Published online Apr 21, 2016. doi: 10.3748/wjg.v22.i15.3978
Peer-review started: December 10, 2015
First decision: December 30, 2015
Revised: January 14, 2016
Accepted: February 20, 2016
Article in press: February 22, 2016
Published online: April 21, 2016
Processing time: 79 Days and 19.1 Hours
AIM: To determine if mir-30d inhibits the autophagy response to Helicobacter pylori (H. pylori) invasion and increases H. pylori intracellular survival.
METHODS: The expression of mir-30d was detected by quantitative polymerase chain reaction (PCR), and autophagy level was examined by transmission electron microscopy, western blot, and GFP-LC3 puncta assay in human AGS cells and GES-1 cells. Luciferase reporter assay was applied to confirm the specificity of mir-30d regulation on the expression of several core molecules involved in autophagy pathway. The expression of multiple core proteins were analyzed at both the mRNA and protein level, and the intracellular survival of H. pylori after different treatments was detected by gentamicin protection assay.
RESULTS: Autophagy level was increased in AGS and GES-1 cells in response to H. pylori infection, which was accompanied by upregulation of mir-30d expression (P < 0.05, vs no H. pylori infection). In the two gastric epithelial cell lines, mimic mir-30d was found to repress the autophagy process, whereas mir-30d inhibitor increased autophagy response to H. pylori invasion. mir-30d mimic decreased the luciferase activity of wild type reporter plasmids carrying the 3′ untranslated region (UTR) of all five tested genes (ATG2B, ATG5, ATG12, BECN1, and BNIP3L), whereas it had no effect on the mutant reporter plasmids. These five genes are core genes of autophagy pathway, and their expression was reduced significantly after mir-30d mimic transfection (P < 0.05, vs control cells without mir-30d mimic treatment). Mir-30d mimic transfection and direct inhibition of autophagy increased the intracellular survival of H. pylori in AGS cells.
CONCLUSION: Mir-30d increases intracellular survival of H. pylori in gastric epithelial cells through inhibition of multiple core proteins in the autophagy pathway.
Core tip: In this study, we tested a hypothesis that mir-30d could repress autophagy in response to Helicobacter pylori (H. pylori) invasion by directly targeting multiple core genes of the autophagy pathway, including ATG2B, ATG5, ATG12, BECN1 and BNIP3L in gastric epithelial cells. Inhibition of autophagy increased the intracellular survival of H. pylori in AGS cells, and the repression of autophagy by mir-30d may help the intracellular H. pylori to evade autophagic clearance. These findings provide a novel mechanism for elucidating persistent H. pylori infection and provide a promising target for gastric cancer prevention.
- Citation: Yang XJ, Si RH, Liang YH, Ma BQ, Jiang ZB, Wang B, Gao P. Mir-30d increases intracellular survival of Helicobacter pylori through inhibition of autophagy pathway. World J Gastroenterol 2016; 22(15): 3978-3991
- URL: https://www.wjgnet.com/1007-9327/full/v22/i15/3978.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i15.3978
Gastric cancer is the second leading cause of cancer-related death in the world, and almost two-thirds of the cases occur in Asian countries, especially China and Japan[1,2]. The prognosis of gastric cancer is generally rather poor, and, therefore, prevention is a better choice than cure for patients with gastric cancer.
Helicobacter pylori (H. pylori) is a class I carcinogen, appointed by the International Agency for Research on Cancer in 1994 due to its strong correlation with gastric cancer in humans[3]. One reason for H.pylori’s high resistance to biomedical therapy may be its residence inside host cells[4,5]. Although regarded generally as an extracellular pathogen, the intracellular survival of H. pylori in both gastric epithelial cells and immunocytes allows it to escape from the host immune response and resist destruction from membrane-impermeable antibiotics[6], leading to persistence in the stomach. Up to now, the detailed molecular mechanisms by which H. pylori escape host cell machineries for intracellular survival are remains obscure.
Autophagy is present in mammalian cells at a low basal level. As an evolutionarily conserved cellular activity, it delivers organelles and cellular materials to the lysosome for degradation within double-membraned vacuoles, called autophagosomes[7,8]. Autophagy is considered one of the innate immune effectors against intracellular bacterial infection (e.g., Streptococcus pyogenes)[9,10]. Autophagic proteins act as cytosolic sensors to rapidly launch the autophagic pathway when the innate defense system recognizes invasive bacterial pathogens[11]. However, some intracellular pathogens use highly evolved machinery to deceive autophagic recognition, manipulate the autophagic pathway, and reconstruct the autophagosomal compartment for their own survival[12]. Over the last decade, many studies have reported that H. pylori infection can induce macroautophagy and that H. pylori may evade the autophagic machinery through downregulating the expression of autophagic proteins[6,13-15].
Recently, interest in the study of mir-30 has been growing. The mir-30 microRNA family is extensively expressed in multiple tissues and cell types[16,17]. It has been shown to be involved in a wide range of physiological activities in normal tissues and cancer tissues, including cell differentiation, development, proliferation, apoptosis, senescence, and cancer metastasis[18-22]. mir-30 expression is amplified in more than 30% of human epithelial tumors, including gastric cancer[15,23,24]. There is increasing evidence that mir-30 is a novel oncomir and understanding the mechanism underlying mir-30 function in tumorigenesis would be helpful for developing targeted cancer therapy against this miRNA family. Previously, we demonstrated that mir-30d regulated cellular autophagy by directly targeting multiple genes in the autophagy pathway[25]. Consistent with our finding, another mir-30 family member, mir-30a was found to regulate autophagy via repressing BECN1 expression in tumor cells[26,27]. In addition, compromised autophagy by mir-30b upregulation might benefit the intracellular survival of H. pylori[15]. These results shed light on the potential role of miRNAs on autophagy regulation during gastric tumorigenesis.
Here, we continue our investigation on mir-30d and H.pylori and suggest that mir-30d downregulated the expression of key autophagy genes, including ATG2B, ATG5, ATG12, BECN1 and BNIP3L, and inhibited the autophagy response to H.pylori invasion of gastric epithelial cells, resulting in increased H. pylori intracellular survival.
The green fluorescent protein (GFP)-LC3 and psiCHECK-2 vectors were purchased from Addgene (Cambridge, MA, United States) and Promega (Madison, WI, United States), respectively.
Antibodies against light chain 3 B (LC3B), autophagy related (ATG)2B, ATG5, ATG12, beclin 1 (BECN1), and BNip3-like protein (BNIP3L) were obtained from Cell Signaling Technology (CST, Beverly, MA, United States). 3-methyladenine (3-MA, M9281) and rapamycin (Rapa, R8781) were purchased from Sigma (St. Louis, MO, United States).
AGS cells (a human gastric adenocarcinoma cell-line) were obtained from American Type Culture Collection (Manassas, VA, United States) and cultured in F12 media (Gibco, Carlsbad, CA, United States). Human gastric mucosal epithelial cell line GES-1 (Purchased from Cell bank of Xiangya Medical School, Central South University, Hunan, China) was cultured in Roswell Park Memorial Institute (RPMI)1640 (Cellgro, Manassas, VA, United States) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, United States), and 100 U/mL penicillin/streptomycin (Gibco, 15140-122) in a humidified incubator at 37 °C with 5% CO2. For autophagy induction, cells were either treated with 200 nM rapamycin (Sigma) supplemented in complete medium or serum starved with Hank’s buffer (Stemcell Technologies, Vancouver, Canada), both at 37 °C for 4 h. The wild-type H. pylori strain 26695(700392) was obtained from American Type Culture Collection and cultured as previously described.
Total RNA was extracted using TRIzol reagent (Invitrogen) and reverse-transcribed using a high capacity RNA-cDNA kit (Applied Biosystems, Carlsbad, CA, United States). cDNA was quantified on an ABI Prism 7900 sequence detection system (Applied Biosystems). Polymerase chain reaction was performed using Power SYBR Green polymerase chain reaction (PCR) master mix (Applied Biosystems).
Cells were lysed in mammalian protein extraction reagent (Pierce, Rockford, IL, United States) with protease inhibitor cocktail (Sigma). After centrifugation at 5000 g for 15 min at 4 °C, the protein concentration was measured with bicinchoninic acid (BCA) protein assay kit (Pierce, 23227). Fifteen micrograms of total protein were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, United States). Membranes were blocked in 5% non-fat milk (Bio-Rad, Hercules, CA, United States) and then incubated with the following primary antibodies: anti-ATG2B, anti-ATG5, anti-ATG12, anti-BECN1, anti-BNIP3L, and anti-LC3B. After incubation with a secondary antibody conjugated with horseradish peroxidase (HRP) (Amersham Biosciences, Chalfont St. Giles, United Kingdom) together with an HRP-conjugated primary antibody for b-actin (Sigma), immunoreactive proteins were visualized using the LumiGLO chemiluminescent substrate (Cell Signaling). Densitometric analyses were performed using Scion Image software.
Cells were seeded onto six-well plates and transfected with a GFP-LC3 expression plasmid at approximately 45%-55% confluence using the Lipofectamine RNAiMAX transfection reagent (Invitrogen). After 24 h, the cells were infected with or without H. pylori for 24 h. For observation, cells were fixed with 4% formaldehyde for 15 min and then washed twice in cold phosphate-buffered saline (PBS). Cell nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI).
Pre-mir miRNA precursor and control oligos were purchased from Ambion (Foster City, CA, United States), and miRCURY LNA miRNA inhibitors and control oligos were purchased from Exiqon (Vedbaek, Denmark). Transfections were performed using the Lipofectamine RNAiMAX transfection reagent (Invitrogen) and then cells were incubated in the medium containing the transfection mixture for 24-48 h.
Cells were plated on a 24-well plate 24 h before transfection at 50% confluence. miRNA mimics (30 nmol/L, Ambion) were transfected using Lipofectamine RNAiMAX. Twenty-four hours post-transfection, 0.125 µg of reporter vector was transfected using FuGENE6 transfection reagent (Roche, Basel, Switzerland). Forty-eight hours after reporter vector transfection, cells were harvested, and reporter assays were performed using a dual luciferase reporter assay system (Promega).
AGS and GES-1 cells were digested with 0.25% trypsinase and rinsed twice with PBS. They were then collected, fixed in 2% paraformaldehyde, 0.1% glutaraldehyde in 0.1 mol/L sodium cacodylate for 2 h, postfixed with 1% OsO4 for 1.5 h, washed, and stained for 1 h in 3% aqueous uranyl acetate. The samples were then washed again, dehydrated with graded alcohol, embedded in Epon-Araldite resin (Canemco, Quebec, Canada), and then cut into 0.05 μm thick sections on an ultramicrotome. The cells were observed under JEM-1230 (Jeol Ltd., Tokyo, Japan) electron microscopy.
After bacteria infection, the GES-1 and bacterium co-culture was washed three times with 1 mL of warm PBS per well to remove nonadherent bacteria. To determine the colony-forming unit (CFU) count corresponding to intracellular bacteria, the GES-1 cell monolayers were treated with gentamicin (100 mg/mL; Sigma, G1272) at 37 °C in 5% CO2 for 1 h, washed three times with warm PBS, and then incubated with 1 mL of 0.5% saponin (Sigma, 47036) in PBS at 37 °C for 15 min. The treated monolayers were resuspended thoroughly, diluted, and plated on serum agar. To determine the total CFU corresponding to host associated bacteria, the infected monolayers were incubated with 1 mL of 0.5% saponin in PBS at 37 °C for 15 min without prior treatment with gentamicin. The resulting suspensions were diluted and plated as described above. Both the CFU of intracellular bacteria and the total CFU of cell-associated bacteria were given as CFU per well of GES-1 cells.
miRNA and mRNA expression microarray data were retrieved from a public accessible database, Cell Miner. http://discover.nci.nih.gov/cellminer/. Gene set enrichment analysis (GSEA) algorithm was used to identify the pathways that were significantly enriched between mir-30d low and high tumor cells. http://www. broadinstitute.org/gsea/index.jsp. TargetScan algorithm was used to predict mir-30d targets. http://www.targetscan.org.
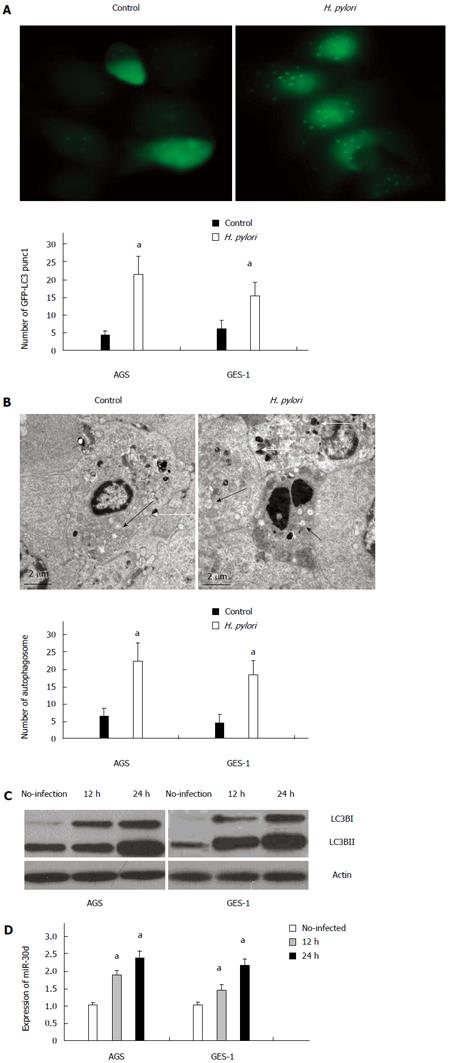
To measure autophagy induction during H. pylori infection, a GFP-LC3 fusion protein expression reporter was used in the assay. Upon autophagy induction, LC3-I, one form of the microtubule-associated protein light chain 3 (LC3), converts to another form LC3-II. LC3-II is accumulated in the autophagosomal membranes, and its amount is correlates to the number of autophagosomes and may serve as a marker for autophagosome formation. Autophagy induction was evaluated by measuring the quantity of GFP-LC3 puncta formed in the tested cell. AGS and GES-1 cells were transfected with GFP-LC3 vector and infected with or without H. pylori for autophagy analysis. Under fluorescence microscopy observation (Figure 1A), the GFP-LC3 puncta was significantly increased in H. pylori infected AGS and GES-1 cells (compared to the control cells without infection) after 24 h infection. This finding indicated that H. pylori infection may induce LC3-II production and autophagosome formation.
Meanwhile, a typical autophagosome, double-limiting membrane, was detectable in the autophagosome (black arrowheads) and autophagolysosome (white arrowheads) examined by transmission electron microscopy (TEM) (Figure 1B). Ultrastructural image analysis showed the presence of double-membrane autophagic vesicles containing H. pylori in the cytoplasm of AGS cells. The number of autophagic vacuoles (AV), including autophagosomes and autophagolysosomes in H. pylori infected AGS cells, was increased (Figure 1B). Similar results were obtained from GES-1 cells (Figure 1B).
To further confirm this finding, western blots for LC3B protein were applied to analyze the conversion of LC3B-I to LC3B-II. At 12 h and 24 h after H. pylori infection, LC3B-II protein level was significantly increased in AGS cells when compared with non-infected cells. A similar pattern was observed in GES-1 cells as well (Figure 1C). The expression of mir-30d in H. pylori infected cells was analyzed by quantitative real-time PCR, and the results showed that the expression of mir-30d was obviously increased at 12 h and 24 h after being infected with H. pylori in both AGS and GES-1 cell lines (P < 0.05, H. pylori infected cells versus without H. pylori infected cells) (Figure 1D).
Taken together, these data demonstrate that H.pylori infection increased the conversion of LC3B-I to LC3B-II (hence higher autophagosome formation), introduced a complete autophagic response, and upregulated mir-30d expression in AGS and GES-1 cell lines.
Transfection of mir-30d mimic in AGS and GES-1 cell lines downregulates autophagy after H. pylori infection
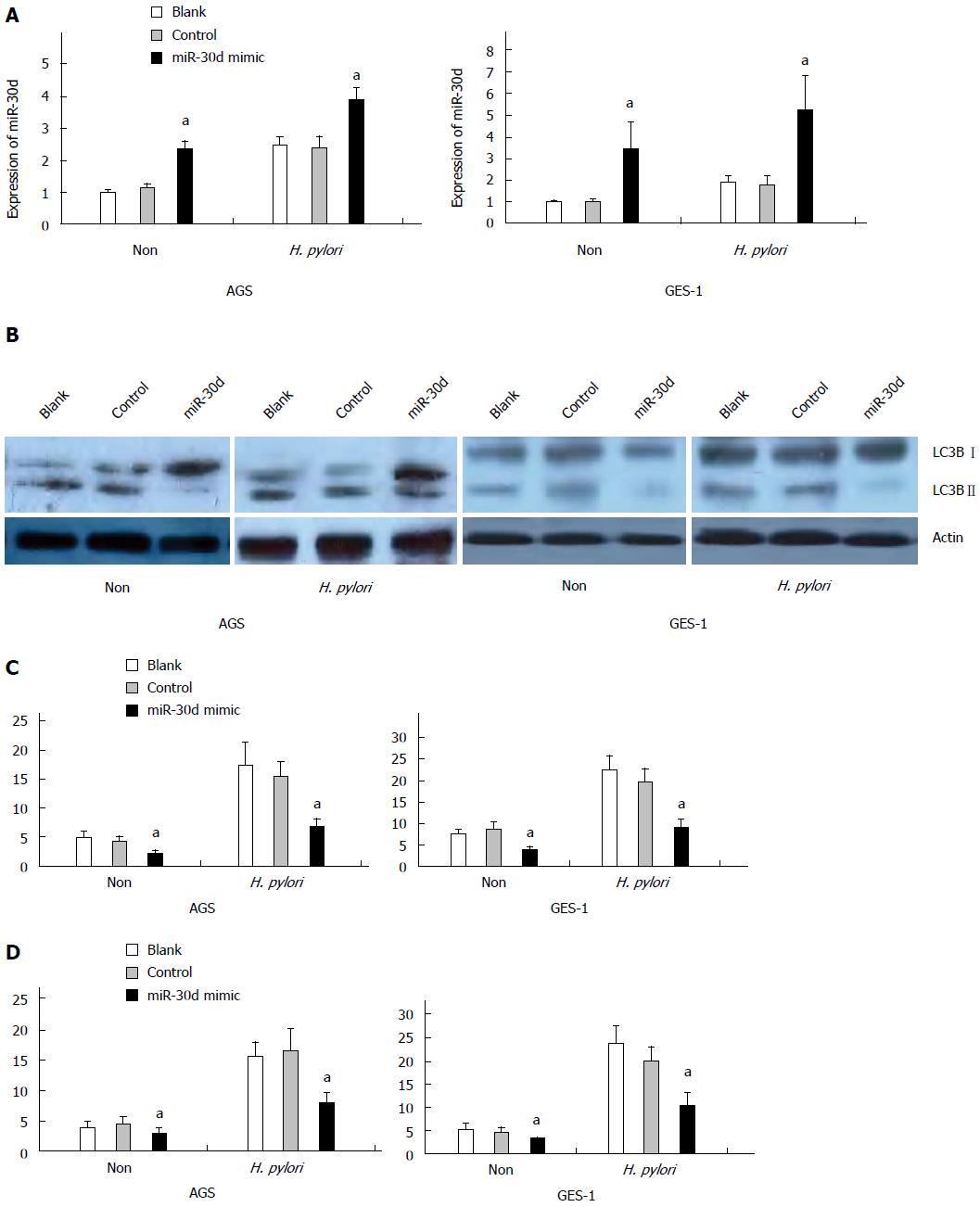
To determine whether mir-30d has a role in the negative regulation of autophagy during H. pylori infection, a mir-30d mimic was transfected into AGS and GES-1 cell lines for 24 h and then infected with H. pylori. The effect of the mir-30d mimic on autophagy was examined in both cell lines by western blotting, GFP-LC3 puncta assay, and TEM at 24 h post-H. pylori infection. Figure 2A showed that the expression of mir-30d was significantly increased in AGS cell lines either with or without H. pylori infection at 48 h after mir-30d mimic transfection (P < 0.05, mimics vs control). The same results were also found in GES-1 cells (P < 0.05 mimic transfected cells vs no mimic transfected control cells).
The results of western blotting revealed that autophagy was enhanced, as evidenced by increased LC3B-II expression, in both two cell lines during H. pylori infection, but transfection of mir-30d mimic significantly downregulated autophagy activity (i.e., attenuated LC3B-II conversion, Figure 2B).
In the GFP-LC3 puncta assay, GFP-LC3 plasmid and mir-30d mimic/control mimic were co-transfected into AGS and GES-1 cells using the lipofectamine RNAiMAX transfection reagent for 24 h and then infected with H. pylori. The treated cells were imaged under confocal laser-scanning microscope 24 h after H. pylori infection. The results showed that GFP-LC3 puncta were significantly increased in AGS and GES-1 cells infected with H. pylori compared to non- infected cells at 24 h after H. pylori infection, but transfection of mir-30d mimic significantly decreased GFP-LC3 positive puncta in AGS and GES-1 cells (P < 0.05, mimics vs control; Figure 2C).
Meanwhile, autophagosome and autophagolysosome were examined by TEM. The number of autophagic vacuoles (AV), including autophagosomes and autophagolysosomes, was increased in H. pylori infected AGS and GES-1 cells compared to non-infected cells at 24 h after H. pylori infection. However, transfection of mir-30d mimic significantly decreased autophagic vacuoles in both cell types (P < 0.05, mimics vs control; Figure 2D).
Transfection of mir-30d inhibitor in AGS and GES-1 cell lines upregulates autophagy after H. pylori infection
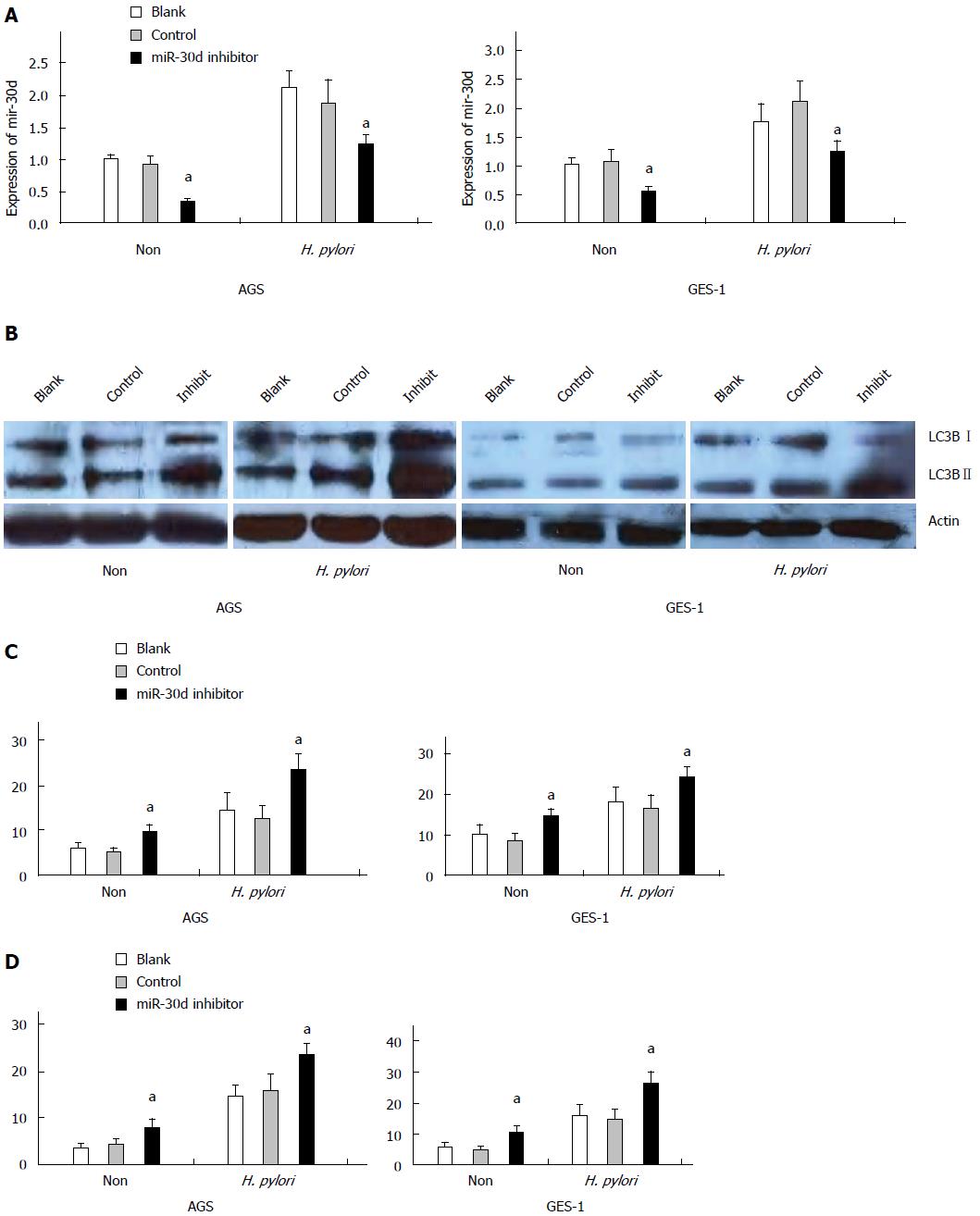
For loss of function experiments, endogenous mir-30d expression was blocked by mir-30d inhibitor in both cell lines and then the cells were infected with H. pylori. The effects of blocked mir-30d expression on autophagy in both cells lines were examined by western blotting, GFP-LC3 puncta assay, and TEM at 24 h after H. pylori infection.
In Figure 3A, the expression of mir-30d was decreased obviously in AGS cell lines with or without H. pylori infection at 48 h after mir-30d inhibitor transfection (P < 0.05, vs control oligos transfected cells). A similar result was found in GES-1 cells (P < 0.05, oligos transfected cells vs control cells). Western blotting showed that autophagy was enhanced (increased LC3B-II expression) in both cell lines during H. pylori infection. After transfection of mir-30d inhibitor into both cell lines, this process was further increased significantly (increased LC3B-II conversion; Figure 3B).
Co-transfection of GFP-LC3 plasmid and mir-30d inhibitor/control oligos into AGS and GES-1 cells was done using the lipofectamine RNAi MAX transfection reagent for 24 h and then the cells were infected with H. pylori. Cells were imaged under confocal laser-scanning microscope 24 h later. The results showed that GFP-LC3 puncta significantly increased in AGS and GES-1 cells infected with H. pylori as compared with non-infected cells. Nevertheless, mir-30d inhibitor significantly increased GFP-LC3 positive puncta in both cell types (P < 0.05, oligos transfected cells vs control cells, Figure 3C).
TEM assay also showed that there was an increased number of autophagic vacuoles (AV) in H. pylori infected AGS and GES-1 cells in contrast to non-infected cells at 24 h after H. pylori infection. After transfection of mir-30d inhibitor into the above cells, autophagic vacuoles were increased in both cell types (P < 0.05, oligos transfected cells vs control; Figure 3D).
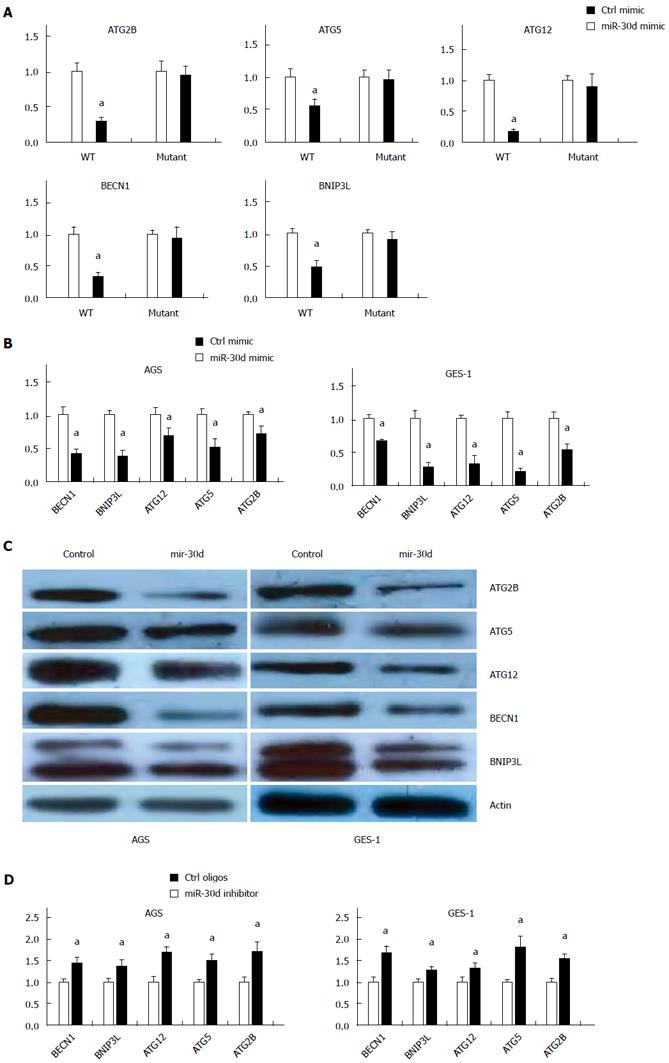
Previously, it was found that mir-30d inhibited the autophagy process in ovarian cancer and breast cancer cell lines by directly targeting multiple genes of the autophagy pathway, including BECN1, BNIP3L, ATG12, ATG5 and ATG2[25]. To test the effect of mir-30d inhibition on gastric epithelial cells infected with H. pylori, we prepared reporter plasmids containing wild type or mutant mir-30d binding sites from 3′untranslated region (UTR) of target genes (ATG2B, ATG5, ATG12, BECN1 and BNIP3L) for luciferase activity assay. Co-transfection of luciferase reporter plasmids and mir-30d mimic or control oligos showed that mir-30d potently decreased the luciferase activity of wild type reporter plasmids that represented all five target genes examined (ATG2B, ATG5, ATG12, BECN1, and BNIP3L), whereas it had no effect on the mutant reporter plasmids (Figure 4A). Perhaps mir-30d suppressed autophagy pathway gene expression by binding to its binding site within the 3′UTR of the target genes in a sequence-specific manner.
To further validate the repression of mir-30d on targeted autophagic genes in the autophagy pathway, mir-30d mimics were transfected in AGS and GES-1 cells, and target gene expression was analyzed with qRT-PCR. The mRNA levels of ATG2B, ATG5, ATG12, BECN1, and BNIP3L were remarkably suppressed by mir-30d mimic transfection compared with mimic control transfected in both AGS and GES-1 cells (Figure 4B). Similar results were obtained using western blots to detect the protein levels of these mir-30d potential targets in the above mir-30d mimic or control mimic treated cells. The protein levels for ATG2B, ATG5, ATG12, BECN1, and BNIP3L were reduced by mir-30d mimic transfection (Figure 4C). These results suggested that mir-30d regulated these autophagic genes at both the mRNA and protein level.
A loss of function experiment was applied with the mir-30d inhibitor. When endogenous mir-30d expression was blocked in the above cell lines (AGS and GES-1), the mRNA levels of ATG2B, ATG5, ATG12, BECN1, and BNIP3L were detected through qRT-PCR. In both cell lines (Figure 4D), the mRNA levels of the above genes were significantly increased by mir-30d inhibitor compared to control oligos.
Mir-30d increases intracellular survival of H. pylori in AGS cells through inhibition of autophagy
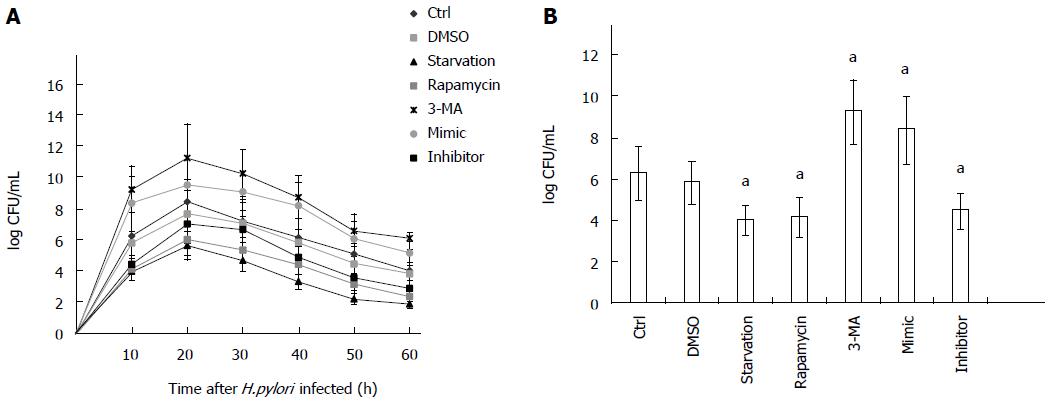
H. pylori invasion of gastric epithelial cells has been reported previously[24,26-28]. To evaluate the number of live internalized H. pylori cells, AGS cells were pretreated with PBS (as control), DMSO, autophagy inhibitor (3-MA), autophagy activators (starvation or rapamycin), mir-30d mimic, and mir-30d inhibitor, respectively, and then, a gentamicin protection assay was performed. The number of H. pylori CFU was increased approximately 10-fold 24 h after infection compared with 3 h of infection in all examined groups, indicating that internalized H. pylori underwent replication. Subsequently, the number of CFU decreased after 24 h in all groups (Figure 5A). However, compared with control (PBS) and DMSO groups, the number of CFU was obviously higher in mir-30d mimic and autophagy inhibitor 3-MA groups and lower in mir-30d inhibitor and autophagy activator (starvation or rapamycin) groups at all time points during a 60 h experiment (Figure 5A). The results from different treatments 24 h after infection with H. pylori are plotted in Figure 5B. These findings suggest that inhibition of autophagy increased the intracellular survival of H. pylori in AGS cells.
H. pylori is a common phenomenon worldwide, reaching nearly one-half of the world’s population. Chronic H. pylori infection is etiologically linked to gastric adenocarcinoma, especially non-cardia type (63% of all stomach cancer or 25% of cancers are associated with infectious etiology)[29]. These discoveries highlight the importance of basic research and clinical research on H. pylori infection and treatment. As resistance to the current proton pump inhibitor-based triple regimens or second-line therapies for the eradication of H. pylori continue to grow, so will the need to search for novel approaches.
It is known that varied autophagy is related to persistent H. pylori infection. Chu and colleagues[6] found that rapamycin, an inducer of autophagy, increased the clearance of H. pylori. However, they also found that many coccoid forms of H. pylori occurred on the membrane of the infected AGS cells. Autophagic vesicles were induced and their maturation was arrested with rapamycin[7], but it was not clear if H. pylori strains were killed inside the autophagic vesicles, as Amano et al[30] had indicated previously.
The role of autophagy in cancer development is important. Autophagy may be tumor-suppressing during the early stages of tumorigenesis, as reduced expression of autophagy proteins was shown to contribute to the development or progression of human breast and other cancers[31-33]. Sometimes, however, autophagy promotes cancer development[25]. In this case, downregulation of autophagy may benefit the intracellular survival of H. pylori, and induction of autophagy may be beneficial indirectly for cancer prevention.
Recently, miRNAs were demonstrated to play a crucial role in autophagy regulation, such as mir-30a, mir-30b, mir-17/20/93/106, mir-204, and mir-10b[15,26,34-36]. Tang et al[15] found that compromised autophagy by mir-30b led to a failure to clear intracellular H. pylori, resulting in persistent H. pylori infection and proliferation in the host cells. Kobayashi et al[37] suggested that mir-30d is a prognostic maker for prostate cancer. Our previous study showed that mir-30d regulated the autophagy process by directly targeting multiple autophagic genes in the autophagy pathway[25].
In this study, we confirmed a hypothesis that mir-30d could inhibit autophagy in gastric epithelial cells induced by H. pylori invasion by down-regulating autophagy-related gene expression, resulting in increased H. pylori intracellular survival. The results obtained from GFP-LC3 puncta assay, TEM, and western blot demonstrated that autophagy could be induced in AGS and GES-1 cells in response to H. pylori infection. The expression of mir-30d was upregulated in both cells after H. pylori infection in our experiments. This event appeared to be unique to H. pylori infection, but it must be repeated with other pathogens to demonstrate its specificity. Study on mir-30b found that infection with other pathogens (E. coli DH5α and O157:H7) or autophagy modulators (e.g., rapamycin and 3-methyladenine) had no effect on mir-30b expression[15]. We found that autophagy was upregulated in both cell lines after H. pylori infection, but upregulation of mir-30d significantly inhibited this process. In contrast, when mir-30d expression was blocked by mir-30d inhibitor, autophagy was obviously increased by downregulation of mir-30d. Mir-30d also repressed the autophagy process by directly targeting multiple core genes (ATG2B, ATG5, ATG12, BECN1, and BNIP3L). A gentamicin protection assay indicated that inhibition of autophagy increased the intracellular survival of H. pylori in AGS cells.
Although overexpression of mir-30d could decrease autophagy by inhibiting the expression of multiple core genes of the autophagy pathway in gastric epithelial cells, the regulation event might have happened after H. pylori-induced autophagy. Moreover, downregulation of autophagy by mir-30d may not be sufficient to block the autophagy induced by H. pylori. In assayed AGS and GES-1 cells with exogenously added mir-30d mimic, autophagy in H. pylori infection was more than that in uninfected cells (Figure 2B, C and D). Given the complexity of H. pylori infection in vivo, other factors may also contribute to autophagy inhibition. As one of the factors, overexpression of mir-30d may slightly continue to inhibit autophagy pathway for a long time, leading to subversion of host autophagic responses for their survival or growth.
Based on the above results, we concluded that repression of autophagy by mir-30d may help intracellular H. pylori evade autophagic clearance through targeting ATG2B, ATG5, ATG12, BECN1, and BNIP3L. These findings provide a novel molecular mechanism for persistent H. pylori occupancy. Although much remains to be studied on the regulation of autophagy in gastric cancer, the current study provides a promising target for gastric cancer prevention. We suggest that enhanced autophagy by mir-30d inhibition may be protective against H. pylori-related gastric cancer.
Preventive measures for gastric cancer must include tertiary prevention and effective treatment of H. pylori infections. The long-term decline in gastric cancer mortality in developed countries has resulted, in part, from interrupting H. pylori transmission through provision of improved basic sanitation, housing, and socioeconomic status. However, secondary prevention may be attempted where simple diagnostic tests, follow-up treatment (urea breath test), and effective, short-term eradication treatment are available to mitigate individual risk[38].
In summary, our report indicates a novel molecular mechanism for the inhibition of autophagy by mir-30d by increasing the intracellular survival of H. pylori. Although a detailed mechanism for H. pylori persistence remains to be elucidated, the present study establishes a basis that will be helpful for future evaluation of mir-30d in H. pylori infections.
Helicobacter pylori (H. pylori) was designated as a class I carcinogen by the International Agency for Research on Cancer in 1994 due to its strong correlation with gastric cancer in humans. One possible hypothesis for the relatively high resistance to therapy may be the ability of H. pylori to reside inside host cells.
Over the last decade, several research groups have independently reported that infection by H. pylori can induce macroautophagy. However, H. pylori has been reported to evade the autophagic machinery by downregulating the expression of autophagic proteins.
In this study, the authors confirmed that mir-30d could represses autophagy in response to H. pylori invasion by directly targeting multiple core genes of the autophagy pathway in gastric epithelial cells, including ATG2B, ATG5, ATG12, BECN1 and BNIP3L. Inhibition of autophagy increased the intracellular survival of H. pylori in AGS cells.
These findings may provide a novel mechanism for elucidating persistent H. pylori infection, and it appears to provide a promising target for gastric cancer prevention. Although the mechanism of H. pylori infection persistence remains to be fully determined, the present study provides the basis for future evaluations of mir-30d in H. pylori infections.
Autophagy, which is present in cells at a low level basally, is an evolutionarily conserved process for delivering cellular materials and organelles to lysosome for degradation within double-membraned vacuoles, called autophagosomes. Autophagy is also regarded as one of the innate immunity effectors against intracellular bacterial infection.
This is a very good study on mir-30d and H. pylori in gastric epithelial cells. Mir-30d was shown to inhibit multiple core proteins in the autophagy pathway. The link between mir-30d in gastric cancer and H. pylori is novel, as no other study has indicated previously this relationship in the literature.
P- Reviewer: Kocazeybek B, Romo-Gonzalez C S- Editor: Qi Y L- Editor: Filipodia E- Editor: Liu XM
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2651] [Cited by in RCA: 2598] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 2. | Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 3. | Vogiatzi P, Cassone M, Luzzi I, Lucchetti C, Otvos L, Giordano A. Helicobacter pylori as a class I carcinogen: physiopathology and management strategies. J Cell Biochem. 2007;102:264-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 4. | Wang YH, Wu JJ, Lei HY. The autophagic induction in Helicobacter pylori-infected macrophage. Exp Biol Med (Maywood). 2009;234:171-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Allen LA. Phagocytosis and persistence of Helicobacter pylori. Cell Microbiol. 2007;9:817-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 72] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Chu YT, Wang YH, Wu JJ, Lei HY. Invasion and multiplication of Helicobacter pylori in gastric epithelial cells and implications for antibiotic resistance. Infect Immun. 2010;78:4157-4165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 7. | Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344-1348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1632] [Cited by in RCA: 1584] [Article Influence: 105.6] [Reference Citation Analysis (0)] |
| 8. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5765] [Cited by in RCA: 5746] [Article Influence: 338.0] [Reference Citation Analysis (1)] |
| 9. | Yoshimori T, Amano A. Group a Streptococcus: a loser in the battle with autophagy. Curr Top Microbiol Immunol. 2009;335:217-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 831] [Cited by in RCA: 892] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 11. | Ogawa M, Sasakawa C. Shigella and autophagy. Autophagy. 2006;2:171-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Ogawa M, Mimuro H, Yoshikawa Y, Ashida H, Sasakawa C. Manipulation of autophagy by bacteria for their own benefit. Microbiol Immunol. 2011;55:459-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Wang YH, Gorvel JP, Chu YT, Wu JJ, Lei HY. Helicobacter pylori impairs murine dendritic cell responses to infection. PLoS One. 2010;5:e10844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Terebiznik MR, Raju D, Vázquez CL, Torbricki K, Kulkarni R, Blanke SR, Yoshimori T, Colombo MI, Jones NL. Effect of Helicobacter pylori’s vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 15. | Tang B, Li N, Gu J, Zhuang Y, Li Q, Wang HG, Fang Y, Yu B, Zhang JY, Xie QH. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. Autophagy. 2012;8:1045-1057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 16. | Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496-2505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 241] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 17. | Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA, Hayward NK. Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS One. 2010;5:e9685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 162] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 18. | Bridge G, Monteiro R, Henderson S, Emuss V, Lagos D, Georgopoulou D, Patient R, Boshoff C. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood. 2012;120:5063-5072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 19. | Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, Delattre O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene. 2013;32:3915-3921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 20. | Quintavalle C, Donnarumma E, Iaboni M, Roscigno G, Garofalo M, Romano G, Fiore D, De Marinis P, Croce CM, Condorelli G. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene. 2013;32:4001-4008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 21. | Martinez I, Cazalla D, Almstead LL, Steitz JA, DiMaio D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci USA. 2011;108:522-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 22. | Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, Zöllner H, Munding J, Klein-Scory S, Reinacher-Schick A, Schwarte-Waldhoff I, Schmiegel W, Hahn SA. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis. 2012;33:732-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 23. | Li N, Kaur S, Greshock J, Lassus H, Zhong X, Wang Y, Leminen A, Shao Z, Hu X, Liang S. A combined array-based comparative genomic hybridization and functional library screening approach identifies mir-30d as an oncomir in cancer. Cancer Res. 2012;72:154-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 24. | Lu Y, Ryan SL, Elliott DJ, Bignell GR, Futreal PA, Ellison DW, Bailey S, Clifford SC. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PLoS One. 2009;4:e6159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 25. | Yang X, Zhong X, Tanyi JL, Shen J, Xu C, Gao P, Zheng TM, DeMichele A, Zhang L. mir-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochem Biophys Res Commun. 2013;431:617-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 26. | Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 369] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 27. | Zou Z, Wu L, Ding H, Wang Y, Zhang Y, Chen X, Chen X, Zhang CY, Zhang Q, Zen K. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem. 2012;287:4148-4156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 159] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 28. | Xiao B, Li W, Guo G, Li B, Liu Z, Jia K, Guo Y, Mao X, Zou Q. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Curr Microbiol. 2009;58:258-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 29. | Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030-3044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1946] [Cited by in RCA: 1973] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 30. | Amano A, Nakagawa I, Yoshimori T. Autophagy in innate immunity against intracellular bacteria. J Biochem. 2006;140:161-166. [PubMed] |
| 31. | Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2446] [Cited by in RCA: 2700] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 32. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1059] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 33. | Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077-15082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1534] [Cited by in RCA: 1721] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 34. | Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru AH, van Adrichem AJ. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 127] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 35. | Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang Z, Ni X. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J Biomed Sci. 2011;18:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 135] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 36. | Gabriely G, Teplyuk NM, Krichevsky AM. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy. 2011;7:1384-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 37. | Kobayashi N, Uemura H, Nagahama K, Okudela K, Furuya M, Ino Y, Ito Y, Hirano H, Inayama Y, Aoki I. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012;3:1455-1471. [PubMed] |
| 38. | Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008;9:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 647] [Article Influence: 38.1] [Reference Citation Analysis (0)] |