated adenoviruses. J Clin Microbiol. 2004;42:3232-3239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Pripuzova N, Wang R, Tsai S, Li B, Hung GC, Ptak RG, Lo SC. Development of real-time PCR array for simultaneous detection of eight human blood-borne viral pathogens. PLoS One. 2012;7:e43246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Ito K, Shimizu N, Watanabe K, Saito T, Yoshioka Y, Sakane E, Tsunemine H, Akasaka H, Kodaka T, Takahashi T. Analysis of viral infection by multiplex polymerase chain reaction assays in patients with liver dysfunction. Intern Med. 2013;52:201-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Zhou L, Gong R, Lu X, Zhang Y, Tang J. Development of a Multiplex Real-Time PCR Assay for the Detection of Treponema pallidum, HCV, HIV-1, and HBV. Jpn J Infect Dis. 2015;68:481-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Qiu F, Cao J, Su Q, Yi Y, Bi S. Multiplex hydrolysis probe real-time PCR for simultaneous detection of hepatitis A virus and hepatitis E virus. Int J Mol Sci. 2014;15:9780-9788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Wang X, Seed B. High-throughput primer and probe design. In: Dorak MT, editor. Real-time PCR. Taylor and Francis Group 2006; 93-106. |
| 24. | Bustin SA, Nolan T. Primers and probes. A-Z of quantitative PCR. La Jolla, CA: International University Line 2004; 279-326. |
| 25. | Navarro E, Serrano-Heras G, Castaño MJ, Solera J. Real-time PCR detection chemistry. Clin Chim Acta. 2015;439:231-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 252] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 26. | Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4152] [Cited by in RCA: 3921] [Article Influence: 135.2] [Reference Citation Analysis (1)] |
| 27. | Kostrikis LG, Tyagi S, Mhlanga MM, Ho DD, Kramer FR. Spectral genotyping of human alleles. Science. 1998;279:1228-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 218] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 28. | Nazarenko IA, Bhatnagar SK, Hohman RJ. A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res. 1997;25:2516-2521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 29. | Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17:804-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 523] [Cited by in RCA: 457] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 30. | Isacsson J, Cao H, Ohlsson L, Nordgren S, Svanvik N, Westman G, Kubista M, Sjöback R, Sehlstedt U. Rapid and specific detection of PCR products using light-up probes. Mol Cell Probes. 2000;14:321-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (N Y). 1992;10:413-417. [PubMed] |
| 32. | Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130-131, 134-138. [PubMed] |
| 33. | Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1127] [Cited by in RCA: 1022] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 34. | Giglio S, Monis PT, Saint CP. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 2003;31:e136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 157] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 35. | Bengtsson M, Karlsson HJ, Westman G, Kubista M. A new minor groove binding asymmetric cyanine reporter dye for real-time PCR. Nucleic Acids Res. 2003;31:e45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 36. | Ishiguro T, Saitoh J, Yawata H, Yamagishi H, Iwasaki S, Mitoma Y. Homogeneous quantitative assay of hepatitis C virus RNA by polymerase chain reaction in the presence of a fluorescent intercalater. Anal Biochem. 1995;229:207-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 37. | Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 942] [Cited by in RCA: 861] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 38. | Monis PT, Giglio S, Saint CP. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal Biochem. 2005;340:24-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 209] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 39. | Giglio S, Monis PT, Saint CP. Legionella confirmation using real-time PCR and SYTO9 is an alternative to current methodology. Appl Environ Microbiol. 2005;71:8944-8948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Gudnason H, Dufva M, Bang DD, Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35:e127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 206] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 41. | Gruhn CC, Wedemeyer H, Bremer B, Heckmann M, Steimer M, Carman WF. Clinical evaluation of a novel Fast-track diagnostics multiplex real-time PCR assay for detection and quantification of Hepatitis E virus. J Clin Virol. 2015;70:S124-S125. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 42. | Kodani M, Mixson-Hayden T, Drobeniuc J, Kamili S. Rapid and sensitive approach to simultaneous detection of genomes of hepatitis A, B, C, D and E viruses. J Clin Virol. 2014;61:260-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 43. | Taranta A, Rogalska-Taranta M, Gutierrez R, Manns MP, Bock M, Wursthorn K. Rapid hepatitis B and hepatitis Delta virus RNA quantification from small-sized liver tissue samples. J Clin Virol. 2014;61:286-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 44. | Fuentes C, Guix S, Pérez-Rodriguez FJ, Fuster N, Carol M, Pintó RM, Bosch A. Standardized multiplex one-step qRT-PCR for hepatitis A virus, norovirus GI and GII quantification in bivalve mollusks and water. Food Microbiol. 2014;40:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 45. | Jia S, Wang F, Li F, Chang K, Yang S, Zhang K, Jiang W, Shang Y, Deng S, Chen M. Rapid detection of hepatitis B virus variants associated with lamivudine and adefovir resistance by multiplex ligation-dependent probe amplification combined with real-time PCR. J Clin Microbiol. 2014;52:460-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 46. | Zhang X, Li A, Shuai J, Dai Y, Zhu Z, Wu S, He Y. Validation of an internally controlled multiplex real time RT-PCR for detection and typing of HEV genotype 3 and 4. J Virol Methods. 2013;193:432-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Becker CE, Kretzmann NA, Mattos AA, Veiga AB. Melting curve analysis for the screening of hepatitis B virus genotypes A, D and F in patients from a general hospital in southern Brazil. Arq Gastroenterol. 2013;50:219-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 48. | Kang LH, Oh SH, Park JW, Won YJ, Ryu S, Paik SY. Simultaneous detection of waterborne viruses by multiplex real-time PCR. J Microbiol. 2013;51:671-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 49. | Marova AA, Oksanich AS, Kaira AN, Meskina ER, Medvedeva EA, Ivanova OE, Lukashev AN, Kyuregian KK, Kalinkina MA, Egorova OV. [Experience of application of multiplex qPCR for differential diagnostics of intestinal viral infections]. Zh Mikrobiol Epidemiol Immunobiol. 2012;39-45. [PubMed] |
| 50. | Ynk Y, Rahmathulla S, Madhavi C, Ramachandra VV, Vishnupriya S, Habeeb MA, Khaja MN. Simultaneous detection of Hepatitis B virus and Hepatitis C virus in human plasma using Taq-man chemistry. J Med Allied Sci. 2011;1:69-73. |
| 51. | Sun S, Meng S, Zhang R, Zhang K, Wang L, Li J. Development of a new duplex real-time polymerase chain reaction assay for hepatitis B viral DNA detection. Virol J. 2011;8:227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 52. | Lee YM, Chen YJ, Lee CM, Kuo LH, Wong WW, Chen YM. Detection of hepatitis C virus subtypes 6a, 6n, 6w and mixed infections using a modified multiplex real-time polymerase chain reaction protocol. J Formos Med Assoc. 2011;110:762-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 53. | Désiré N, Sanchis T, Ben Moussa F, Stitou H, Katlama C, Thibault V. [Development and validation of a specific method for relative HBV-genotype G (G-HBV) quantification in the context of co-infection with other genotypes]. Pathol Biol (Paris). 2011;59:e13-e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 54. | De Crignis E, Re MC, Cimatti L, Zecchi L, Gibellini D. HIV-1 and HCV detection in dried blood spots by SYBR Green multiplex real-time RT-PCR. J Virol Methods. 2010;165:51-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 55. | Meng S, Li J. A novel duplex real-time reverse transcriptase-polymerase chain reaction assay for the detection of hepatitis C viral RNA with armored RNA as internal control. Virol J. 2010;7:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 56. | Morales-Rayas R, Wolffs PF, Griffiths MW. Simultaneous separation and detection of hepatitis A virus and norovirus in produce. Int J Food Microbiol. 2010;139:48-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 57. | Blaise-Boisseau S, Hennechart-Collette C, Guillier L, Perelle S. Duplex real-time qRT-PCR for the detection of hepatitis A virus in water and raspberries using the MS2 bacteriophage as a process control. J Virol Methods. 2010;166:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 58. | Leblanc D, Poitras E, Gagné MJ, Ward P, Houde A. Hepatitis E virus load in swine organs and tissues at slaughterhouse determined by real-time RT-PCR. Int J Food Microbiol. 2010;139:206-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 59. | Malmström S, Berglin-Enquist I, Lindh M. Novel method for genotyping hepatitis B virus on the basis of TaqMan real-time PCR. J Clin Microbiol. 2010;48:1105-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 60. | Tanic N, Stanojevic B, Tanic N, Schaefer S, Niesters HG, Bozic M, Dimitrijevic B. Concurrent quantitation of the A and D genotypes of hepatitis B virus. J Virol Methods. 2009;161:265-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 61. | Ward P, Poitras E, Leblanc D, Letellier A, Brassard J, Plante D, Houde A. Comparative analysis of different TaqMan real-time RT-PCR assays for the detection of swine Hepatitis E virus and integration of Feline calicivirus as internal control. J Appl Microbiol. 2009;106:1360-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 62. | Clancy A, Crowley B, Niesters H, Herra C. The development of a qualitative real-time RT-PCR assay for the detection of hepatitis C virus. Eur J Clin Microbiol Infect Dis. 2008;27:1177-1182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 63. | Compston LI, Sarkobie F, Li C, Candotti D, Opare-Sem O, Allain JP. Multiplex real-time PCR for the detection and quantification of latent and persistent viral genomes in cellular or plasma blood fractions. J Virol Methods. 2008;151:47-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 64. | Sun Z, Zhou L, Zeng H, Chen Z, Zhu H. Multiplex locked nucleic acid probes for analysis of hepatitis B virus mutants using real-time PCR. Genomics. 2007;89:151-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 65. | Hsia CC, Chizhikov VE, Yang AX, Selvapandiyan A, Hewlett I, Duncan R, Puri RK, Nakhasi HL, Kaplan GG. Microarray multiplex assay for the simultaneous detection and discrimination of hepatitis B, hepatitis C, and human immunodeficiency type-1 viruses in human blood samples. Biochem Biophys Res Commun. 2007;356:1017-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 66. | Oksanich AS, Faĭzuloev EB, Nikonova AA, Kashirin VI, Lotte VD, Ivanova OE, Zverev VV. [Real-time multiplex PCR for rapid detection of enteroviruses, adenoviruses and hepatitis A virus in clinical specimens]. Zh Mikrobiol Epidemiol Immunobiol. 2007;65-70. [PubMed] |
| 67. | Gibellini D, Gardini F, Vitone F, Schiavone P, Furlini G, Re MC. Simultaneous detection of HCV and HIV-1 by SYBR Green real time multiplex RT-PCR technique in plasma samples. Mol Cell Probes. 2006;20:223-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 68. | Pugnale P, Latorre P, Rossi C, Crovatto K, Pazienza V, Gottardi AD, Negro F. Real-time multiplex PCR assay to quantify hepatitis C virus RNA in peripheral blood mononuclear cells. J Virol Methods. 2006;133:195-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 69. | Cook L, Sullivan K, Krantz EM, Bagabag A, Jerome KR. Multiplex real-time reverse transcription-PCR assay for determination of hepatitis C virus genotypes. J Clin Microbiol. 2006;44:4149-4156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 70. | Candotti D, Temple J, Owusu-Ofori S, Allain JP. Multiplex real-time quantitative RT-PCR assay for hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1. J Virol Methods. 2004;118:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 71. | Meng Q, Wong C, Rangachari A, Tamatsukuri S, Sasaki M, Fiss E, Cheng L, Ramankutty T, Clarke D, Yawata H. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2001;39:2937-2945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 72. | Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5’----3’ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276-7280. [PubMed] |
| 73. | Lee LG, Connell CR, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761-3766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 520] [Cited by in RCA: 475] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 74. | Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357-362. [PubMed] |
| 75. | Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995-1001. [PubMed] |
| 76. | Crisalli P, Kool ET. Multi-path quenchers: efficient quenching of common fluorophores. Bioconjug Chem. 2011;22:2345-2354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 77. | Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3216] [Cited by in RCA: 2888] [Article Influence: 99.6] [Reference Citation Analysis (0)] |
| 78. | Bonetta L. Prime time for real-time PCR. Nat Methods. 2005;2:305-312. [RCA] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 79. | Cheng J, Zhang Y, Li Q. Real-time PCR genotyping using displacing probes. Nucleic Acids Res. 2004;32:e61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 80. | Zhang DY, Chen SX, Yin P. Optimizing the specificity of nucleic acid hybridization. Nat Chem. 2012;4:208-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 314] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 81. | Tyagi S, Marras SAE, Vet JAM, Kramer FR. Molecular beacons: Hybridization probes for detection of nucleic acids in homogeneous solutions. Nonradioactive Analysis of Biomolecules. 2nd ed. Berlin: Springer-Verlag 2000; 606-616. |
| 82. | Murray JL, Hu P, Shafer DA. Seven novel probe systems for real-time PCR provide absolute single-base discrimination, higher signaling, and generic components. J Mol Diagn. 2014;16:627-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 83. | Bustin S, Bergkvist A, Nolan T. In silico tools for qPCR assay design and data analysis. Methods Mol Biol. 2011;760:283-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 84. | Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365-386. [PubMed] |
| 85. | Kalendar R, Lee D, Schulman AH. FastPCR software for PCR, in silico PCR, and oligonucleotide assembly and analysis. Methods Mol Biol. 2014;1116:271-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 86. | Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471-2472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 466] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 87. | Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163-W169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 293] [Cited by in RCA: 340] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 88. | Bekaert M, Teeling EC. UniPrime: a workflow-based platform for improved universal primer design. Nucleic Acids Res. 2008;36:e56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 89. | Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3185] [Cited by in RCA: 3897] [Article Influence: 299.8] [Reference Citation Analysis (0)] |
| 90. | Shen Z, Qu W, Wang W, Lu Y, Wu Y, Li Z, Hang X, Wang X, Zhao D, Zhang C. MPprimer: a program for reliable multiplex PCR primer design. BMC Bioinformatics. 2010;11:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 91. | Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9915] [Cited by in RCA: 11125] [Article Influence: 695.3] [Reference Citation Analysis (0)] |
| 92. | Balashov SV, Gardiner R, Park S, Perlin DS. Rapid, high-throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J Clin Microbiol. 2005;43:214-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 93. | Tuffaha MSA. Phenotypic and genotypic diagnosis of malignancies: An immunohistochemical and molecular approach. : Wiley-Blackwell 2008; . [DOI] [Full Text] |
| 94. | Acharya SK, Madan K, Dattagupta S, Panda SK. Viral hepatitis in India. Natl Med J India. 2006;19:203-217. [PubMed] |
| 95. | Irshad M, Acharya SK. Hepatitis D virus (HDV) infection in severe forms of liver diseases in north India. Eur J Gastroenterol Hepatol. 1996;8:995-998. [PubMed] |
| 96. | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16283] [Cited by in RCA: 19185] [Article Influence: 1128.5] [Reference Citation Analysis (0)] |
| 97. | Caraguel CG, Stryhn H, Gagné N, Dohoo IR, Hammell KL. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: analytical and epidemiologic approaches. J Vet Diagn Invest. 2011;23:2-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 158] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 98. | McCormick MK, Dockter J, Linnen JM, Kolk D, Wu Y, Giachetti C. Evaluation of a new molecular assay for detection of human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA. J Clin Virol. 2006;36:166-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 99. | Defoort JP, Martin M, Casano B, Prato S, Camilla C, Fert V. Simultaneous detection of multiplex-amplified human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA using a flow cytometer microsphere-based hybridization assay. J Clin Microbiol. 2000;38:1066-1071. [PubMed] |
| 100. | Stramer SL, Krysztof DE, Brodsky JP, Fickett TA, Reynolds B, Dodd RY, Kleinman SH. Comparative analysis of triplex nucleic acid test assays in United States blood donors. Transfusion. 2013;53:2525-2537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 101. | Deiman B, Jay C, Zintilini C, Vermeer S, van Strijp D, Venema F, van de Wiel P. Efficient amplification with NASBA of hepatitis B virus, herpes simplex virus and methicillin resistant Staphylococcus aureus DNA. J Virol Methods. 2008;151:283-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (1)] |
| 102. | Parker J, Fowler N, Walmsley ML, Schmidt T, Scharrer J, Kowaleski J, Grimes T, Hoyos S, Chen J. Analytical Sensitivity Comparison between Singleplex Real-Time PCR and a Multiplex PCR Platform for Detecting Respiratory Viruses. PLoS One. 2015;10:e0143164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 103. | Pabinger S, Rödiger S, Kriegner A, Vierlinger K, Weinhäusel A. A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol Detect Quantif. 2014;1:23-33. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |