Published online Mar 7, 2015. doi: 10.3748/wjg.v21.i9.2759
Peer-review started: April 15, 2014
First decision: June 10, 2014
Revised: July 12, 2014
Accepted: August 13, 2014
Article in press: August 28, 2014
Published online: March 7, 2015
Processing time: 328 Days and 16.6 Hours
AIM: To investigate differences in microbes and short chain fatty acid (SCFA) levels in stool samples from Hispanic and non-Hispanic African American, American Indian, and White participants.
METHODS: Stool samples from twenty participants were subjected to analysis for relative levels of viable bacteria and for SCFA levels. Additionally, the samples were subjected to 16S rRNA gene pyrosequencing for identification of bacteria present in the stool. We used a metagenome functional prediction technique to analyze genome copy numbers and estimate the abundance of butyrate kinase in all samples.
RESULTS: We found that African Americans had significantly lower levels of acetate, butyrate, and total SCFAs than all other racial/ethnic groups. We also found that participant microbial profiles differed by racial/ethnic group. African Americans had significantly more Firmicutes than Whites, with enriched Ruminococcaceae. The Firmicutes/Bacteroidetes ratio was also significantly higher for African Americans than for Whites (P = 0.049). We found Clostridium levels to be significantly and inversely related to total SCFA levels (P = 0.019) and we found Bacteroides to be positively associated (P = 0.027) and Clostridium to be negatively associated (P = 0.012) with levels of butyrate. We also identified a correlation between copy number for a butyrate kinase predicted from 16S rRNA gene abundance and levels of butyrate in stool.
CONCLUSION: The identified differences in gut flora and SCFA levels may relate to colorectal cancer mortality differentials and may be useful as targets for future clinical and behavioral interventions.
Core tip: This brief report describes analysis of stool samples from 20 adult participants aged 50 years and above using 16S rRNA pyrosequencing. We found significantly lower short chain fatty acid levels and significantly different microbial profiles in African Americans vs Whites. We also found a significant correlation between the predicted butyrate kinase levels based on 16S rRNA gene abundance and stool levels of butyrate. These results should be useful in future analysis of colorectal cancer incidence and mortality differentials across racial/ethnic groups.
- Citation: Hester CM, Jala VR, Langille MG, Umar S, Greiner KA, Haribabu B. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J Gastroenterol 2015; 21(9): 2759-2769
- URL: https://www.wjgnet.com/1007-9327/full/v21/i9/2759.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i9.2759
Colorectal cancer (CRC) is preventable and curable[1-6] but remains the second most common cause of cancer death in the United States[7]. African Americans suffer greater incidence and mortality due to CRC than other racial/ethnic groups in the United States and have a much lower five-year survival rate than whites (56% vs 65% for whites)[8-12]. African Americans are diagnosed with CRC at later stages than others, despite nearly equal rates of CRC screening. Although biological factors may play an important contribution to CRC disparities, there is a lack of information about the identity and relative contribution of these biological factors, especially microbial factors, to CRC in this group.
Bacteria in the human intestinal tract break down food into useable nutrients[13,14], modulate the immune system[15-17], and protect the intestinal epithelium from infection by pathogens[18]. They also contribute to disease, both directly and indirectly. Recent studies have begun to examine differences in gut bacteria profiles between individuals with and without CRC[19-24]. Sobhani et al[20] reported higher levels of Bacteroides and Prevotella in the stool of patients with CRC than in the stool of patients with normal colonoscopy. Recent studies have found Fusobacterium associated with colorectal cancer tissue and not normal colon tissue[21,22]. The exact role of bacteria in CRC development is still unclear, but the evidence supports both protective and harmful roles for both bacteria and their metabolites in CRC[24].
In terms of bacteria playing a positive role in the colon, bacteria such as Lactobacillus and Bifidobacter species have beneficial effects in the gut. The byproducts of their fermentation activities are short chain fatty acids (SCFAs)[25], such as acetate, n-butyrate, propionate, and valerate. These SCFAs are utilized by epithelial cells in the gut and/or excreted in stool. Butyrate has anti-proliferative properties in vitro and anti-cancerous properties in mouse models[26]. Thus, the influence of diet on the composition of the microbiota as well as the exposure to metabolites produced by gut bacteria (such as SCFAs), thereby influences the intestinal epithelium in ways that could reduce or increase CRC risk[27-29]. Improved understanding of SCFA differences among individuals should aid future approaches to understanding and reducing CRC risk in various population groups.
Research to assess the interplay among human behavior, environment, gut flora, bacterial metabolites, and genetics will promote understanding of cancer development and will be valuable for uncovering approaches for eliminating racial/ethnic colorectal cancer disparities. This pilot study was designed to collect preliminary information to determine whether microbial and/or microbial metabolite (SCFA) differences exist across a racially/ethnically mixed sample of adults over age 50 years. We report findings for short chain fatty acids and for 16S pyrosequencing from stool samples in self-identified Hispanics and non-Hispanic African Americans, American Indians, and Whites.
This study was reviewed and approved by the University of Kansas Medical Center Human Subjects Committee. Written, informed consent was obtained from all participants prior to engaging in any study activities.
This study was funded by a small pilot award from the University of Kansas Cancer Center. The available funds limited our sample size to a convenience sample of twenty participants. Participants were recruited through databases maintained by the University of Kansas Medical Center Department of Family Medicine Research Division. Eligibility criteria included age 50-70 years and willing to perform study requirements. Exclusion criteria included acute medical illness, current gastrointestinal bleed, history of adenomatous polyps, CRC, first degree relative with CRC < age 60 years, inherited polyposis/non-polyposis syndrome, inflammatory bowel disease, cognitive impairment or inappropriate affect or behavior.
Twenty participants were recruited, five from each of four different racial/ethnic groups: Hispanics and non-Hispanic African Americans, American Indians, and Whites. We used these four self-declared categories as Hispanics all selected “other” for race, whereas non-Hispanic African Americans, American Indians, and Whites all declared a listed racial category. We combined race and ethnicity into race/ethnicity to indicate that both ethnicity and race are taken into account in the categorization of participants.
The participants were administered informed written consent; given a brief demographic and truncated food frequency survey (i.e., How often do you eat vegetables? Daily, Weekly, Monthly, Rarely, or Never; How many servings of vegetables and/or vegetable juices do you usually have during a single day? None, 1, 2-3, 4 or more); and were given a kit to collect stool to be sent for comprehensive stool analysis (CSA). Participants were instructed to collect their stool within two to three days of recruitment and send it to Doctor’s Data Laboratory, Inc. The CSA kit contained gloves, a collection container, and two vials, one empty and one containing a preservative. The participant collected his or her stool in the container, then used spoons attached to the tube caps to transfer stool to both vials. The samples were then submitted to Doctor’s Data Laboratory, Inc, for analysis.
The Doctor’s Data CSA assesses a number of variables including species of bacteria and yeast that could be cultured from the sample. For this study, we recorded SCFA information (proportions of acetate, propionate, butyrate and valerate; the level of n-butyrate; and the total level of detected SCFAs in mg/mL of stool); stool pH; and the quintile values of the following bacteria: Bacteroides fragilis group, Bifidobacterium species, Clostridium species, Enterococcus species, Escherichia coli, and Lactobacillus species. The CSA panel also includes a guaiac based test for fecal occult blood. Their quantitation methods detect SCFAs by gas chromatography and viable bacteria by culture based methods. Cultured bacterial isolates are identified using Vitek-2 or MALDI-TOF mass spectrometry. Detection of viable, culturable bacteria was done to complement the results of total bacterial identification by pyrosequencing.
The fecal samples from 19 diverse participants (Hispanic, n = 4; non-Hispanic African American, n = 5; non-Hispanic American Indian, n = 5; non-Hispanic White, n = 5) were collected, and the fecal bacterial genomic DNA was isolated using maxwell tissue DNA isolation kit (Promega). (The quantity of stool provided by one of the Hispanic participants was insufficient for analysis by pyrosequencing, reducing the number from 20 to 19 total participants.) The 16S ribosomal RNA gene was amplified using 16S rRNA specific primers (v1-v3), 27f (AGAGTTTGATCCTGGCTCAG) and 534r (ATTACCGCGGCTGCTGG) on the isolated genomic DNA (10 ng). These primers were anchored with adapters and Multiplex Identifiers (MIDs; 10 bp long) to distinguish various samples in a single 454 pyro sequencing reaction. An average of 3250 high quality sequences per sample were obtained and the microbial classification was performed using GreenGenes reference data base (gg_otus-4feb2011) using QIIME tools (http://www.qiime.org)[30]. Briefly, the sequences were rarified (to standardize the sequences across the samples with uneven sampling) at 1500 randomly selected sequences per sample. The sequences reference picked into 97% OTUs using the GreenGenes reference dataset gg_otus_4feb2011. The OTUs were classified taxonomically by using the GreenGenes reference database at various taxonomic ranks (phylum, order, class, family, genus, and species).
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to predict metagenome functional content from the 16S rRNA gene surveys (http://picrust.github.com)[31]. PICRUSt uses information from reference genome databases and ancestral state reconstruction to predict functional categories based on 16S rRNA gene sequences. The predicted copy number abundance of K00929, butyrate kinase, which catalyzed one of the last metabolic steps in butanoate metabolism, ko00650, was correlated with the amount of actual butyrate detected in the stool samples using Spearman correlation.
Because our sample size was limited, we determined the power of our sample with respect to our pre-determined sample size to detect differences in total SCFA levels. The sample size of 20 participants provides 72% power for one sided test and 61% power for a two sided test at 5% significance to detect an effect size of 0.5 with a standard deviation of 4 mg/mL. Because population values for total SCFA levels are not available, the midrange of the Doctor’s Data, Inc., reference range of total SCFA levels, 9 mg/mL, was used as the “population average” for calculating power. The reference range is 4-14 mg/mL; an average of 9 ± 4 mg/mL is consistent with average total SCFAs from 60 individuals having undergone CSA in studies performed since the completion of this one (Hester and Greiner, unpublished data).
Statistical analysis of participant survey and CSA data was performed with SPSS version 20 software. Mean SCFA levels (individual and total) and pH, by racial/ethnic group, were compared by one way ANOVA. Additionally, the mean SCFA levels and pH for African Americans vs all others were compared by independent samples t test. Relationships between SCFAs and bacterial levels detected by CSA were identified by backwards stepwise regression analysis. Bacterial levels determined by CSA, the food frequency questions, and servings per day were analyzed by race using χ2 with Fisher’s exact test.
The percentages of phylum from sequences were plotted using GraphPad prism, and statistical analysis was performed using unpaired two-tailed t test using GraphPad Prism 4.0 software. Spearman correlations for the PICRUSt analysis were also performed with GraphPad Prism 4.0 software.
Participant characteristics are listed in Table 1. Twenty participants completed the intake survey and submitted stool for CSA. By one way ANOVA, there were no statistically significant differences in mean age or BMI among the four racial/ethnic groups represented. By χ2 analysis with Fisher’s exact test, there were no statistically significant differences among the groups for sex, marital status, or insurance status. There was a statistically significant difference in education level among the racial/ethnic groups identified by χ2 with Fisher’s exact test with Hispanics having the lowest level of educational attainment (P = 0.008).
| Characteristic | AA | AI | Hispanic | White | All groups |
| Age, mean (range)12 | 61.8 (50-72) | 59.4 (50-75) | 54.4 (50-59) | 63.8 (57-74) | 59.9 (50-75) |
| Age group, yr | |||||
| 50-59 | 2 | 3 | 5 | 1 | 11 (55) |
| 60-69 | 1 | 1 | 0 | 3 | 5 (25) |
| 70-79 | 2 | 1 | 0 | 1 | 4 (20) |
| BMI, mean ± SD12 | 33.6 ± 7.7 | 29.8 ± 4.4 | 32.2 ± 8.2 | 34.8 ± 4.0 | 32.7 (22.8-43.9) |
| Sex | |||||
| Male | 1 | 2 | 2 | 1 | 6 (30) |
| Female | 4 | 3 | 3 | 4 | 14 (70) |
| Marital status3 | |||||
| Married | 4 | 4 | 3 | 2 | 13 (65) |
| Widowed | 0 | 1 | 1 | 1 | 3 (15) |
| Never married/other | 1 | 0 | 1 | 2 | 4 (20) |
| Education level4 | |||||
| Some high school and below | 0 | 0 | 2 | 0 | 2 (10) |
| High school grad/GED | 0 | 0 | 3 | 0 | 3 (15) |
| Some college or tech school | 2 | 3 | 0 | 4 | 9 (45) |
| College grad and above | 3 | 2 | 0 | 1 | 6 (30) |
| Insurance5 | |||||
| Yes | 4 | 4 | 1 | 4 | 14 (70) |
| No | 1 | 1 | 4 | 1 | 6 (30) |
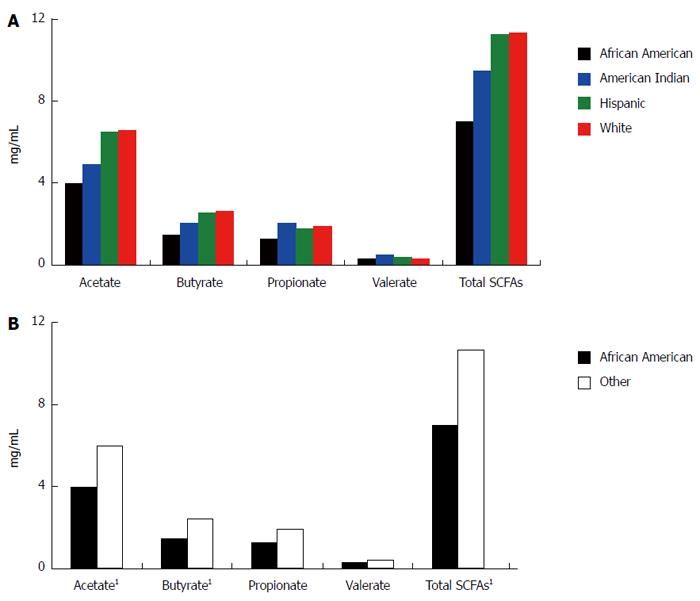
By one way ANOVA, there was no significant difference in SCFA levels, either individual or total, among all four racial/ethnic groups (Figure 1A). However, we identified a statistically significant difference in mean acetate, n-butyrate, and total SCFA levels between African Americans and all others (Figure 1B). The four participants testing positive for occult blood in stool were referred for follow up (no significant difference in FOBT positive by racial/ethnic group or African Americans and all others).
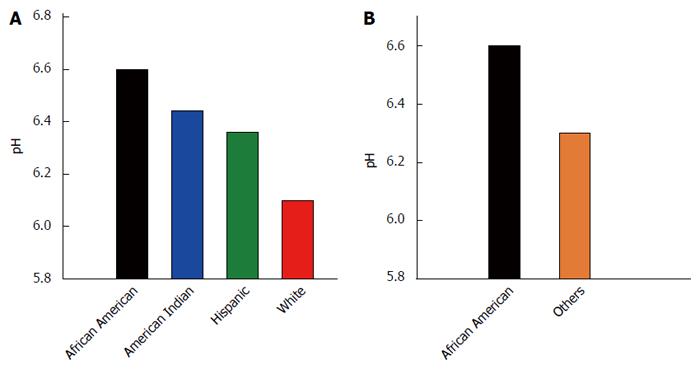
Figure 2 depicts the mean pH by race/ethnicity and for African Americans vs others. The mean pH of the samples among the racial/ethnic groups was not found to differ statistically by ANOVA (P = 0.275; Figure 2A), although African Americans had the highest mean pH of all groups. The mean stool pH of African Americans was higher than that for all others, but this difference was also not statistically significantly different by independent samples t test, equal variances not assumed (P = 0.082; Figure 2B).
Backwards stepwise regression yielded some bacterial variables that were associated with total and individual SCFA levels (Table 2). With total SCFA (mg/mL stool) as the dependent variable, and the quintile levels (0-4) of Bacteroides, Bifidobacteria, Clostridium, Escherichia coli, Enterococci, and Lactobacillus as the independent variables, only Clostridium remained in the model as significant (P = 0.019, B = -3.164). For each unit of increase in Clostridium, total SCFA levels decreased by 3.164 mg/mL. With butyrate as the dependent variable and the same bacterial levels listed previously as the independent variables, Bacteroides (P = 0.027, B = 0.347) and Clostridium (P = 0.012, B = -0.880) remained in the model as significant. For each unit increase in Bacteroides, there is a corresponding increase of 0.347 mg/mL of butyrate, and for each unit increase of Clostridium, there is a decrease of 0.88 mg/mL of butyrate. Finally, for the dependent variable of acetate, with the bacterial levels as independent variables, only Clostridium remained in the model (P = 0.001, B = -2.294). For each unit of increase of Clostridium, there is a decrease of 2.294 mg/mL of acetate.
Levels of viable bacteria detected by CSA were compared using χ2 with Fisher’s exact test. There were no significant differences in any bacterial level detected in the analysis by race or by African Americans vs others (data not shown).
In Table 3, differences in food intake are reported among the different racial/ethnic groups and between African Americans and all others. The percent of participants reporting most frequent intake of vegetables, fruit, cultured foods, and probiotic containing foods (percent reporting daily intake shown; weekly, monthly, rarely, or never are not shown) and highest number of servings of fruit and vegetables per day are also shown (percent reporting consumption of 4 or more daily servings shown; 0, 1, or 2-3 servings are not shown). There was a statistically significant difference in vegetable intake frequency among racial/ethnic groups (P = 0.003) and in vegetable servings reported by African Americans vs others (P = 0.027). There was no significant difference in reported fruit intake frequency or servings among racial/ethnic groups (P = 0.664 and P = 0.375 respectively). While African Americans and others did not differ in fruit intake frequency (P = 0.736), African Americans reported significantly more fruit servings per day than all others (P = 0.031). We also evaluated whether there were differences in reports of frequency of intake of cultured foods (i.e., yogurt, sauer kraut) or foods labeled as containing probiotics. For frequency of cultured food intake, there was no statistically significant difference revealed among groups (P = 0.450) or between African Americans vs others (P = 0.128). For foods containing probiotics, there was a statistically significant difference in reported frequency of intake among racial/ethnic groups (P = 0.047), but there was no difference between African Americans and all others (P = 1.000).
| Food frequency question | Percent reporting “daily” intake (frequency) or≥4 servings/d (servings) | Race/ethnicityP value1 | AA or all othersP value1 | |||
| AA | AI | Hispanic | White | |||
| Frequency2 | ||||||
| Vegetables | 100 | 0 | 40 | 80 | 0.0034 | 0.205 |
| Fruit | 80 | 40 | 40 | 40 | 0.664 | 0.736 |
| Cultured Food | 0 | 20 | 20 | 40 | 0.450 | 0.128 |
| Foods with probiotics | 0 | 0 | 20 | 0 | 0.0474 | 1.000 |
| Servings3 | ||||||
| Vegetables | 40 | 0 | 0 | 0 | 0.375 | 0.0274 |
| Fruit | 40 | 0 | 0 | 0 | 0.119 | 0.0314 |
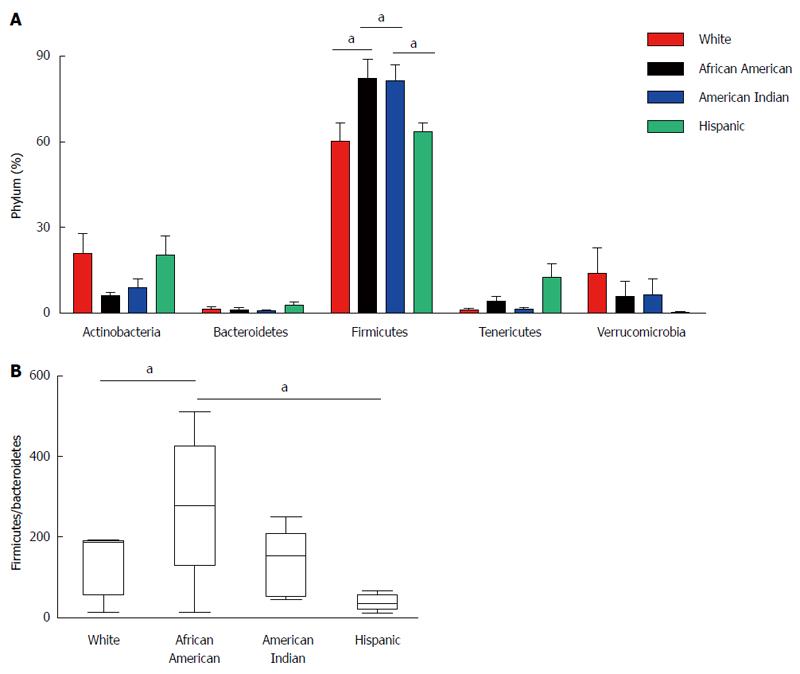
To evaluate the microbiota composition of each of the stool samples, 16S ribosomal RNA (v1-v3 region) sequencing was performed using the Roche-454 Junior sequencing platform. High quality 16S rRNA sequences from all 19 samples were obtained with an average of 3250 sequences per sample. These were analyzed using QIIME 1.5.0 pipeline as described in the methods. The 16S rRNA gene analyses showed substantial inter individual variability among different ethnic groups. Most of the gut microflora in the analyzed samples was composed of Firmicutes. However, there were significant differences in microbial content among participants of different racial/ethnic groups at phylum level (Figure 3A) despite the limited number of samples. The Firmicutes phylum was significantly increased in African Americans compared to Whites (P = 0.0443). Among Firmicutes, bacteria belonging to Ruminococcaceae were increased in African Americans. The data revealed a significant difference (increase) in the ratio of Firmicutes/Bacteroidetes phylum in African Americans compared to Whites (P = 0.0439) (Figure 3B). We observed high levels of Subdoligranulum genus (belongs to Ruminococcaceae) in two African American subjects (AM012559: 71% and TT041061: 61%). Interestingly, we also observed high levels of Akkermansia muciniphila (mucin degrading bacteria) in two White participants (MB071445: 43% and SW041237: 24%), whereas it was completely absent in the Hispanic participants.
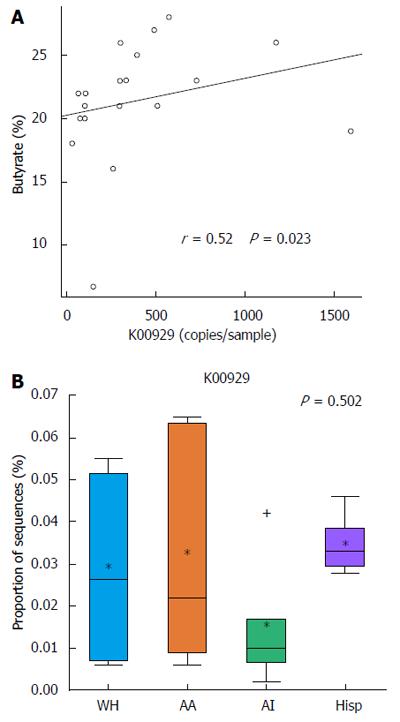
PICRUSt was used to predict metagenomes from 16s rRNA sequence data and to correlate actual metabolite levels from the stool with the predicted abundances of genes involved in butanoate metabolism across all of the samples. The analysis with PICRUSt revealed that KO K00929, a butyrate kinase that completes the last reaction in the butanoate metabolism pathway, correlated in its predicted copy number across the sampled subjects with the experimentally identified stool levels of butyrate by metabolomics (Spearman r = 0.52, P = 0.023) (Figure 4A). In addition, when grouping the samples by race/ethnicity, the median abundance of KO K00929 was lowest in African Americans and American Indians, coinciding with the actual measured butyrate levels (Figure 4B).
The current study was a pilot study to analyze SCFA and microbiota composition in a small group of individuals over age 50 and from four racial/ethnic groups. The mean total SCFA levels were significantly lower in African Americans compared to other groups. This may be important given that a recent study by Ou et al[29], found that SCFAs (acetic acid, propionic acid, butyric acid) were significantly reduced in African Americans compared to native Africans. There is a wealth of evidence supporting the beneficial effects of butyrate in reducing colon cancer risk with anti-inflammatory, immunomodulatory effects and down regulating Wnt signaling events[28,32]. There is also strong epidemiological evidence linking high fat consumption to high risk of colon cancer. Epidemiological data continues to suggest that African Americans have higher risk of developing colon cancer than others. It is possible that lower SCFAs in African Americans might explain why African Americans have higher risk of colon cancer and higher CRC mortality as well. Additionally, SCFAs reduce the pH of stool, and a recent study by Ohigashi et al[33] found that the stool from CRC patients had lower levels of acetic acid, butyric acid, propionic acid, and valeric acid and significantly higher pH than the stool of healthy controls. Interestingly, we observed here that the stool SCFA content is lower and pH is higher for African Americans than for participants of other races/ethnicities (Figure 2).
The microbial analysis from this study supports the notion that substantial variation exists in microbial composition among individual subjects. Defining the composition of a “healthy microbiota” has become a major topic of discussion in microbiota research. It is possible that a commensal for one individual can be a pathogen for another based on factors such as general health, environment, and genetic background. The main goal of this study was to evaluate the variation in microbiota and SCFA levels among different racial/ethnic groups. The microbial analysis suggests a significant increase in Firmicutes in African Americans compared to Whites and Hispanics. The microbial genome analysis also revealed that members of the Lachnospiraceae and Ruminococcaceae families play an important role in degrading cellulose and hemicellulose components of plant material in the gut environment[34]. These bacteria decompose the substrates such that the host can easily digest, ferment, and convert into SCFAs for absorption.
The mean average Ruminococcaceae family was increased in African Americans, while the Lachnospiraceae family was decreased compared to Whites. The major reported bacteria in the Ruminococcaceae family are Ruminococcus, Faecalibaterium, Anaerotruncus and Subdoligranulum. In our studies, we observed increased mean average of Subdoligranulum genus in African Americans compared to whites. Subdoligranulum bacteria have been significantly associated with colon tumor tissue[19], in some cases. It is important to note that bacteria belonging to Lachnospiraceae family such as Eubacterium rectale, Eubacterium ventriosum, Coprococcus sp. and Roseburia sp. have been associated with the production of butyrate necessary for the health of colonic epithelial tissue[35,36] and shown to be present at very low levels in inflammatory bowel disease[37]. The results presented here suggest that decreased (mean average) levels of Lachnospiraceae and butyrate levels in African Americans might offer an explanation for their increased risk for developing colon cancer.
The ratio of Firmicutes/Bacteriodetes has been associated with obesity and age. Studies have reported that the Firmicutes/Bacteroidetes ratio is significantly different between infants and adults (0.4 and 10.9 respectively) and between adults and elderly (10.9 and 0.6 respectively)[38]. Previously, the ratio of Firmicutes/Bacteroidetes was found to be significantly increased in obese adults and subjects with type 2 diabetes[39], suggesting it could be the result of dysbiosis arising from adaptation of individual microbial communities to long-term metabolic dysfunction[40]. Our studies suggest a significant increase in the ratio of Firmicutes/Bacteroidetes in African Americans compared to Whites and Hispanics, perhaps providing clues as to the risk factors for obesity linked colon cancer and the influence of the microbiota on these disorders in African Americans. [In the current study, there was not a significant difference in BMI (Table 1) or diabetes (data not shown) among groups or between African Americans and others. The gut flora could be one of the risk factors for CRC in African Americans; however, with substantial variation in individual samples, one must be cautious in interpreting microflora data and its association with disease states. Further studies with large cohorts will be needed to establish the correlations and causative links between race/ethnicity and health status.
The PICRUSt analysis was used to take microbial data and predict expected metabolite pathways based on 16S rRNA sequences. Results corroborated findings from stool analysis butyrate levels, suggesting validity of the methodology and that increased butyrate levels could be partially caused by changes in the microbial organisms within the sample. The observed variation between butyrate levels and butyrate metabolism gene copy abundances is likely due to not only PICRUSt estimations, but that butyrate levels can be changed by gene regulation within the same group of organisms. Future studies that utilize metatranscriptomics could lead to an improved understanding of whether gene regulation or selection of different organisms has a greater effect on microbial butyrate production. Further applications of the PICRUSt methodology will be useful in efforts to identify metabolic pathways and their correlation with 16S rRNA sequences in groups suffering high CRC incidence and mortality.
Limitations of this pilot study include the small sample size and the convenience sampling frame that resulted in younger Hispanic participants who had significantly lower educational attainment than the participants in other groups. Also, 70% of the participants in this study (across all groups) were female, and future studies should be more sex-balanced. Unfortunately, we did not collect information about the socioeconomic status of our participants. As education and socioeconomic status can drive dietary and health decisions, it will be important to collect this information in future studies. Despite these limitations, this pilot study was primarily performed to begin to investigate whether or not differences exist in the stool SCFA and bacteria levels in the different racial/ethnic groups represented herein, and we contend that our data suggest that further study of the differences we have observed here is warranted.
The results presented here suggest that racial/ethnic variations in the colonic environment resulting from differences in microbiota and the production of metabolites by these bacteria could be important factors related to and possibly influencing long-term CRC risk. Our preliminary evidence for racial/ethnic differences in these factors in our small sample will need to be followed up by larger-scale longitudinal studies designed to untangle the relationships among diet, race/ethnicity, the intestinal microbiota and the compounds produced by these organisms, allowing for the ultimate development of novel interventions and therapies that can be targeted to those with the highest colorectal cancer risk.
The authors would like to thank the following individuals for their support of this work: Kristin Young, PhD, MSCR, and Philip Hardwidge, PhD, for helpful discussion; Kristina Bridges, PhD, for her assistance with survey development; Angela Watson, MBA, Megan Eckles, MPH, Heraclio Perez, Marina Carrizosa-Ramos, Angelia Cully, and Lance Cully (posthumously) for their efforts in recruitment of participants to this study and administration of surveys to participants; and Aaron Epp for assistance with statistical analysis. The authors would also like to thank the Center for American Indian Community Health (CAICH; PI: Christine Daley, PhD) and Juntos at The University of Kansas Medical Center for their support of the project.
This pilot study investigated racial/ethnic differences in levels of short chain fatty acids (SCFAs) and in the microbiota of study participants from four racial/ethnic groups: Hispanics and non-Hispanic African Americans, American Indians, and Whites. The results indicate that the African Americans in our study, who are at a higher risk for development of colorectal cancer on a population level, display some of the differences that have been shown to be related to colorectal cancer such as significantly decreased stool SCFA levels and higher stool pH. African Americans also had increased levels of bacteria that have been found to be associated with colon tumor tissue and reduced levels of bacteria known to produce the SCFA, butyrate. The authors also predicted metagenome functional content and found that the predicted copy number of a gene for a butyrate kinase corresponded to the experimentally identified levels of butyrate in the stool of participants. Overall, their findings are consistent with the stool of African Americans bearing evidence of a number of indicators related to increased colorectal cancer risk, which may be informative in terms of the increased colorectal cancer morbidity and mortality faced by African Americans.
Specific bacterial genera and species have been associated with colorectal cancer (CRC), while others have beneficial effects in the gut which are mediated by the byproducts of their fermentation activities, SCFAs. The SCFA, butyrate, has anti-proliferative properties in vitro and anti-cancerous properties in mouse models. African Americans have been found to have lower levels of SCFAs than native Africans who have reduced risk for CRC. Thus, diet influences the composition of the microbiota as well as the exposure to metabolites produced by gut bacteria (such as SCFAs), thereby influencing the intestinal epithelium in ways that could reduce or increase CRC risk.
CRC is the second most common cause of cancer death in the United States. African Americans suffer greater incidence and mortality due to CRC than other racial/ethnic groups in the United States and have a much lower five-year survival rate than whites (56% vs 65% for whites). Although biological factors may play an important contribution to CRC disparities, there is a lack of information about the identity and relative contribution of these biological factors, especially microbial factors, to CRC in this group. Here, the authors provide preliminary evidence of racial/ethnic differences in the levels of microbial, metabolite, and bacterial genetic elements that could contribute to CRC disparities faced by African Americans.
Identification of differences in the microbiota and microbial metabolites in racial/ethnic groups will be important for informing the development of CRC disparities reduction and prevention strategies across the board.
Understanding racial/ethnic differences in the intestinal microflora as they relate to CRC disparities is relevant and important. This study provides initial insight into differences that may exist; however, conclusions are limited by the small sample size. This small study effectively draws attention to the need for future studies with larger sample sizes will be critical to further examine these differences as well as the factors (including dietary, demographic, and socioeconomic) that influence observed differences.
P- Reviewer: Gao CM, Kapischke M, Soares RLS, Suarez J S- Editor: Qi Y L- Editor: A E- Editor: Ma S
| 1. | Winawer SJ, Fletcher RH, Miller L, Godlee F, Stolar MH, Mulrow CD, Woolf SH, Glick SN, Ganiats TG, Bond JH. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology. 1997;112:594-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1353] [Cited by in RCA: 1246] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 2. | Mandel JS, Church TR, Ederer F, Bond JH. Colorectal cancer mortality: effectiveness of biennial screening for fecal occult blood. J Natl Cancer Inst. 1999;91:434-437. [PubMed] |
| 3. | Mandel JS, Church TR, Bond JH, Ederer F, Geisser MS, Mongin SJ, Snover DC, Schuman LM. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000;343:1603-1607. [PubMed] |
| 4. | Atkin WS, Hart A, Edwards R, McIntyre P, Aubrey R, Wardle J, Sutton S, Cuzick J, Northover JM. Uptake, yield of neoplasia, and adverse effects of flexible sigmoidoscopy screening. Gut. 1998;42:560-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 95] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 5. | Winawer SJ, Zauber AG. Colonoscopic polypectomy and the incidence of colorectal cancer. Gut. 2001;48:753-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 6. | Müller AD, Sonnenberg A. Prevention of colorectal cancer by flexible endoscopy and polypectomy. A case-control study of 32,702 veterans. Ann Intern Med. 1995;123:904-910. [PubMed] |
| 7. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10002] [Cited by in RCA: 10453] [Article Influence: 696.9] [Reference Citation Analysis (0)] |
| 8. | Kim J, Artinyan A, Mailey B, Christopher S, Lee W, McKenzie S, Chen SL, Bhatia S, Pigazzi A, Garcia-Aguilar J. An interaction of race and ethnicity with socioeconomic status in rectal cancer outcomes. Ann Surg. 2011;253:647-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 9. | Day GE, Provost E, Lanier AP. Alaska native mortality rates and trends. Public Health Rep. 2009;124:54-64. [PubMed] |
| 10. | Lanier AP, Day GE, Kelly JJ, Provost E. Disparities in cancer mortality among Alaska Native people, 1994-2003. Alaska Med. 2008;49:120-125. [PubMed] |
| 11. | Yothers G, Sargent DJ, Wolmark N, Goldberg RM, O’Connell MJ, Benedetti JK, Saltz LB, Dignam JJ, Blackstock AW. Outcomes among black patients with stage II and III colon cancer receiving chemotherapy: an analysis of ACCENT adjuvant trials. J Natl Cancer Inst. 2011;103:1498-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Howlader NA, Krapcho M, Neyman N, Aminou R, Waldron W, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H. SEER Cancer Statistics Review, 1975-2008. Bethesda, MD: National Cancer Institute; cited 2011; October 24 Available from: http://seer.cancer.gov/csr/1975_2008/. |
| 13. | Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1088] [Cited by in RCA: 1156] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 14. | Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci (Landmark Ed). 2011;16:1768-1786. [PubMed] |
| 15. | Kosiewicz MM, Zirnheld AL, Alard P. Gut microbiota, immunity, and disease: a complex relationship. Front Microbiol. 2011;2:180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 16. | DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8:523-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 229] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 17. | Mueller C, Macpherson AJ. Layers of mutualism with commensal bacteria protect us from intestinal inflammation. Gut. 2006;55:276-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Corr SC, Hill C, Gahan CG. Understanding the mechanisms by which probiotics inhibit gastrointestinal pathogens. Adv Food Nutr Res. 2009;56:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 19. | Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 442] [Cited by in RCA: 430] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 20. | Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 630] [Cited by in RCA: 622] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 21. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1498] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 22. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1499] [Article Influence: 107.1] [Reference Citation Analysis (0)] |
| 23. | Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 738] [Cited by in RCA: 950] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 24. | Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. MBio. 2013;4:e00692-e00613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 433] [Cited by in RCA: 535] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 25. | O'Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol. 2008;24:51-58. [PubMed] |
| 26. | Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time? Nutr Clin Pract. 2006;21:351-366. [PubMed] |
| 27. | O'Keefe SJ, Ou J, Aufreiter S, O’Connor D, Sharma S, Sepulveda J, Fukuwatari T, Shibata K, Mawhinney T. Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J Nutr. 2009;139:2044-2048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 28. | Vipperla K, O’Keefe SJ. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr Clin Pract. 2012;27:624-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 29. | Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, Gaskins HR, O’Keefe SJ. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 463] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 30. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23751] [Article Influence: 1583.4] [Reference Citation Analysis (0)] |
| 31. | Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814-821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5694] [Cited by in RCA: 6304] [Article Influence: 525.3] [Reference Citation Analysis (0)] |
| 32. | Bordonaro M, Lazarova DL, Sartorelli AC. Butyrate and Wnt signaling: a possible solution to the puzzle of dietary fiber and colon cancer risk? Cell Cycle. 2008;7:1178-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 33. | Ohigashi S, Sudo K, Kobayashi D, Takahashi O, Takahashi T, Asahara T, Nomoto K, Onodera H. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig Dis Sci. 2013;58:1717-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 34. | Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity. 2013;5:627-640. [RCA] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 809] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 35. | Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654-1661. [PubMed] |
| 36. | Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA): acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68:5186-5190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 512] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 37. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780-13785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3731] [Cited by in RCA: 3432] [Article Influence: 190.7] [Reference Citation Analysis (1)] |
| 38. | Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 997] [Cited by in RCA: 1246] [Article Influence: 77.9] [Reference Citation Analysis (0)] |
| 39. | Remely M, Dworzak S, Hippe B, Zwielehner J, Aumüller E, Brath H, Haslberger A. Abundance and Diversity of Microbiota in Type 2 Diabetes and Obesity. J Diabetes Metab. 2013;4:253. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Payne AN, Chassard C, Zimmermann M, Müller P, Stinca S, Lacroix C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr Diabetes. 2011;1:e12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |